Analysis of Solid Formulates Using UV-Visible Diffused Reflectance Spectroscopy with Multivariate Data Processing Based on Net Analyte Signal and Standard Additions Method
Abstract
1. Introduction
2. Materials and Methods
2.1. Samples Preparation
2.1.1. Laboratory Sample
2.1.2. Real Samples
2.2. Instrumentation
2.2.1. UV-Vis Diffuse Reflectance
2.2.2. HPLC-DAD
2.3. Data Processing
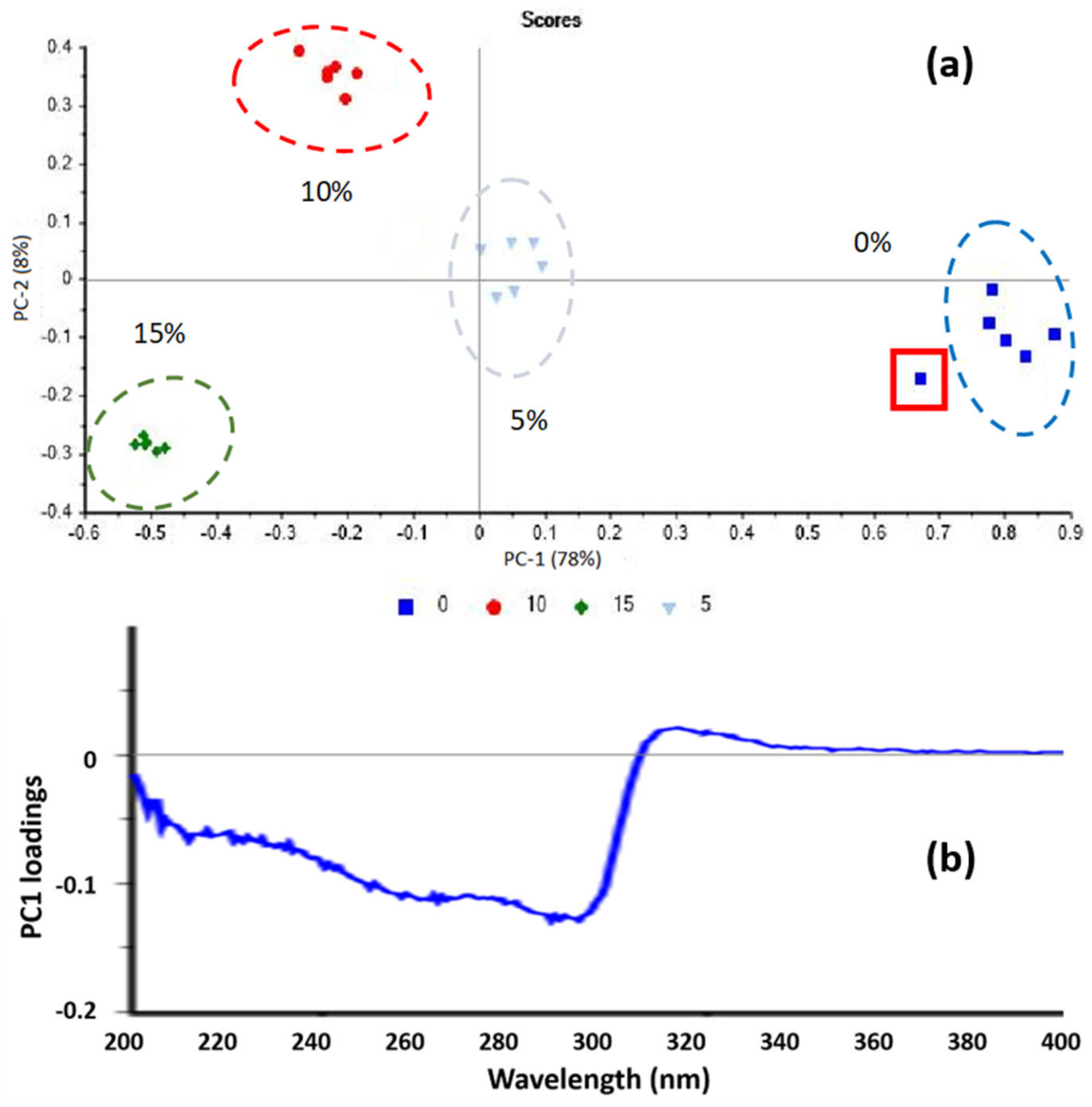
2.3.1. Principal Components Analysis (PCA) and Dataset Pretreatment
2.3.2. Partial Least Squares (PLS)
2.3.3. Net Analyte Signal (NAS)
3. Results and Discussion
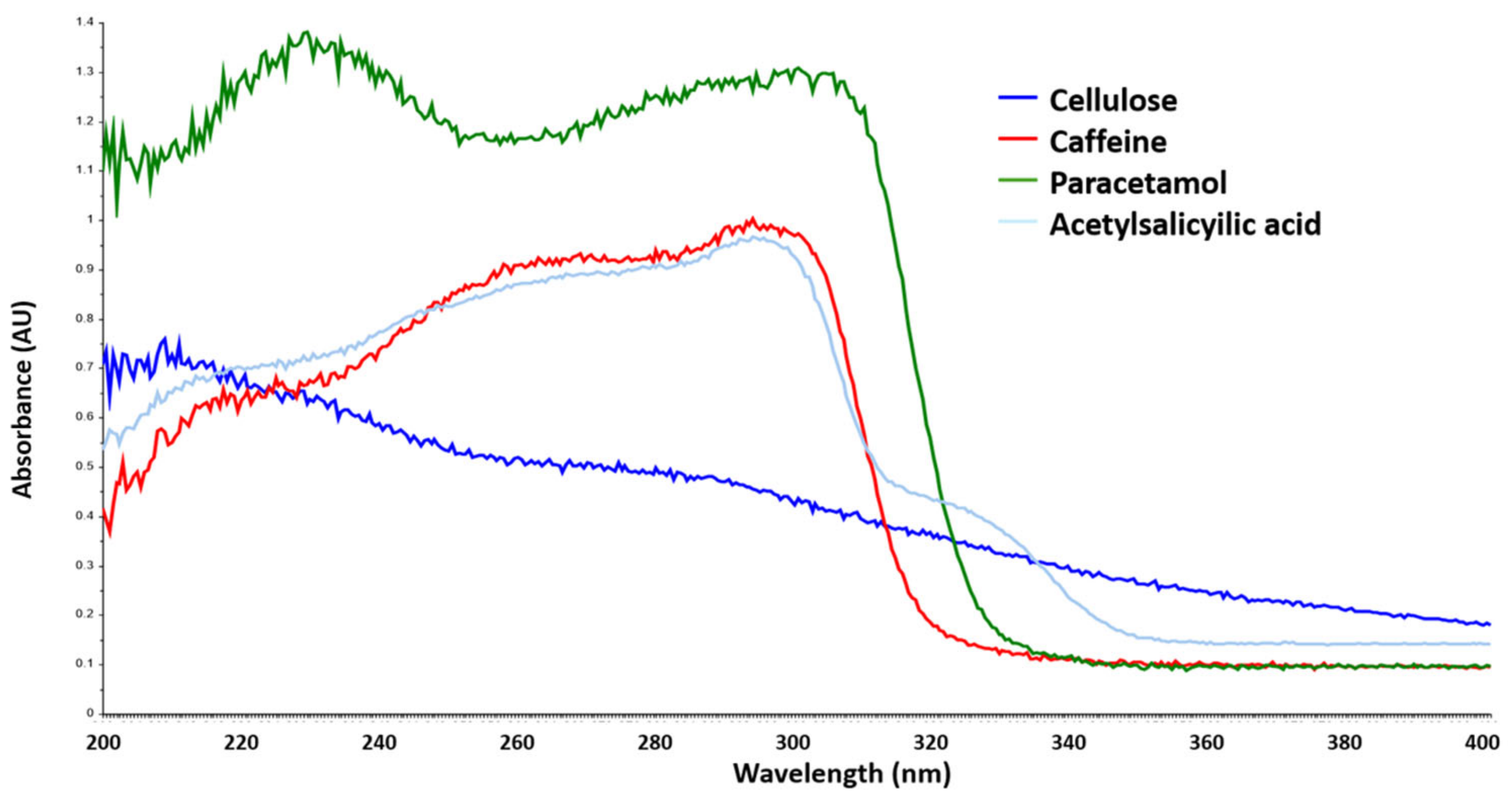
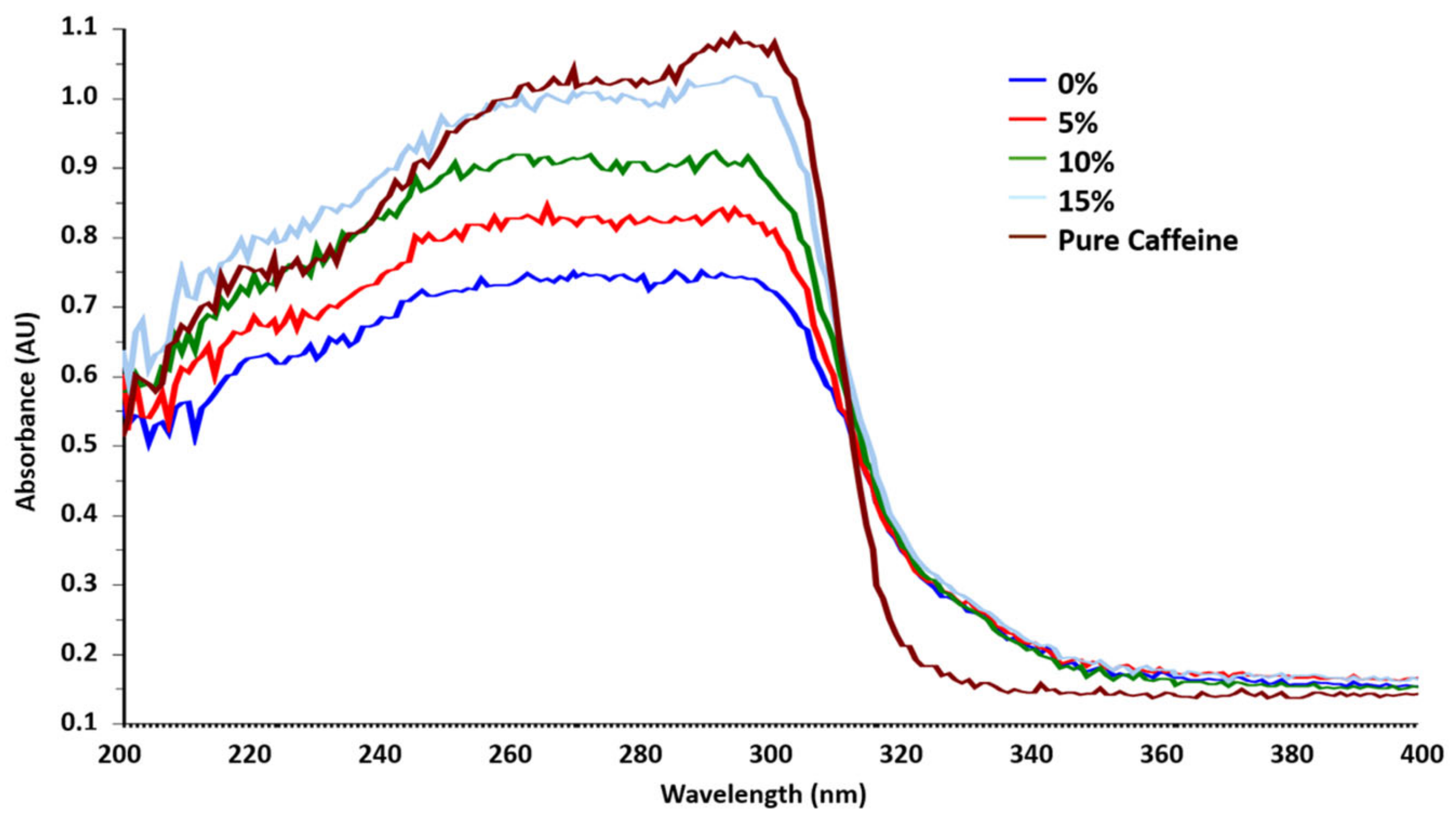
3.1. Solid Standard Solutions
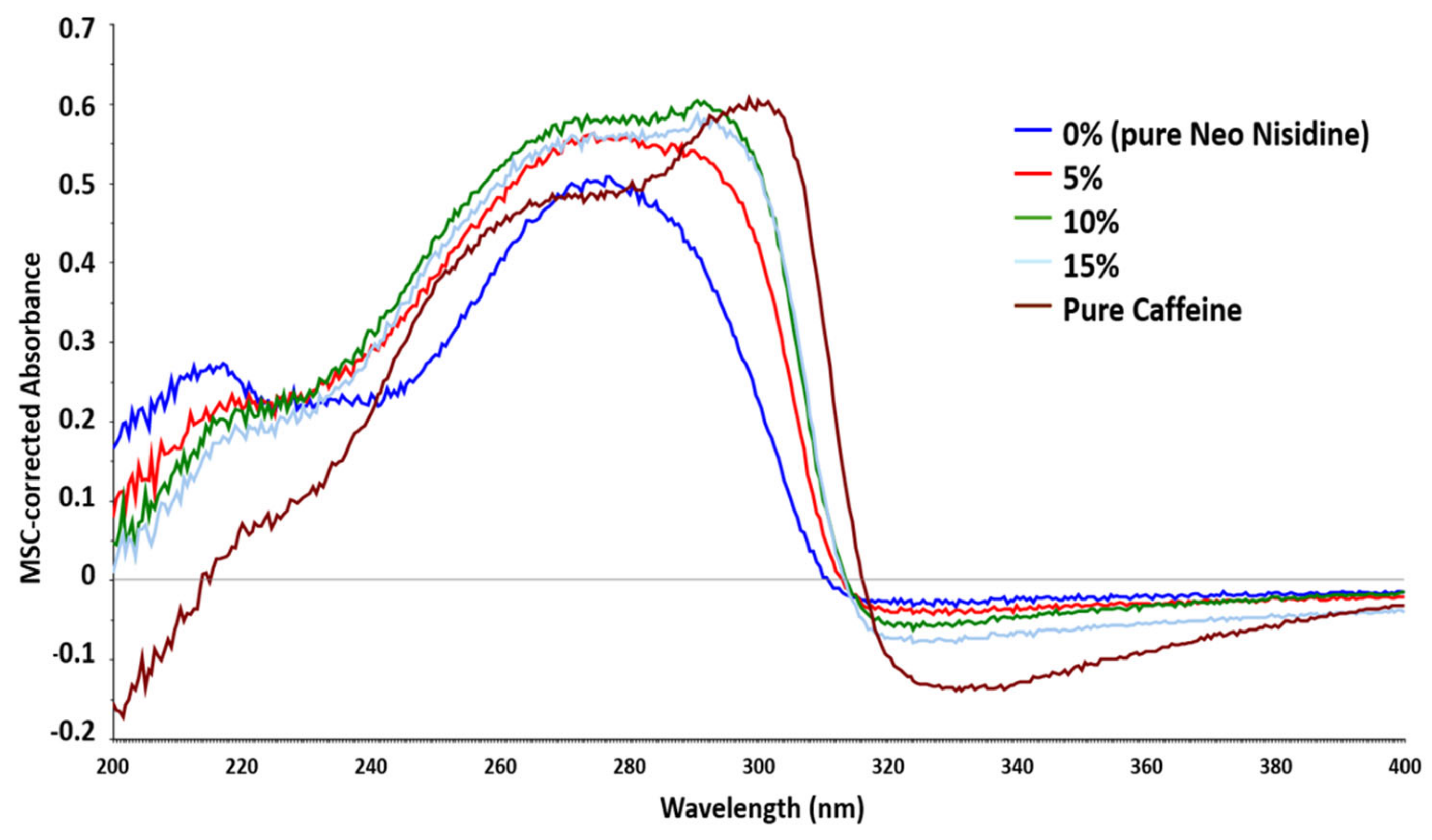
Neo Nisidine® Real Sample
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mrugalska, B.; Tytyk, E. Quality Control Methods for Product Reliability and Safety. Procedia Manuf. 2015, 3, 2730–2737. [Google Scholar] [CrossRef][Green Version]
- Macchietti, L.; Melucci, D.; Menarini, L.; Consoli, F.; Zappi, A. Analytical Comparison between Batch and Continuous Direct Compression Processes for Pharmaceutical Manufacturing Using an Innovative UV–Vis Reflectance Method and Chemometrics. Int. J. Pharm. 2024, 656, 124090. [Google Scholar] [CrossRef] [PubMed]
- Karpinski, P.H. Polymorphism of Active Pharmaceutical Ingredients. Chem. Eng. Technol. 2006, 29, 233–237. [Google Scholar] [CrossRef]
