Abstract
Hoechst dyes are among the most popular fluorophores used to stain DNA in living and fixed cells. Moreover, their high affinity and specificity towards DNA make Hoechst dyes excellent targeting moieties, which can be conjugated to various other molecules in order to tether them to DNA. The recent developments in the fields of microscopy and flow cytometry have sparked interest in such composite molecules, whose applications range from investigating nucleus microenvironment to drug delivery into tumours. Here we provide an overview of the properties of Hoechst dyes and discuss recent developments in Hoechst-based composite probes.
1. Introduction
DNA content, its distribution and the morphology of the cell nucleus serve as indicators for cell cycle progression, allowing for discrimination between different cell types and identification of mutation- or drug-induced phenotypes. This is most often achieved by fluorescent microscopy and cell-sorting techniques widely employed in academic research, commercial production and medical diagnostics. There is a long list of methods used to fluorescently label total DNA or specific genomic loci in living or fixed cells [1,2], but the staining of cells with DNA-specific blue fluorescent dyes DAPI and Hoechst remains the method of choice for many applications, due to the simplicity, low cost and no requirement for genetic modification. Generally, Hoechst is preferred, since it offers greater cell permeability and lower cytotoxicity. In this review, we summarize the properties of Hoechst dyes and provide an overview of their less conventional applications in intracellular targeting and in developing new molecular sensors.
2. Properties of Hoechst Dyes
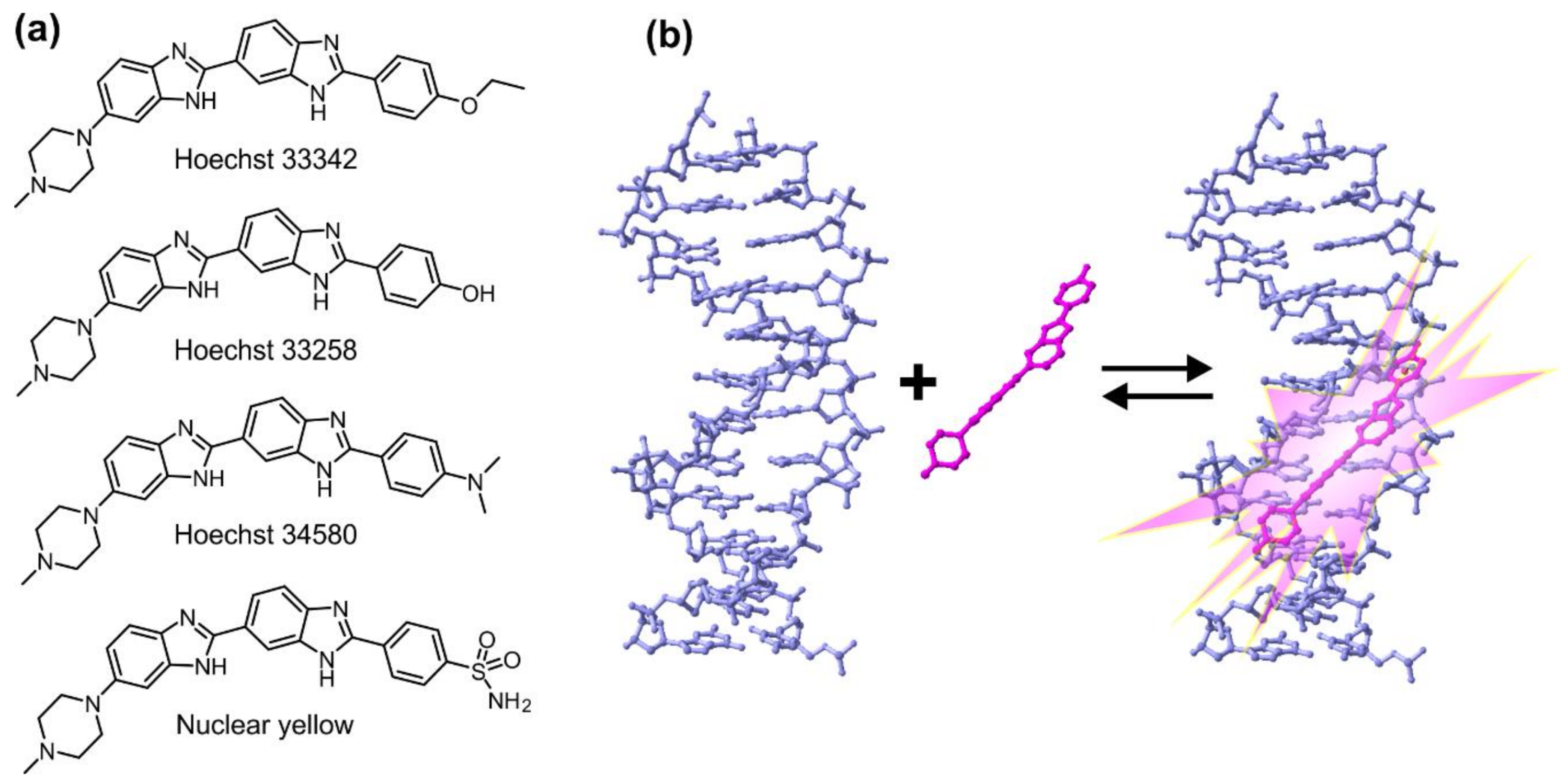
Although often overlooked in the biomedical literature, the generic name “Hoechst” can refer to any of related bisbenzimide dyes (Hoechst 33258, Hoechst 33342 and Hoechst 34580 (Figure 1a and Table 1) [3,4]. They were developed by the German company Hoechst AG in the early 1970s and are being used for DNA staining ever since. Hoechst dyes are excited by UV light (~360 nm) of xenon or mercury-arc lamps or UV lasers and emit a broad spectrum of blue light with a maximum in the 460 nm region. Upon DNA binding, their fluorescence increases ~30-fold, ensuring a good signal-to-noise ratio. This fluorescence enhancement results from the suppression of rotational relaxation and hydratation reduction upon DNA binding [5,6,7]. Hoechst are non-intercalating dyes that bind the DNA minor groove at A–T-rich regions (Figure 1b). Hoechst dyes stain the DNA of both living and fixed cells and are compatible with immunohistochemistry applications. Binding of Hoechst 33342 to DNA induces minimal cytotoxicity, a property exploited in sex preselection flow-cytometry-based assays [8].
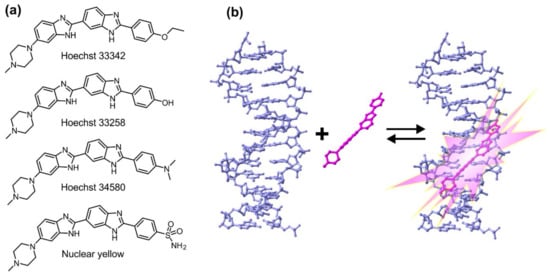
Figure 1.
DNA binding mode and chemical structures of Hoechst dyes. (a) Structures of Hoechst dyes used for DNA staining; (b) Hoechst dyes bind the minor groove of B-DNA, which results in an approximately 30-fold increase in their fluorescence (pdb ID: 8BNA).

