An Aptamer-Array-Based Sample-to-Answer Biosensor for Ochratoxin A Detection via Fluorescence Resonance Energy Transfer
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals, Reagents, and Instruments
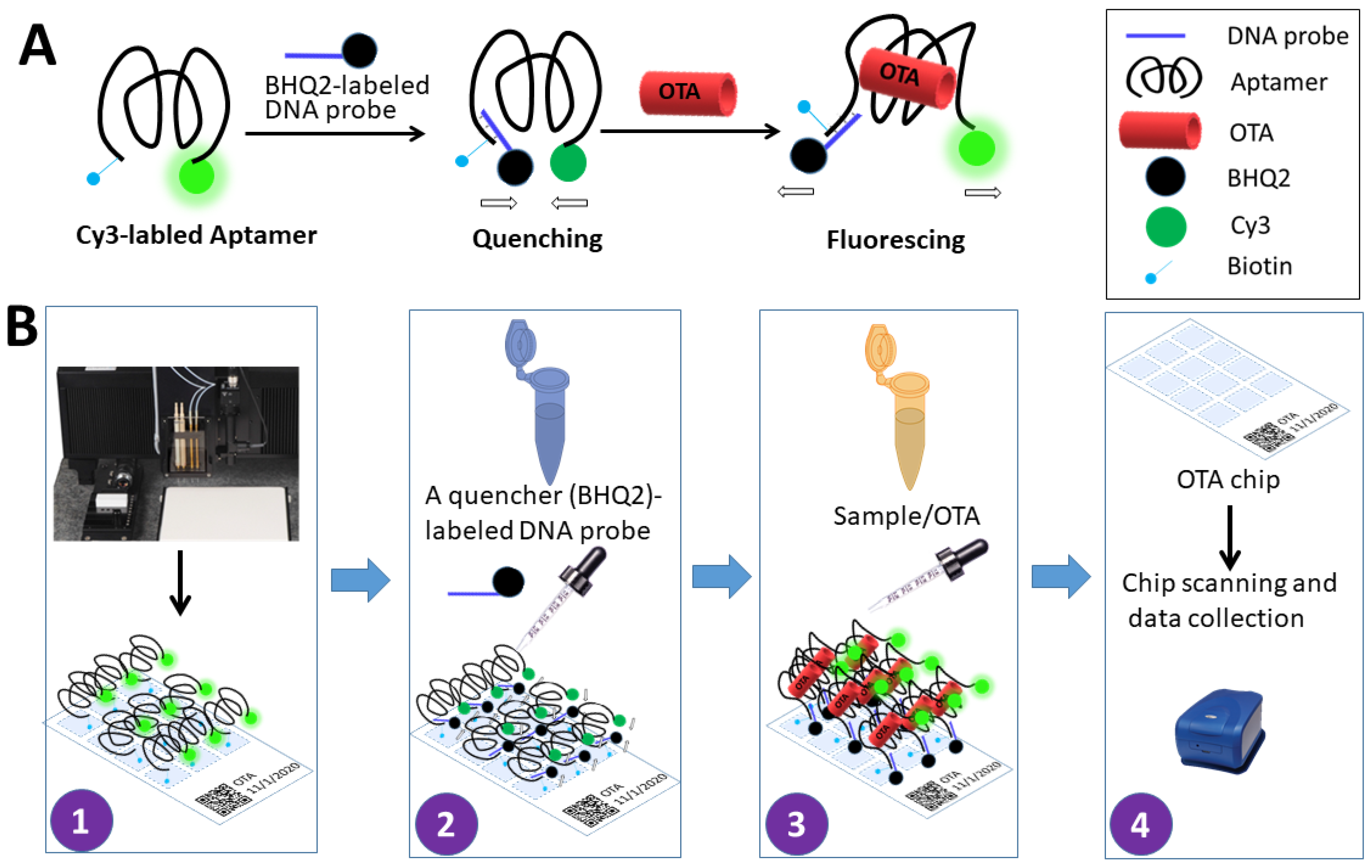
2.2. Development of Array Sensor and Streptavidin Coating
2.3. Immobilization and Optimization of OTA Aptamer Concentrations on the Chip
2.4. Optimization of the Complementary Strand Concentration
2.5. Standard Curve for Determination of OTA
2.6. Specificity Test
2.7. Recovery Test of OTA in Artificially Contaminated Rice Samples
3. Results
3.1. Streptavidin Coating and Optimization of Streptavidin Concentration
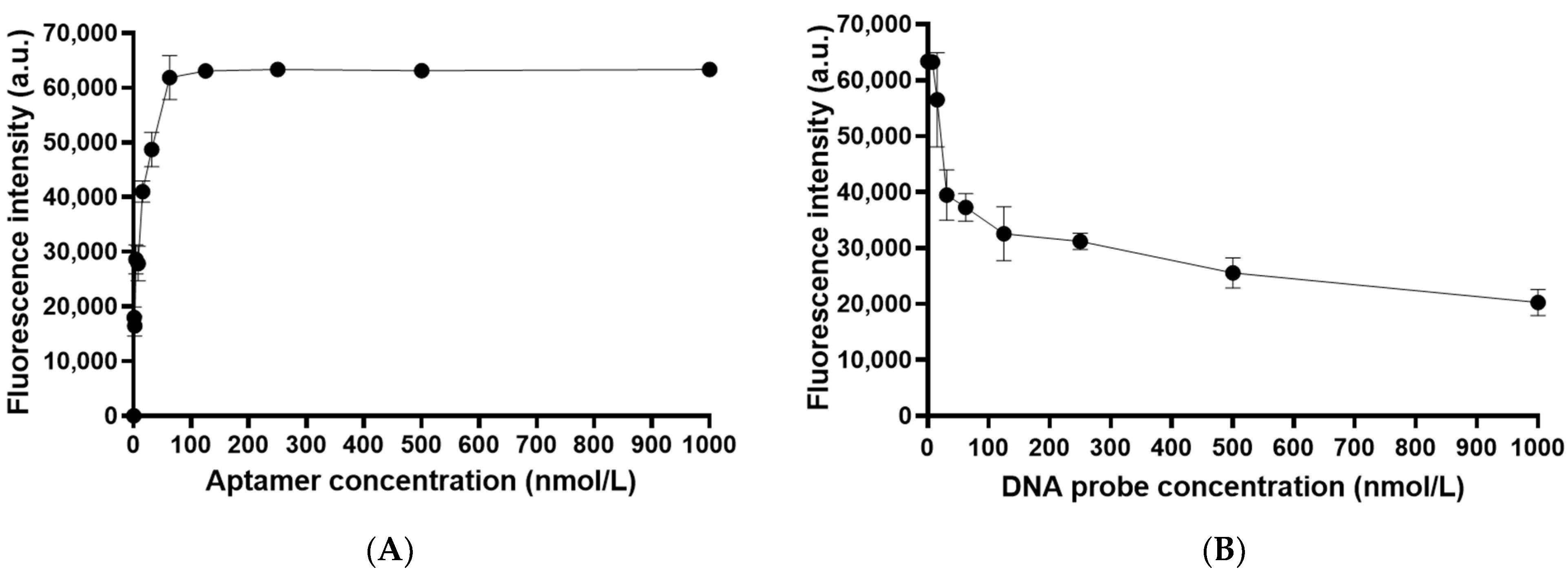
3.2. Immobilization of Cy3-Labled OTA Aptamer on the Chip and Optimization of OTA Aptamer Concentration
3.3. Optimization of the Concentration of BHQ2-Labeled DNA Probe
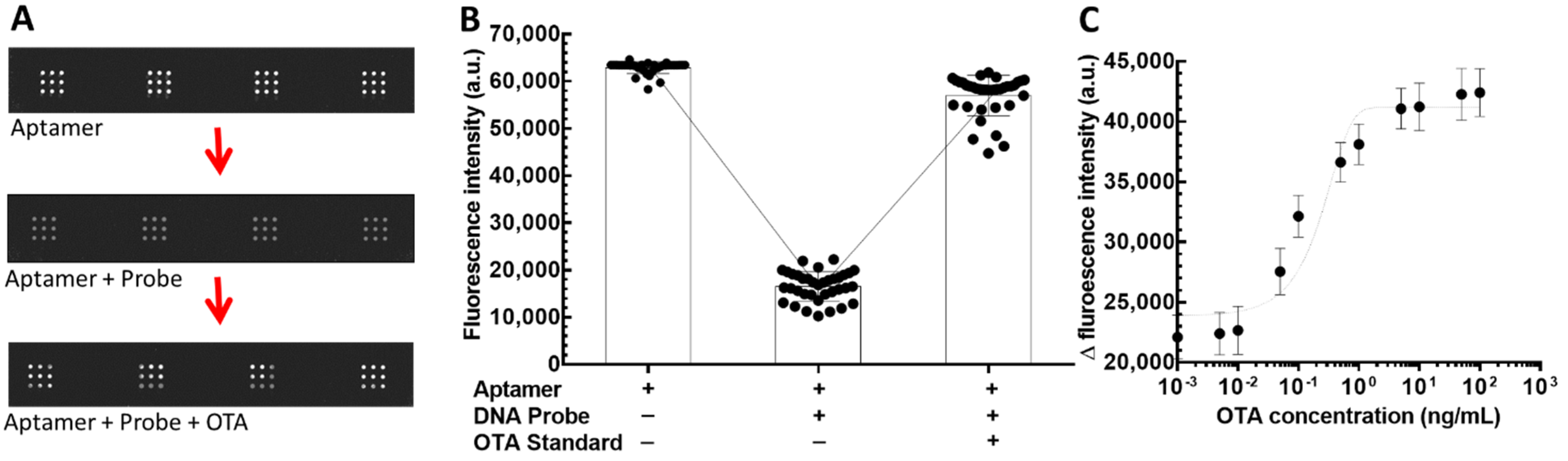
3.4. Detectability and Sensitivity of ACSB for Ochratoxin A (OTA)
3.5. Specificity of ACSB in Detecting OTA
3.6. Recovery Test of OTA in Artificially Contaminated Rice Samples
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bennett, J.W.; Klich, M. Mycotoxins. Clin. Microbiol. Rev. 2003, 16, 497–516. [Google Scholar] [CrossRef] [PubMed]
- Bui-Klimke, T.R.; Wu, F. Ochratoxin A and human health risk: A review of the evidence. Crit. Rev. Food Sci. Nutr. 2015, 55, 1860–1869. [Google Scholar] [CrossRef]
- Duarte, S.C.; Pena, A.; Lino, C.M. A review on ochratoxin A occurrence and effects of processing of cereal and cereal derived food products. Food. Microbiol. 2010, 27, 187–198. [Google Scholar] [CrossRef]
- Covarelli, L.; Beccari, G.; Marini, A.; Tosi, L. A review on the occurrence and control of ochratoxigenic fungal species and ochratoxin A in dehydrated grapes, non-fortified dessert wines and dried vine fruit in the Mediterranean area. Food Control 2012, 26, 347–356. [Google Scholar] [CrossRef]