- Dogra, R.; Kumar, M.; Kumar, A.; Roverso, M.; Bogialli, S.; Pastore, P.; Mandal, U.K. Derivatization, an Applicable Asset for Conventional HPLC Systems without MS Detection in Food and Miscellaneous Analysis. Crit. Rev. Anal. Chem. 2022, 53, 1807–1827. [Google Scholar] [CrossRef] [PubMed]
- European Medicines Agency. Good Manufacturing Practice. Available online: https://www.ema.europa.eu/en/human-regulatory-overview/research-and-development/compliance-research-and-development/good-manufacturing-practice (accessed on 6 December 2023).
- Mali, A.; Jagtap, M.; Karekar, P.; Maruska, A. A Brief Review on Process Analytical Technology (PAT) Review Article. Available online: https://www.researchgate.net/publication/294053255_A_BRIEF_REVIEW_ON_PROCESS_ANALYTICAL_TECHNOLOGY_PAT_Review_Article#fullTextFileContent (accessed on 6 December 2023).
- Krämer, K.; Ebel, S. Application of NIR Reflectance Spectroscopy for the Identification of Pharmaceutical Excipients. Anal. Chim. Acta 2000, 420, 155–161. [Google Scholar] [CrossRef]
- Roggo, Y.; Chalus, P.; Maurer, L.; Lema-Martinez, C.; Edmond, A.; Jent, N. A Review of near Infrared Spectroscopy and Chemometrics in Pharmaceutical Technologies. J. Pharm. Biomed. Anal. 2007, 44, 683–700. [Google Scholar] [CrossRef]
- Kumar, M.; Bhatia, R.; Rawal, R.K. Applications of Various Analytical Techniques in Quality Control of Pharmaceutical Excipients. J. Pharm. Biomed. Anal. 2018, 157, 122–136. [Google Scholar] [CrossRef]