Table 1.
Properties of Hoechst-based DNA stains.
All three Hoechst dyes are used in similar applications, but their properties are slightly different. Compared to Hoechst 33258, Hoechst 33342 is significantly more cell-permeable due to the addition of a lipophilic ethyl group and is usually preferred for living cell staining. Hoechst 34580 possesses a dimethylamine group instead of the phenol, which shifts its emission maximum to 490 nm compared to the 461 nm of Hoechst 33258 and Hoechst 33342. Emission of yet another dye of the family, Nuclear yellow (Hoechst S769121), is further shifted to the yellow region (495 nm), which allows its use together with a popular retrograde tracer True Blue for mapping branched neurons in animal brains [15,16,17,18].
Hoechst is widely used in co-staining applications that simultaneously visualize DNA and other cellular structures or specific proteins. Its excitation/emission spectra do not overlap with other commonly used small-molecule fluorophores and fluorescent proteins that emit in the green-red range. In addition, there have been recent reports that exposure of Hoechst 33258 and Hoechst 33342 to UV light results not only in bleaching, but also in photoconversion to species with excitation/emission in the blue/green and green/red range [19,20]. This property was employed in single-molecule localization microscopy to obtain higher resolution DNA maps [21]. Although photoconversion required prolonged UV illumination (from several seconds to minutes), which is not characteristic to a typical experiment, it can nevertheless lead to errors in co-localizations assays when using GFP-tagged protein and Hoechst. If the protein of interest is poorly expressed, the fluorescence of photoconverted Hoechst in the green channel might be mistaken for GFP fluorescence and misinterpreted as nuclear localization of the protein of interest. Therefore, when using Hoechst dyes in co-localization experiments, appropriate controls are essential and the UV exposure of the samples must be minimized.
Hoechst 33258 binds to DNA in two different modes: the high affinity (Kd 1–10 nM) binding results from the specific interaction with B-DNA minor groove and the low affinity (Kd ~1000 nM) reflects the nonspecific interaction with DNA sugar–phosphate backbone [22]. The optimum binding site is AAA/TTT sequence and the B-DNA structure is not affected by interaction with the dye [23]. There have been numerous attempts to increase the binding affinity with reports of the synthesis and binding evaluation of trisbenzimidazoles [24,25] and furamidine–benzimidazoles [26,27]. However, Hoechst 33258 is still the most commonly used dye for DNA staining and as a starting compound for the synthesis of more complex sensors or probes. It possesses a phenolic (-OH) group, which can be used for the attachment of the linker by standard Williamson ether synthesis conditions or can be readily transformed to triflate and further coupled to a moiety of interest by a transition metal catalysed cross-coupling reaction [28].
3. Visible/Infrared DNA Probes
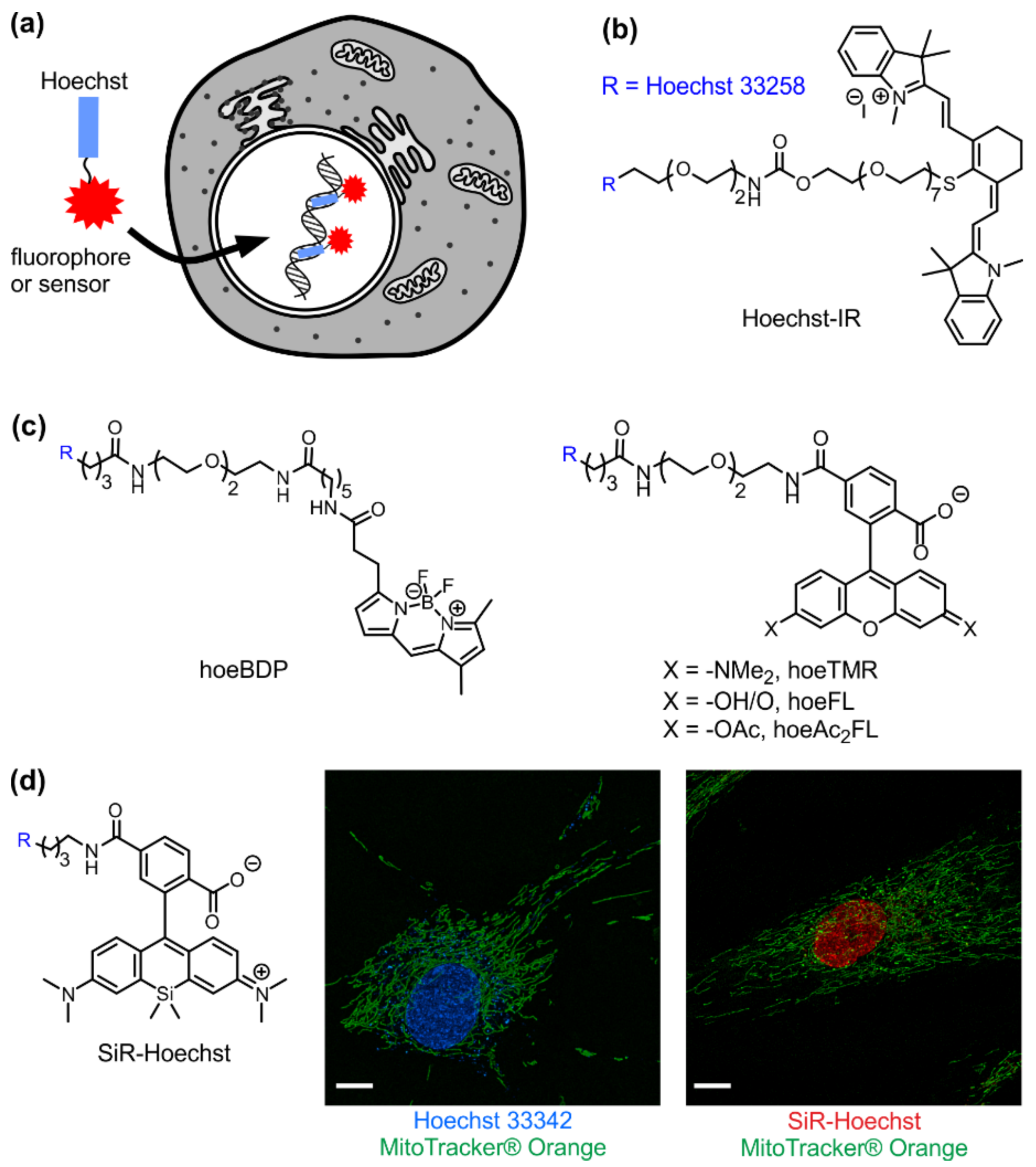
Although characterized by a good signal-to-noise ratio, low cost and simplicity of use, imaging with Hoechst in the UV/blue region (Table 1) has serious disadvantages of interfering tissue autofluorescence and phototoxicity of UV light [29,30,31]. This hampers Hoechst applications in living cells, tissues and animals and may pose a major problem when detecting relatively small amounts of non-nuclear DNA. Therefore, in long-term imaging of living cells, imaging of whole animals and tissue slices, imaging in the visible or infrared region are better choices. Therefore, synthesis of composite probes, where Hoechst serves as a DNA-targeting module, is gaining momentum (Figure 2a and Table 1).