- JECFA. Safety Evaluation f Certain Mycotoxins in Food—Prepared by the 56th Meeting of the JECFA (Joint FAO/WHO Expert Committee on Food Additives)—FAO Food and Nutrition Paper 74/WHO Foods Additives Series 47. 2001. Available online: https://inchem.org/documents/jecfa/jecmono/v47je01.htm (accessed on 16 September 2021).
- Petzinger, E.; Ziegler, K. Ochratoxin A from a toxicological perspective. J. Vet. Pharmacol. Ther. 2000, 23, 91–98. [Google Scholar] [CrossRef]
- Santos, E.A.; Vargas, E.A. Immunoaffinity column clean-up and thin layer chromatography for determination of ochratoxin A in green coffee. Food Addit. Contam. 2002, 19, 447–458. [Google Scholar] [CrossRef]
- Degelmann, P.; Becker, M.; Herderich, M.; Humpf, H.U. Determination of ochratoxin A in beer by high-performance liquid chromatography. Chromatographia 1999, 49, 543–546. [Google Scholar] [CrossRef]
- Tessini, C.; Mardones, C.; von Baer, D.; Vega, M.; Herlitz, E.; Saelzer, R.; Silva, J.; Torres, O. Alternatives for sample pre-treatment and HPLC determination of Ochratoxin A in red wine using fluorescence detection. Anal. Chim. Acta 2010, 660, 119–126. [Google Scholar] [CrossRef] [PubMed]
- Reinsch, M.; Topfer, A.; Lehmann, A.; Nehls, I.; Panne, U. Determination of ochratoxin A in beer by LC-MS/MS ion trap detection. Food Chem. 2007, 100, 312–317. [Google Scholar] [CrossRef]
- Olsson, J.; Borjesson, T.; Lundstedt, T.; Schnurer, J. Detection and quantification of ochratoxin A and deoxynivalenol in barley grains by GC-MS and electronic nose. Int. J. Food Microbiol. 2002, 72, 203–214. [Google Scholar] [CrossRef]
- Yu, F.Y.; Chi, T.F.; Liu, B.H.; Su, C.C. Development of a sensitive enzyme-linked immunosorbent assay for the determination of ochratoxin A. J. Agric. Food Chem. 2005, 53, 6947–6953. [Google Scholar] [CrossRef]
- Wei, M.; Wang, C.L.; Xu, E.S.; Chen, J.; Xu, X.L.; Wei, W.; Liu, S.Q. A simple and sensitive electrochemiluminescence aptasensor for determination of ochratoxin A based on a nicking endonuclease-powered DNA walking machine. Food Chem. 2019, 282, 141–146. [Google Scholar] [CrossRef] [PubMed]