- Kwok, C.S.; Muntean, E.A.; Foster, W.; Mallen, C.D. Patient Pathways in Cardiology: Should Pharmaceutical and Medical Device Companies Care? Crit. Pathw. Cardiol. 2022, 21, 57–60. [Google Scholar] [CrossRef]
- Pereira, L.S.A.; Carneiro, M.F.; Botelho, B.G.; Sena, M.M. Calibration Transfer from Powder Mixtures to Intact Tablets: A New Use in Pharmaceutical Analysis for a Known Tool. Talanta 2016, 147, 351–357. [Google Scholar] [CrossRef]
- Schönbichler, S.A.; Bittner, L.K.H.; Weiss, A.K.H.; Griesser, U.J.; Pallua, J.D.; Huck, C.W. Comparison of NIR Chemical Imaging with Conventional NIR, Raman and ATR-IR Spectroscopy for Quantification of Furosemide Crystal Polymorphs in Ternary Powder Mixtures. Eur. J. Pharm. Biopharm. 2013, 84, 616–625. [Google Scholar] [CrossRef]
- Zappi, A.; Marassi, V.; Giordani, S.; Kassouf, N.; Roda, B.; Zattoni, A.; Reschiglian, P.; Melucci, D. Extracting Information and Enhancing the Quality of Separation Data: A Review on Chemometrics-Assisted Analysis of Volatile, Soluble and Colloidal Samples. Chemosensors 2023, 11, 45. [Google Scholar] [CrossRef]
- Bouhsain, Z.; Garrigues, S.; de la Guardia, M. PLS-UV Spectrophotometric method for the simultaneous determination of paracetamol, acetylsalicylic acid and caffeine in pharmaceutical formulations. Fresenius J. Anal. Chem. 1997, 357, 973–976. [Google Scholar] [CrossRef]
- Blanco, M.; Bautista, M.; Alcalá, M. Preparing Calibration Sets for Use in Pharmaceutical Analysis by NIR Spectroscopy. J. Pharm. Sci. 2008, 97, 1236–1245. [Google Scholar] [CrossRef] [PubMed]