Figure 2.
Hoechst as a component of DNA dyes with fluorescence in the visible and infrared spectrum. (a) Principle of Hoechst tagging: fluorophore or other small molecule of interest is fused to Hoechst via short flexible linker. Such composite probe enters the cell and accumulates inside the nucleus because of the high affinity of Hoechst moiety to DNA; (b,c) Structures of composite DNA probes with fluorescence spectrum in the visible or infrared spectrum; (d) On the left, structure of SiR-Hoechst. On the right, staining of nucleus of a living cell with UV light excitable Hoechst 33342 (blue) or far red light excitable SiR-Hoechst (red). In addition, mitochondria stained with MitoTracker Orange (green), Scale bar 10 µm.
In 2010 an infrared probe Hoechst-IR (Figure 2b and Table 1) for imaging necrotic tissue in vivo by binding to extracellular DNA was described (Figure 2b) [11]. Necrotic cell death is a hallmark of many diseases, including cancer, and results in DNA release to the extracellular space. Hoechst-IR was designed by conjugating Hoechst 33258 to the near-infrared dye IR-786, enabling DNA detection in the near-infrared spectrum. A long hydrophilic 11-PEG linker between Hoechst and IR-786 was chosen to lower cell permeability so that the probe would bind only extracellular DNA. Interestingly, Hoechst-IR (Kd = 0.2 nM) showed higher binding affinity than free Hoechst 33258 (Kd = 1–3 nM), which was attributed to the known moderate affinity of cyanine dyes to DNA.
Later Tsukiji et al. introduced a modular strategy to design synthetic fluorescent probes for live-cell imaging of nucleus and named it Hoechst tagging (Figure 2a) [13]. It was demonstrated that simple fluorescent dyes, fluorescein (FL), tetramethylrhodamine (TMR) and BODIPY (BDP), could be converted to nucleus-selective imaging probes by conjugating them to DNA-binding Hoechst (hoe) via a flexible linker (Figure 2c and Table 1). All three conjugates showed a turn-on fluorescence response upon binding to DNA, although their fluorescence enhancement ratios and Kd values were different. HoeBDP showed the best affinity (28 nM) and 5-fold fluorescence increase upon binding to DNA. The affinities of hoeFL (2.5 µM) and hoeTMR (1.8 µM) to DNA were by two orders of magnitude lower, but they showed much higher fluorescence enhancement—94- and 27-fold, respectively. It has been proposed that the fluorescence of the free probe is quenched by the interaction (π-π stacking) between the two fluorophores. Binding of Hoechst moiety to DNA supresses this interaction, which leads to fluorescence increase upon DNA binding. All these probes specifically stained the nucleus of living cells and were less cytotoxic than free Hoechst, thus demonstrating the applicability of Hoechst tagging strategy. It should be noted, that in the experiments with living cells hoeFL was substituted with hoeAc2FL, containing fluorescein diacetate in order to increase cell permeability of the probe. Once inside the cell, the acetyl groups are hydrolysed by intracellular esterases to regenerate hoeFL. Recently, the utility of hoeAc2FL for plant cell imaging has been demonstrated by staining nucleus of Arabidopsis thaliana guard cells [32]. This is important, as the choice of dyes and probes for plant cells is limited by autofluorescence of chlorophyl and other cellular components [33].
Lukinavičius et al. introduced a far-red DNA stain for live-cell imaging SiR-Hoechst by conjugating Hoechst to silicon rhodamine (Figure 2d and Table 1) [18]. In contrast to previously reported conjugates, this one is compatible with STED super-resolution microscopy at a standard 775 nm wavelength [14]. SiR-Hoechst binding to DNA is reduced by approximately 1000-fold compared to the parent compound, resulting in minimal cytotoxicity even when applied for many hours. Furthermore, SiR-Hoechst keeps fluorogenic properties and shows 50-fold fluorescence intensity increase at 670 nm upon binding to DNA. The proposed mechanism of fluorogenic behaviour is different from the Hoechst 33342 itself and involves a shift of the silicon-rhodamine equilibrium from the non-fluorescent spirolactone to the fluorescent zwitterion. A set of favourable spectroscopic properties, namely excellent biocompatibility and applicability to STED microscopy, makes SiR-Hoechst a unique probe amongst all other nucleus selective dyes, allowing us to study mitosis events in cultured cells and tissues by time-lapse microscopy.
4. Sensors of Nucleus Microenvironment
It turns out that hoeFL (Figure 2c and Table 1) can be used to measure intranuclear pH. Upon excitation at 405 nm, the hoeFL–DNA complex displays two fluorescence bands at ~460 nm and ~520 nm, corresponding to Hoechst and fluorescein fluorescence, respectively [34]. When the pH is decreased from 8.3 to 5.5, the fluorescein is protonated, resulting in a significant decrease in the fluorescence intensity ratio F520/F460, which allows for reliable pH measurements in ratiometric mode. The proof-of-principle experiment was performed on nigericin-treated and intact living human cells by ratiometric fluorescence imaging. In contrast to currently available small-molecule fluorescent pH probes, hoeFL localizes to the nucleus and thus allows for the specific visualization of nuclear pH without interference from other compartments, including the cytoplasm. In principle, the Hoechst tagging strategy should be applicable to other small-molecule pH sensors, such as SNARF dye, in order to expand the toolkit of synthetic fluorescent probes that allow nucleus-specific pH imaging with various photophysical properties.