- Kaur, N.; Bharti, A.; Batra, S.; Rana, S.; Rana, S.; Bhalla, A.; Prabhakar, N. An electrochemical aptasensor based on graphene doped chitosan nanocomposites for determination of Ochratoxin A. Microchem. J. 2019, 144, 102–109. [Google Scholar] [CrossRef]
- Wei, M.; Zhang, W.Y. The determination of Ochratoxin A based on the electrochemical aptasensor by carbon aerogels and methylene blue assisted signal amplification. Chem. Cent. J. 2018, 12, 45. [Google Scholar] [CrossRef]
- Zhou, L.; Gan, N.; Zhou, Y.; Li, T.; Cao, Y.; Chen, Y. A label-free and universal platform for antibiotics detection based on microchip electrophoresis using aptamer probes. Talanta 2017, 167, 544–549. [Google Scholar] [CrossRef]
- Guo, X.; Wen, F.; Zheng, N.; Saive, M.; Fauconnier, M.L.; Wang, J. Aptamer-Based Biosensor for Detection of Mycotoxins. Front. Chem. 2020, 8, 195. [Google Scholar] [CrossRef] [PubMed]
- Jung, Y.; Lee, C.Y.; Park, K.S.; Park, H.G. Sensitive and specific detection of proteins based on target-responsive DNA polymerase activity. Anal. Chim. Acta 2019, 1059, 80–85. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Yu, Y.; Jiang, F.; Zhou, J.; Li, Y.; Liang, C.; Dang, L.; Lu, A.; Zhang, G. Development of Cell-SELEX Technology and Its Application in Cancer Diagnosis and Therapy. Int. J. Mol. Sci. 2016, 17, 2079. [Google Scholar] [CrossRef]
- Chen, X.J.; Bai, X.J.; Li, H.Y.; Zhang, B.L. Aptamer-based microcantilever array biosensor for detection of fumonisin B-1. RSC Adv. 2015, 5, 35448–35452. [Google Scholar] [CrossRef]
- Costantini, F.; Sberna, C.; Petrucci, G.; Reverberi, M.; Domenici, F.; Fanelli, C.; Manetti, C.; de Cesare, G.; DeRosa, M.; Nascetti, A.; et al. Aptamer-based sandwich assay for on chip detection of Ochratoxin A by an array of amorphous silicon photosensors. Sens. Actuators B Chem. 2016, 230, 31–39. [Google Scholar] [CrossRef]
- Cruz-Aguado, J.A.; Penner, G. Determination of Ochratoxin A with a DNA Aptamer. J. Agric. Food Chem. 2008, 56, 10456–10461. [Google Scholar] [CrossRef]
- Chen, L.; Wen, F.; Li, M.; Guo, X.; Li, S.; Zheng, N.; Wang, J. A simple aptamer-based fluorescent assay for the detection of Aflatoxin B1 in infant rice cereal. Food Chem. 2017, 215, 377–382. [Google Scholar] [CrossRef] [PubMed]