- Sarraguça, M.C.; Lopes, J.A. The Use of Net Analyte Signal (NAS) in near Infrared Spectroscopy Pharmaceutical Applications: Interpretability and Figures of Merit. Anal. Chim. Acta 2009, 642, 179–185. [Google Scholar] [CrossRef] [PubMed]
- de Jong, S. SIMPLS: An Alternative Approach to Partial Least Squares Regression. Chemom. Intell. Lab. Syst. 1993, 18, 251–263. [Google Scholar] [CrossRef]
- Zappi, A.; Maini, L.; Galimberti, G.; Caliandro, R.; Melucci, D. Quantifying API Polymorphs in Formulations Using X-Ray Powder Diffraction and Multivariate Standard Addition Method Combined with Net Analyte Signal Analysis. Eur. J. Pharm. Sci. 2019, 130, 36–43. [Google Scholar] [CrossRef]
- Johnson, T.J.; Bernacki, B.E.; Redding, R.L.; Su, Y.F.; Brauer, C.S.; Myers, T.L.; Stephan, E.G. Intensity-Value Corrections for Integrating Sphere Measurements of Solid Samples Measured behind Glass. Appl. Spectrosc. 2014, 68, 1224–1234. [Google Scholar] [CrossRef]
- Morozzi, P.; Ballarin, B.; Arcozzi, S.; Brattich, E.; Lucarelli, F.; Nava, S.; Gómez-Cascales, P.J.; Orza, J.A.G.; Tositti, L. Ultraviolet–Visible Diffuse Reflectance Spectroscopy (UV–Vis DRS), a Rapid and Non-Destructive Analytical Tool for the Identification of Saharan Dust Events in Particulate Matter Filters. Atmos. Environ. 2021, 252, 118297. [Google Scholar] [CrossRef]