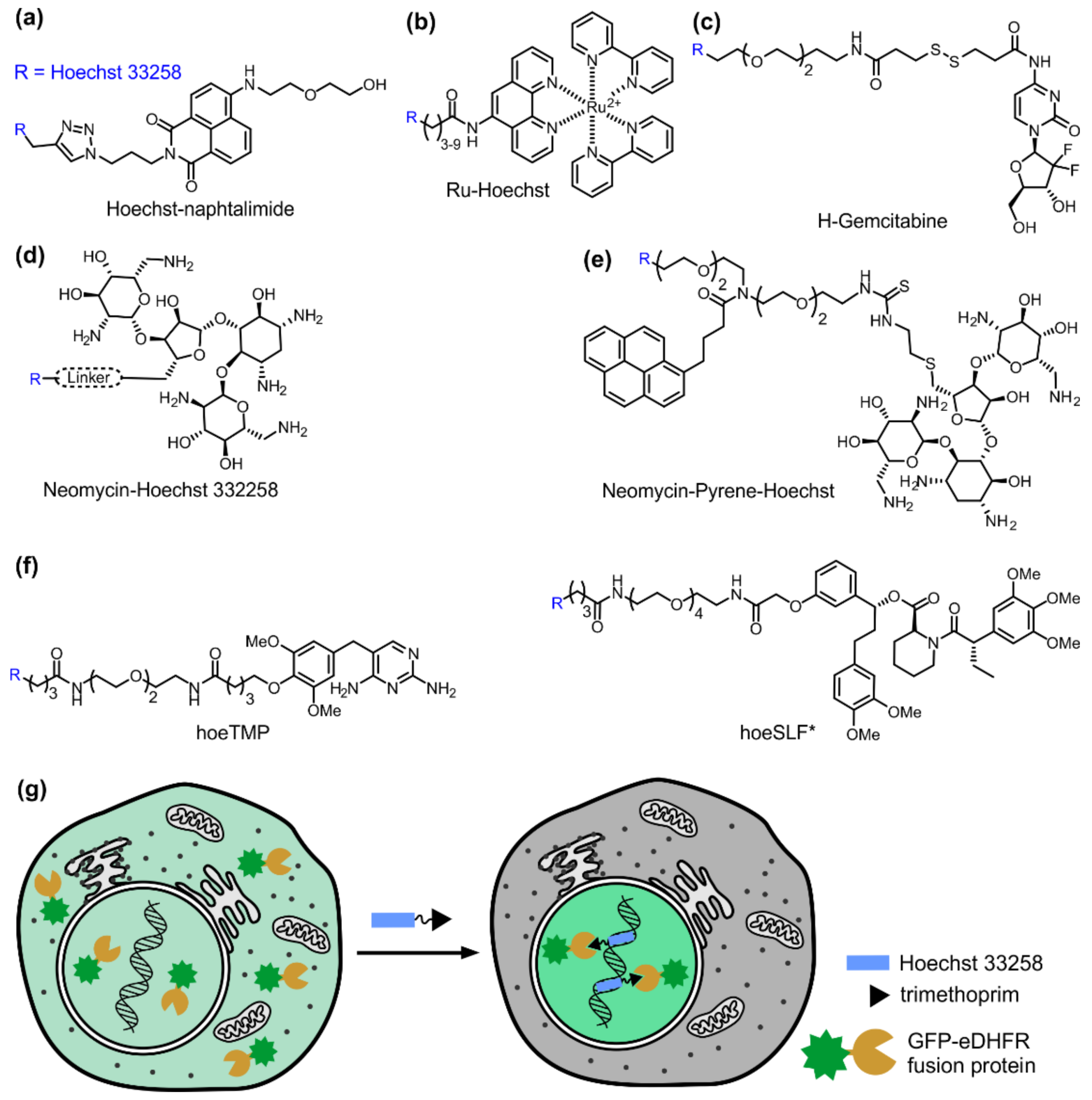
Yang et al. reported a Hoechst–naphtalimide dyad (Hoe-NI) with dual emissions as a ratiometric probe for DNA damage by similarly exploiting the ratio of fluorescence of Hoechst (450 nm) and naphthalimide (505 nm) units (Figure 3a) [32]. In solution, fluorescence intensity ratio F505/F450 increased from 0.52 to 1.76 with the concentrations of calf thymus DNA changing from 0 to 25 µM which allowed using ratiometric fluorescence response for DNA quantification. It was proposed that the fluorescence is quenched via intramolecular π-π stacking between Hoechst and naphthalimide in the free sensor. After DNA binding, Hoechst and naphthalimide are separated leading to the increase of fluorescence intensity and F505/F450 ratio change. In this system, the decreasing DNA content was taken as a read-out for the DNA damage and it was demonstrated that it is possible to detect DNA damage within the nuclei of living cells: the F505/F450 ratio dropped by 30% after exposing living cells to OH radicals for 2 h.

Figure 3.
Hoechst as a DNA-targeting moiety for drugs and proteins. (a) Hoechst-naphatalimide functions as a ratiometric sensor for DNA; (b) Ru-Hoechst contains a ruthenium complex, which is phosphorescent; (c) H-gemcitabine, anti-cancer prodrug targeted into tumours; (d,e) Conjugation of Hoechst to other nucleic acid-binding molecule, neomycin, increases binding affinity and changes specificity; (f) Ligands of eDHFR (hoeTMP) and FKBP36V (SLF*) that can be used to induce protein accumulation inside the nucleus; (g) Principle of Hoechst-targeted protein localization inside the nucleus. In the absence of ligand, GFP-eDHFR fusion protein is evenly distributed inside the cell. After the addition of the Hoechst–trimethoprim conjugate, the trimethoprim moiety is bound by eDHFR, while Hoechst anchors the whole complex to DNA, resulting in the retention of the fusion protein inside the nucleus.
Ru-Hoechst (Figure 3b) represents yet another example of Hoechst-tagged sensors, this time designed to monitor oxygen inside the nucleus [35]. In these sensors Hoechst 33258 is linked to a phosphorescent ruthenium complex. The phosphorescence is a photon emission from the triplet excited state, which can be efficiently quenched by oxygen. As a consequence, the light intensity emitted by Ru-Hoechst is inversely proportional to the concentration of oxygen in the environment.
5. Hoechst as a Targeting Module for Drug Delivery
The high affinity to DNA, moderate cytotoxicity and good cell permeability of Hoechst derivatives open the way to exploit them as DNA-targeting modules in drug development. For example, the delivery of anti-cancer drug gemcitabine into the necrotic core of tumours is enhanced by conjugating it with Hoechst 33258 (Figure 3c) [36]. Gemcitabine itself has tremendous clinical potential, but limited efficacy due to its high toxicity and inactivation in serum. A prodrug H-gemcitabine was designed by conjugating gemcitabine via a cleavable linker to Hoechst. This protects the drug from inactivation in the blood stream and allows its accumulation inside tumours by binding to the extracellular DNA. Free gemcitabine is then released by slow hydrolysis of the linker and it is able to locally kill the tumour cells. H-gemcitabine is membrane-impermeable, which results in less systemic toxicity. It showed a similar affinity to DNA as free Hoechst (Kd = 11 nM), and a wider therapeutic window and greater efficacy than unmodified gemcitabine.
Another example of a drug–Hoechst conjugate is Neomycin–Hoechst 332258 (Figure 3d). Neomycin is an aminoglycoside that binds to different nucleic acid structures with high affinity to A-RNA form, where neomycin binds to the narrow major groove [37]. First reports on Hoechst–Neomycin conjugates revealed that DNA binding by these conjugates was still largely controlled by the Hoechst moiety, but Neomycin could be forced into the wide major groove of a B- form DNA [38,39]. Neomycin contributed to the affinity of these hybrid molecules, as the conjugates stabilized DNA duplex to a higher extent than free Hoechst. Interestingly, the Neomycin–Hoechst 332258 conjugate shows good antifungal activity and minimal cytotoxicity to mammalian cells [40].