- Goud, K.Y.; Hayat, A.; Satyanarayana, M.; Kumar, V.S.; Catanante, G.; Gobi, K.V.; Marty, J.L. Aptamer-based zearalenone assay based on the use of a fluorescein label and a functional graphene oxide as a quencher. Microchim. Acta 2017, 184, 4401–4408. [Google Scholar] [CrossRef]
- Ruscito, A.; Smith, M.; Goudreau, D.N.; DeRosa, M.C. Current Status and Future Prospects for Aptamer-Based Mycotoxin Detection. J. AOAC Int. 2016, 99, 865–877. [Google Scholar] [CrossRef]
- Bertone, P.; Snyder, M. Advances in functional protein microarray technology. FEBS J. 2005, 272, 5400–5411. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Li, P.; Hu, X.; Zhang, Q.; Ding, X.; Zhang, W. Microarray technology for major chemical contaminants analysis in food: Current status and prospects. Sensors 2012, 12, 9234–9252. [Google Scholar] [CrossRef]
- Zhao, H.; Xiang, X.Y.; Chen, M.J.; Ma, C.B. Aptamer-Based Fluorometric Ochratoxin A Assay Based on Photoinduced Electron Transfer. Toxins 2019, 11, 65. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Wang, H.; Lin, Z.-T.; Hong, X.; Heon, M.; Wu, T. Protein arrays II: Antigen arrays. In Functional Genomics; Springer: Berlin/Heidelberg, Germany, 2017; pp. 271–277. [Google Scholar]
- Wu, T.; Du, Y.; Han, J.; Singh, S.; Xie, C.; Guo, Y.; Zhou, X.J.; Ahn, C.; Saxena, R.; Mohan, C. Urinary angiostatin—A novel putative marker of renal pathology chronicity in lupus nephritis. Mol. Cell. Proteom. 2013, 12, 1170–1179. [Google Scholar] [CrossRef]
- Yuan, Y.; Qiu, J.; Lin, Z.T.; Li, W.; Haley, C.; Mui, U.N.; Ning, J.; Tyring, S.K.; Wu, T. Identification of Novel Autoantibodies Associated with Psoriatic Arthritis. Arthritis Rheumatol. 2019, 71, 941–951. [Google Scholar] [CrossRef]