- Franeta, J.T.; Agbaba, D.; Eric, S.; Pavkov, S.; Aleksic, M.; Vladimirov, S. HPLC Assay of Acetylsalicylic Acid, Paracetamol, Caffeine and Phenobarbital in Tablets. Il Farm. 2002, 57, 709–713. [Google Scholar] [CrossRef]
- Dvořák, J.; Hájková, R.; Matysová, L.; Nováková, L.; Koupparis, M.A.; Solich, P. Simultaneous HPLC Determination of Ketoprofen and Its Degradation Products in the Presence of Preservatives in Pharmaceuticals. J. Pharm. Biomed. Anal. 2004, 36, 625–629. [Google Scholar] [CrossRef]
- Brereton, R.G. One-class classifiers. J. Chemom. 2011, 25, 225–246. [Google Scholar] [CrossRef]
- Louen, C.; Ding, S.X. Distribution Independent Threshold Setting Based on One-Class Support Vector Machine. IFAC-PapersOnLine 2020, 53, 11307–11312. [Google Scholar] [CrossRef]
- Seliya, N.; Abdollah Zadeh, A.; Khoshgoftaar, T.M. A Literature Review on One-Class Classification and Its Potential Applications in Big Data. J. Big Data 2021, 8, 122. [Google Scholar] [CrossRef]
- Rinnan, Å.; Berg, F.V.D.; Engelsen, S.B. Review of the Most Common Pre-Processing Techniques for near-Infrared Spectra. TrAC Trends Anal. Chem. 2009, 28, 1201–1222. [Google Scholar] [CrossRef]
- Rajalahti, T.; Kvalheim, O.M. Multivariate Data Analysis in Pharmaceutics: A Tutorial Review. Int. J. Pharm. 2011, 417, 280–290. [Google Scholar] [CrossRef]
- Wold, S.; Sjöström, M.; Eriksson, L. PLS-Regression: A Basic Tool of Chemometrics. Chemom. Intell. Lab. Syst. 2001, 58, 109–130. [Google Scholar] [CrossRef]