DNA recognition was extended even more by incorporating an intercalating Pyrene moiety into the structure [41]. The Neomycin–Pyrene–Hoechst (NHP) (Figure 3e) triple recognition conjugate showed an increase in DNA stabilisation, suggesting that all three components contribute to the binding. The optimal binding site for the conjugate was determined to be a contiguous stretch of nine A–T base pairs with higher binding constants for NHP than for Hoechst itself. Recently, Arya et al. showed that the Neomycin–Hoechst conjugate also shows improved duplex RNA recognition in the dual binding mode [42]. Furthermore, it was revealed that DNA versus RNA selectivity might be achieved by varying the length of the linker: conjugates with longer linkers stabilised the RNA duplex better than the DNA duplex, and vice versa [43]. Despite the fact that, in theory, selective recognition of RNA versus DNA may be achieved by very similar conjugates, no further in vivo experiments were carried out.
6. Targeting Proteins to the Nucleus
The use of Hoechst as a nucleus/DNA-targeting module is not limited to small molecules. In 2013 Tsukiji et al. introduced the concept of self-localizing ligands, designed to localize to specific subcellular regions and relocate their target proteins there, serving as synthetic protein translocators [44]. For nuclear targeting, hoeTMP and hoeSLF probes were synthesized by conjugating Hoechst with antibiotic trimethoprim (TMP) and Synthetic Ligand of protein FKBP F36V (SLF*), respectively (Figure 3f). TMP binds to E. coli dihydrofolate reductase (eDHFR) with nanomolar affinity and exhibits >1000-fold selectivity over its mammalian counterparts. The addition of hoeTMP to mammalian cells expressing eDHFR-GFP resulted in accumulation of fusion protein in the nucleus within 90 min (Figure 3g). SLF* binds specifically to F36V mutant of FKBP12 (FKBP36V). HoeSLF* was shown to induce nuclear accumulation of an FKB36V-mCherry fusion. TMP and SLF* systems are orthogonal; therefore, it is possible to simultaneously control nuclear localization of two proteins. However, it is important to note that these ligands act by retention of nuclear pools of target proteins rather than inducing protein transport. Therefore, this approach might not be applicable for proteins that are too large to freely diffuse through the nuclear pore or are excluded from the nucleus by active export.
7. Proteomics of the Nucleus
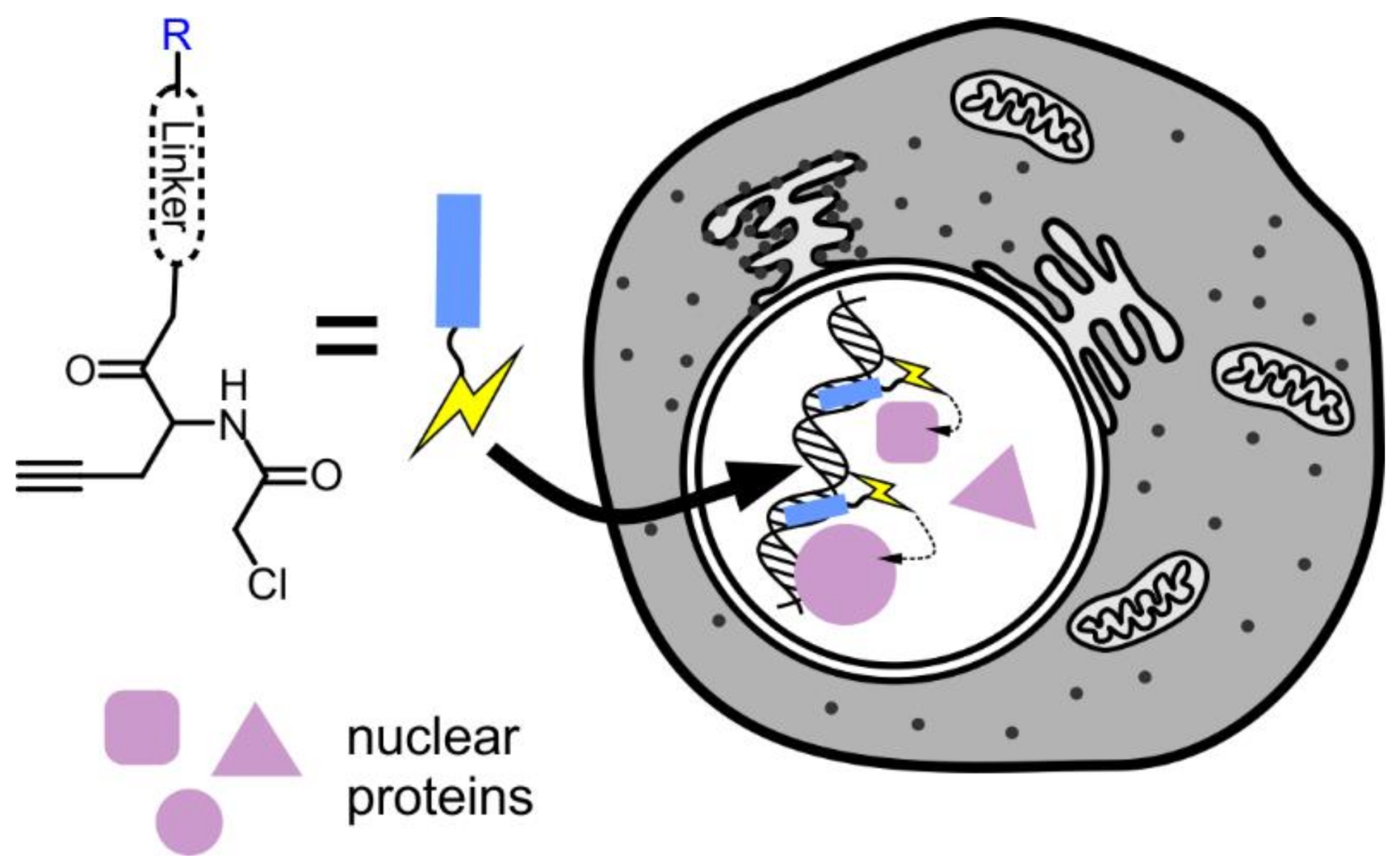
Hamachi et al. reported a novel chemical proteomic approach to labelling nuclear proteins in living cells [45]. In this strategy, Hoechst was conjugated to the reactive chloroacetyl motif (Figure 4). Such conjugates efficiently accumulate in the nucleus, where the chloroacetyl motif can react with the thiol groups of cysteines in close proximity. The linker between the two modules determines the distance between the reactive group and target protein. Three linkers were examined during this study, and it was found that the medium-length hydrophilic PEG3 linker performed best. A C3 aliphatic linker did not allow efficient labelling, while the longer hydrophilic linker interfered with cell permeability. An alkyne moiety was introduced in the structure as the handle for copper catalysed azide–alkyne cycloaddition for conjugation of affinity purification tags, but the efficiency of this reaction was found to be too low for practical use. After labelling, proteins were recovered by immunoprecipitation with anti-Hoechst antibody and analysed by mass spectrometry. Out of 67 recovered proteins, 58 were previously reported to localize in the nucleus, thus confirming the validity of this approach for nucleus selective proteomics.

Figure 4.
Hoechst-based probe for nuclear proteomics. Chloroacetyl group is targeted to the nucleus by conjugating it to Hoechst 33258 (R). There, the probe reacts with proteins in the immediate vicinity. The modified proteins can be isolated by affinity purification and identified by mass spectrometry.
Successful design of Hoechst-based drugs or probes usually requires screening a number of linkers in order to achieve optimum activity. The importance of the linker composition and length to live-cell uptake and B-DNA binding of alkynyl Hoechst 33258 have been recently systematically examined by the Dev. P. Arya group [46]. It was found that linkers that are too long weaken the binding. Moreover, only probes with short linkers (up to eight atoms) localize inside the nucleus, whilst probes with longer linkers were detected in both the nucleus and the extranuclear space. A similar trend was observed years ago for the synthesis of cell-impermeable probes such as IR-Hoechst. However, this was the first detailed study of linker impact to binding and the cell permeability of Hoechst probes.
8. Hoechst–DNA Interaction as a Read-Out in Aptamer-Based Sensors
Hoechst dyes significantly enhance their fluorescence upon binding to AT-rich double stranded DNA. This enabled us to use a combination of Hoechst dyes and specifically designed functional DNA sequences, also known as aptamers, as “label-free” (non-covalently labelled) biomolecule sensors. Aptamers are DNA molecules designed to specifically bind proteins or low-molecular mass inorganic or organic substrates. The first label-free aptamer and Hoechst 33258-based sensor was synthesized by incorporating a Hoechst 33258 binding site (AATT sequence) into the double strand stem of l-argininamide binding aptamer [47,48]. In the absence of ligand, there is an equilibrium between the single stranded and hairpin conformation of aptamer DNA, and thus an aptamer cannot form a stable Watson–Crick stem. In the presence of l-argininamide, an aptamer-ligand complex is formed and the Watson–Crick stem becomes stable, which results in the enhancement of Hoechst 33258 binding and the significant increase of fluorescence (Figure 5). A linear response of fluorescence intensity versus l-argininamide concentration was obtained in the range of 0–0.1 mM, with a detection limit of 2.5 µM. Later, this label-free aptamer strategy was expanded to more targets and dyes [49]. By employing a similar strategy, a label-free fluorescent assay was developed for “light-up” detection of ATP (adenosine triphosphate) [50]. The designed aptamer could assemble a hairpin-like structure in the presence of ATP, thus the ATP-directed formation of a hairpin provided a binding site for Hoechst and an increase of fluorescence was observed. ATP concentrations as low as 50 µM could be detected.
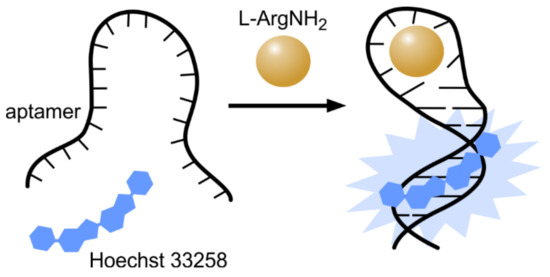
Figure 5.
Hoechst as a non-covalent reporter for ligand binding by aptamer. Hoechst binding site (AATT) was incorporated into the double-stranded stem of l-argininamide aptamer, which is not involved in ligand recognition. Binding of the ligand stabilizes the hairpin structure of the aptamer and creates a stable binding site for Hoechst, which can be detected by measuring the increase in fluorescence.
Other promising designs of label-free sensors are DNA-metal base pair optical sensors [51]. Thymine is a specific ligand for Hg2+, forming a T–Hg2+–T complex with strong affinity and high selectivity. This was used to design AT-rich single-stranded DNA, which can be directed to form a stem-loop hairpin with an Hg2+ induced T–Hg2+–T complex. This in turn provides a binding site for Hoechst and results in fluorescence increase. The designed fluorescence turn-on probe showed an Hg2+ detection range of 0–7 µM, with an outstanding detection limit of 5 nM. In measurements of real aqueous samples with added known amounts of Hg2+, an impressive accuracy of 95% was reached. Moreover, the formation of fluorescent complex can be reversed by thiols that compete with thymines for Hg2+ binding. Thus the preformed Hg2+–ssDNA–Hoechst complex can be used sensitively and selectively to monitor biothiols such as cysteine, gluthathione or homocysteine. The fluorescence turn-off of T–Hg2+–T–hairpin–Hoechst gives a detection limit of 0.1 µM for cysteine and can generally be applied for direct measurements of cysteine in real samples, including urine and serum. Based on a similar DNA–metal base pair principle, an OFF/ON recoverable probe for Hg2+ detection was designed [52]. In this case, the addition of Hg2+ to a preformed DNA–Hoechst complex results in the quenching of up to 87% of the fluorescence, which allows Hg2+ detection with a limit of 0.87 µM. In general, this mix-and-measure label-free fluorescent probe design offers many advantages such as simplicity of preparation and manipulation compared with methods that require labelled aptamers/DNA strands.
9. Conclusions
Forty-five years after their original synthesis, Hoechst 33342 and 33258 fluorophores remain in use in research and diagnostics. Although their importance in imaging applications might diminish over time, giving way to probes with more favourable photophysical properties, their importance as DNA-targeting moieties is only beginning to emerge. Further developments in Hoechst-based probes hold great potential for revealing the fine details of the processes inside the cell nucleus.
Acknowledgments
Gražvydas Lukinavičius is grateful to the Max Planck Society for a Nobel Laureate Fellowship.
Author Contributions
Gražvydas Lukinavičius conceived the general idea, all authors analysed the literature and wrote the manuscript.
Conflicts of Interest
Gražvydas Lukinavičius has filed a patent application on SiR derivatives.