- Hawkins, D.M.; Basak, S.C.; Mills, D. Assessing model fit by cross-validation. J. Chem. Inf. Comput. Sci. 2003, 43, 579–586. [Google Scholar] [CrossRef]
- Ni, W.; Brown, S.D.; Man, R. The Relationship between Net Analyte Signal/Preprocessing and Orthogonal Signal Correction Algorithms. Chemom. Intell. Lab. Syst. 2009, 98, 97–107. [Google Scholar] [CrossRef]
- Ferré, J.; Faber, N.M. Net Analyte Signal Calculation for Multivariate Calibration. Chemom. Intell. Lab. Syst. 2003, 69, 123–136. [Google Scholar] [CrossRef]
- Bro, R.; Andersen, C.M. Theory of Net Analyte Signal Vectors in Inverse Regression. J. Chemom. 2003, 17, 646–652. [Google Scholar] [CrossRef]
- Lorber, A. Error Propagation and Figures of Merit for Quantification by Solving Matrix Equations. Anal. Chem. 1986, 58, 1167–1172. [Google Scholar] [CrossRef]
- Stute, W. The Statistical Analysis of Kaplan-Meier Integrals. IMS Lect. Notes Monogr. Ser. 1995, 27, 231–254. [Google Scholar] [CrossRef]
- Kupiec, T.C.; Vu, N.; Branscum, D. Quality-control analytical methods: Homogeneity of dosage forms. Int. J. Pharm. Compd. 2008, 12, 340–343. [Google Scholar]

| Pretreatment | SNV | First der. | MSC | MSC + SNV |
|---|---|---|---|---|
| RMSEP | 1.11 | 0.839 | 0.0540 | 0.0650 |
| R2 | 0.8863 | 0.9010 | 0.9957 | 0.9522 |
| Number of PLS factors | 5 | 9 | 6 | 6 |
| NAS prediction (% w/w) | 2.32 | 3.93 | 1.54 | 1.88 |
| Case Study | AAS (% w/w) | PAR (% w/w) | CAF (% w/w) |