References
- Mazzini, G.; Danova, M. Fluorochromes for DNA staining and quantitation. Methods Mol. Biol. 2017, 1560, 239–259. [Google Scholar] [PubMed]
- Gottfried, A.; Weinhold, E. Sequence-specific covalent labelling of DNA. Biochem. Soc. Trans. 2011, 39, 623–628. [Google Scholar] [CrossRef] [PubMed]
- Bontemps, J.; Houssier, C.; Fredericq, E. Physico-chemical study of the complexes of “33258 hoechst” with DNA and nucleohistone. Nucleic Acids Res. 1975, 2, 971–984. [Google Scholar] [CrossRef] [PubMed]
- Weisblum, B.; Haenssler, E. Fluorometric properties of the bibenzimidazole derivative hoechst 33258, a fluorescent probe specific for at concentration in chromosomal DNA. Chromosoma 1974, 46, 255–260. [Google Scholar] [CrossRef] [PubMed]
- Han, F.; Taulier, N.; Chalikian, T.V. Association of the minor groove binding drug hoechst 33258 with d(cgcgaattcgcg)2: Volumetric, calorimetric, and spectroscopic characterizations. Biochemistry 2005, 44, 9785–9794. [Google Scholar] [CrossRef] [PubMed]
- Pal, S.K.; Zhao, L.; Zewail, A.H. Water at DNA surfaces: Ultrafast dynamics in minor groove recognition. Proc. Natl. Acad. Sci. USA 2003, 100, 8113–8118. [Google Scholar] [CrossRef] [PubMed]
- Adhikary, A.; Buschmann, V.; Muller, C.; Sauer, M. Ensemble and single-molecule fluorescence spectroscopic study of the binding modes of the bis-benzimidazole derivative hoechst 33258 with DNA. Nucleic Acids Res. 2003, 31, 2178–2186. [Google Scholar] [CrossRef] [PubMed]
- Watkins, A.M.; Chan, P.J.; Kalugdan, T.H.; Patton, W.C.; Jacobson, J.D.; King, A. Analysis of the flow cytometer stain hoechst 33342 on human spermatozoa. Mol. Hum. Reprod. 1996, 2, 709–712. [Google Scholar] [CrossRef] [PubMed]
- Loontiens, F.G.; Regenfuss, P.; Zechel, A.; Dumortier, L.; Clegg, R.M. Binding characteristics of hoechst 33258 with calf thymus DNA, poly[d(a-t)], and d(ccggaattccgg): Multiple stoichiometries and determination of tight binding with a wide spectrum of site affinities. Biochemistry 1990, 29, 9029–9039. [Google Scholar] [CrossRef] [PubMed]
- Sandhu, L.C.; Warters, R.L.; Dethlefsen, L.A. Fluorescence studies of hoechst 33342 with supercoiled and relaxed plasmid pbr322 DNA. Cytometry 1985, 6, 191–194. [Google Scholar] [CrossRef] [PubMed]
- Dasari, M.; Lee, S.; Sy, J.; Kim, D.; Lee, S.; Brown, M.; Davis, M.; Murthy, N. Hoechst-ir: An imaging agent that detects necrotic tissue in vivo by binding extracellular DNA. Org. Lett. 2010, 12, 3300–3303. [Google Scholar] [CrossRef] [PubMed]
- Nakayama, A.; Bianco, A.C.; Zhang, C.Y.; Lowell, B.B.; Frangioni, J.V. Quantitation of brown adipose tissue perfusion in transgenic mice using near-infrared fluorescence imaging. Mol. Imaging 2003, 2, 37–49. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, A.; Takigawa, K.; Kurishita, Y.; Kuwata, K.; Ishida, M.; Shimoda, Y.; Hamachi, I.; Tsukiji, S. Hoechst tagging: A modular strategy to design synthetic fluorescent probes for live-cell nucleus imaging. Chem. Commun. 2014, 50, 6149–6152. [Google Scholar] [CrossRef] [PubMed]
- Lukinavičius, G.; Blaukopf, C.; Pershagen, E.; Schena, A.; Reymond, L.; Derivery, E.; Gonzalez-Gaitan, M.; D’Este, E.; Hell, S.W.; Gerlich, D.W.; et al. Sir-hoechst is a far-red DNA stain for live-cell nanoscopy. Nat. Commun. 2015, 6, 8497. [Google Scholar] [CrossRef] [PubMed]
- Bentivoglio, M.; Kuypers, H.G.; Catsman-Berrevoets, C.E.; Loewe, H.; Dann, O. Two new fluorescent retrograde neuronal tracers which are transported over long distances. Neurosci. Lett. 1980, 18, 25–30. [Google Scholar] [CrossRef]
- Lee, H.S.; Mihailoff, G.A. Fluorescent double-label study of lateral reticular nucleus projections to the spinal cord and periaqueductal gray in the rat. Anat. Rec. 1999, 256, 91–98. [Google Scholar] [CrossRef]
- Katoh, Y.Y.; Arai, R.; Benedek, G. Bifurcating projections from the cerebellar fastigial neurons to the thalamic suprageniculate nucleus and to the superior colliculus. Brain Res. 2000, 864, 308–311. [Google Scholar] [CrossRef]
- Nagatomo, F.; Ishihara, A.; Ohira, Y. Effects of hindlimb unloading at early postnatal growth on cell body size in spinal motoneurons innervating soleus muscle of rats. Int. J. Dev. Neurosci. 2009, 27, 21–26. [Google Scholar] [CrossRef] [PubMed]
- Karg, T.J.; Golic, K.G. Photoconversion of dapi and hoechst dyes to green and red-emitting forms after exposure to uv excitation. Chromosoma 2017. [Google Scholar] [CrossRef] [PubMed]
- Zurek-Biesiada, D.; Kedracka-Krok, S.; Dobrucki, J.W. Uv-activated conversion of hoechst 33258, dapi, and vybrant dyecycle fluorescent dyes into blue-excited, green-emitting protonated forms. Cytometry A 2013, 83, 441–451. [Google Scholar] [CrossRef] [PubMed]
- Szczurek, A.T.; Prakash, K.; Lee, H.K.; Zurek-Biesiada, D.J.; Best, G.; Hagmann, M.; Dobrucki, J.W.; Cremer, C.; Birk, U. Single molecule localization microscopy of the distribution of chromatin using hoechst and dapi fluorescent probes. Nucleus 2014, 5, 331–340. [Google Scholar] [CrossRef] [PubMed]
- Guan, Y.; Shi, R.; Li, X.; Zhao, M.; Li, Y. Multiple binding modes for dicationic hoechst 33258 to DNA. J. Phys. Chem. B 2007, 111, 7336–7344. [Google Scholar] [CrossRef] [PubMed]
- Pjura, P.E.; Grzeskowiak, K.; Dickerson, R.E. Binding of hoechst 33258 to the minor groove of B-DNA. J. Mol. Biol. 1987, 197, 257–271. [Google Scholar] [CrossRef]
- Aymami, J.; Nunn, C.M.; Neidle, S. DNA minor groove recognition of a non-self-complementary at-rich sequence by a tris-benzimidazole ligand. Nucleic Acids Res. 1999, 27, 2691–2698. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Li, T.K.; Kim, J.S.; LaVoie, E.J.; Breslauer, K.J.; Liu, L.F.; Pilch, D.S. DNA minor groove binding-directed poisoning of human DNA topoisomerase i by terbenzimidazoles. Biochemistry 1998, 37, 3558–3566. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Carrasco, C.; Kumar, A.; Stephens, C.E.; Bailly, C.; Boykin, D.W.; Wilson, W.D. Evaluation of the influence of compound structure on stacked-dimer formation in the DNA minor groove. Biochemistry 2001, 40, 2511–2521. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Kumar, A.; Boykin, D.W.; Bailly, C.; Wilson, W.D. Comparative thermodynamics for monomer and dimer sequence-dependent binding of a heterocyclic dication in the DNA minor groove. J. Mol. Biol. 2002, 317, 361–374. [Google Scholar] [CrossRef] [PubMed]