|---|---|---|---|
| UV-Vis | 44 ± 4 | 34 ± 2 | 4.7 ± 0.4 |
| HPLC-DAD | 43.0 ± 0.9 | 35.2 ± 0.8 | 4.2 ± 0.1 |
| Expected | 43.4 | 34.7 | 4.3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kassouf, N.; Zappi, A.; Monticelli, M.; Melucci, D. Analysis of Solid Formulates Using UV-Visible Diffused Reflectance Spectroscopy with Multivariate Data Processing Based on Net Analyte Signal and Standard Additions Method. Chemosensors 2024, 12, 227. https://doi.org/10.3390/chemosensors12110227
Kassouf N, Zappi A, Monticelli M, Melucci D. Analysis of Solid Formulates Using UV-Visible Diffused Reflectance Spectroscopy with Multivariate Data Processing Based on Net Analyte Signal and Standard Additions Method. Chemosensors. 2024; 12(11):227. https://doi.org/10.3390/chemosensors12110227
Chicago/Turabian StyleKassouf, Nicholas, Alessandro Zappi, Michela Monticelli, and Dora Melucci. 2024. "Analysis of Solid Formulates Using UV-Visible Diffused Reflectance Spectroscopy with Multivariate Data Processing Based on Net Analyte Signal and Standard Additions Method" Chemosensors 12, no. 11: 227. https://doi.org/10.3390/chemosensors12110227
APA StyleKassouf, N., Zappi, A., Monticelli, M., & Melucci, D. (2024). Analysis of Solid Formulates Using UV-Visible Diffused Reflectance Spectroscopy with Multivariate Data Processing Based on Net Analyte Signal and Standard Additions Method. Chemosensors, 12(11), 227. https://doi.org/10.3390/chemosensors12110227