- Amirbekyan, K.; Duchemin, N.; Benedetti, E.; Joseph, R.; Colon, A.; Markarian, S.A.; Bethge, L.; Vonhoff, S.; Klussmann, S.; Cossy, J.; et al. Design, synthesis, and binding affinity evaluation of hoechst 33258 derivatives for the development of sequence-specific DNA-based asymmetric catalysts. ACS Catal. 2016, 6, 3096–3105. [Google Scholar] [CrossRef]
- Libbus, B.L.; Perreault, S.D.; Johnson, L.A.; Pinkel, D. Incidence of chromosome aberrations in mammalian sperm stained with hoechst 33342 and uv-laser irradiated during flow sorting. Mutat. Res. 1987, 182, 265–274. [Google Scholar] [CrossRef]
- Wagner, M.; Weber, P.; Bruns, T.; Strauss, W.S.; Wittig, R.; Schneckenburger, H. Light dose is a limiting factor to maintain cell viability in fluorescence microscopy and single molecule detection. Int. J. Mol. Sci. 2010, 11, 956–966. [Google Scholar] [CrossRef] [PubMed]
- Waldchen, S.; Lehmann, J.; Klein, T.; van de Linde, S.; Sauer, M. Light-induced cell damage in live-cell super-resolution microscopy. Sci. Rep. 2015, 5, 15348. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Wang, C.; Wang, L.; Ye, Z.-W.; Song, X.-B.; Xiao, Y. Hoechst-naphthalimide dyad with dual emissions as specific and ratiometric sensor for nucleus DNA damage. Chin. Chem. Lett. 2017, 28, 2019–2022. [Google Scholar] [CrossRef]
- Schoor, S.; Lung, S.-C.; Sigurdson, D.; Chuong, S.D.X. Fluorescent staining of living plant cells. In Plant Microtechniques and Protocols; Yeung, E.C.T., Stasolla, C., Sumner, M.J., Huang, B.Q., Eds.; Springer International Publishing: Cham, Switzerland, 2015; pp. 153–165. [Google Scholar]
- Nakamura, A.; Tsukiji, S. Ratiometric fluorescence imaging of nuclear ph in living cells using hoechst-tagged fluorescein. Bioorg. Med. Chem. Lett. 2017, 27, 3127–3130. [Google Scholar] [CrossRef] [PubMed]
- Tanabe, K.; Hara, D.; Umehara, Y.; Son, A.; Asahi, W.; Misu, S.; Kurihara, R.; Kondo, T. Tracking the oxygen status in the cell nucleus by using a hoechst-tagged phosphorescent ruthenium complex. Chembiochem 2018. [Google Scholar] [CrossRef]
- Dasari, M.; Acharya, A.P.; Kim, D.; Lee, S.; Lee, S.; Rhea, J.; Molinaro, R.; Murthy, N. H-gemcitabine: A new gemcitabine prodrug for treating cancer. Bioconj. Chem. 2013, 24, 4–8. [Google Scholar] [CrossRef] [PubMed]
- Kaul, M.; Pilch, D.S. Thermodynamics of aminoglycoside-rrna recognition: The binding of neomycin-class aminoglycosides to the a site of 16s rrna. Biochemistry 2002, 41, 7695–7706. [Google Scholar] [CrossRef] [PubMed]
- Arya, D.P.; Willis, B. Reaching into the major groove of b-DNA: Synthesis and nucleic acid binding of a neomycin-hoechst 33258 conjugate. J. Am. Chem. Soc. 2003, 125, 12398–12399. [Google Scholar] [CrossRef] [PubMed]
- Willis, B.; Arya, D.P. Recognition of b-DNA by neomycin—Hoechst 33258 conjugates. Biochemistry 2006, 45, 10217–10232. [Google Scholar] [CrossRef] [PubMed]
- Thamban Chandrika, N.; Shrestha, S.K.; Ranjan, N.; Sharma, A.; Arya, D.P.; Garneau-Tsodikova, S. New application of neomycin b-bisbenzimidazole hybrids as antifungal agents. ACS Infect. Dis. 2018, 4, 196–207. [Google Scholar] [CrossRef] [PubMed]
- Willis, B.; Arya, D.P. Triple recognition of B-DNA by a neomycin-hoechst 33258-pyrene conjugate. Biochemistry 2010, 49, 452–469. [Google Scholar] [CrossRef] [PubMed]
- Willis, B.; Arya, D.P. Recognition of rna duplex by a neomycin-hoechst 33258 conjugate. Bioorg. Med. Chem. 2014, 22, 2327–2332. [Google Scholar] [CrossRef] [PubMed]
- Ranjan, N.; Arya, D.P. Linker dependent intercalation of bisbenzimidazole-aminosugars in an rna duplex; selectivity in RNA vs. DNA binding. Bioorg. Med. Chem. Lett. 2016, 26, 5989–5994. [Google Scholar] [CrossRef] [PubMed]
- Ishida, M.; Watanabe, H.; Takigawa, K.; Kurishita, Y.; Oki, C.; Nakamura, A.; Hamachi, I.; Tsukiji, S. Synthetic self-localizing ligands that control the spatial location of proteins in living cells. J. Am. Chem. Soc. 2013, 135, 12684–12689. [Google Scholar] [CrossRef] [PubMed]
- Yasueda, Y.; Tamura, T.; Hamachi, I. Nucleus-selective chemical proteomics using hoechst-tagged reactive molecules. Chem. Lett. 2016, 45, 265–267. [Google Scholar] [CrossRef]
- Ranjan, N.; Kellish, P.; King, A.; Arya, D.P. Impact of linker length and composition on fragment binding and cell permeation: Story of a bisbenzimidazole dye fragment. Biochemistry 2017, 56, 6434–6447. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Yang, C.; Zhou, X.; Qin, J. Label-free aptamer-based sensors for l-argininamide by using nucleic acid minor groove binding dyes. Chem. Commun. 2011, 47, 3192–3194. [Google Scholar] [CrossRef] [PubMed]
- Ozaki, H.; Nishihira, A.; Wakabayashi, M.; Kuwahara, M.; Sawai, H. Biomolecular sensor based on fluorescence-labeled aptamer. Bioorg. Med. Chem. Lett. 2006, 16, 4381–4384. [Google Scholar] [CrossRef] [PubMed]
- Sarpong, K.; Datta, B. Nucleic-acid-binding chromophores as efficient indicators of aptamer-target interactions. J. Nucleic Acids 2012, 2012, 247280. [Google Scholar] [CrossRef] [PubMed]
- Le, H.-N.; Jiang, X.-Q.; Zhang, M.; Ye, B.-C. Label-free fluorescent assay of atp based on an aptamer-assisted light-up of hoechst dyes. Anal. Methods 2014, 6, 2028–2030. [Google Scholar] [CrossRef]
- Zhang, M.; Le, H.N.; Jiang, X.Q.; Ye, B.C. “Molecular beacon”-directed fluorescence of hoechst dyes for visual detection of hg(II) and biothiols and its application for a logic gate. Chem. Commun. 2013, 49, 2133–2135. [Google Scholar] [CrossRef] [PubMed]
- Paramanik, B.; Bain, D.; Patra, A. Making and breaking of DNA-metal base pairs: Hg2+ and au nanocluster based off/on probe. J. Phys. Chem. C 2016, 120, 17127–17135. [Google Scholar] [CrossRef]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).