- Qiu, J.; Yuan, Y.; Li, Y.; Haley, C.; Mui, U.N.; Swali, R.; Mohan, C.; Tyring, S.K.; Wu, T. Discovery of IgG4 Anti-Gliadin Autoantibody as a Potential Biomarker of Psoriasis Using an Autoantigen Array. Proteom. Clin. Appl. 2019, e1800114. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Lin, Z.-T.; Wang, H.; Hong, X.; Heon, M.; Wu, T. Protein arrays I: Antibody arrays. In Functional Genomics; Springer: Berlin/Heidelberg, Germany, 2017; pp. 261–269. [Google Scholar]
- Bulyk, M.L. DNA microarray technologies for measuring protein-DNA interactions. Curr. Opin. Biotechnol. 2006, 17, 422–430. [Google Scholar] [CrossRef][Green Version]
- Liang, L.; Juarez, S.; Nga, T.V.; Dunstan, S.; Nakajima-Sasaki, R.; Davies, D.H.; McSorley, S.; Baker, S.; Felgner, P.L. Immune profiling with a Salmonella Typhi antigen microarray identifies new diagnostic biomarkers of human typhoid. Sci. Rep. 2013, 3, 1043. [Google Scholar] [CrossRef]
- Monsurro, V.; Marincola, F.M. Microarray analysis for a comprehensive immunological-status evaluation during cancer vaccine immune monitoring. J. Biomed. Biotechnol. 2011, 2011, 307297. [Google Scholar] [CrossRef] [PubMed]
- Dang, K.; Zhang, W.; Jiang, S.; Lin, X.; Qian, A. Application of Lectin Microarrays for Biomarker Discovery. ChemistryOpen 2020, 9, 285–300. [Google Scholar] [CrossRef]
- Balashanmugam, M.V.; Shivanandappa, T.B.; Nagarethinam, S.; Vastrad, B.; Vastrad, C. Analysis of Differentially Expressed Genes in Coronary Artery Disease by Integrated Microarray Analysis. Biomolecules 2019, 10, 35. [Google Scholar] [CrossRef]
- Schumacher, S.; Muekusch, S.; Seitz, H. Up-to-Date Applications of Microarrays and Their Way to Commercialization. Microarrays 2015, 4, 196–213. [Google Scholar] [CrossRef] [PubMed]
- Logrieco, A.; Arrigan, D.W.; Brengel-Pesce, K.; Siciliano, P.; Tothill, I. DNA arrays, electronic noses and tongues, biosensors and receptors for rapid detection of toxigenic fungi and mycotoxins: A review. Food Addit. Contam. 2005, 22, 335–344. [Google Scholar] [CrossRef]
- Dorokhin, D.; Haasnoot, W.; Franssen, M.C.; Zuilhof, H.; Nielen, M.W. Imaging surface plasmon resonance for multiplex microassay sensing of mycotoxins. Anal. Bioanal. Chem. 2011, 400, 3005–3011. [Google Scholar] [CrossRef]
- Sauceda-Friebe, J.C.; Karsunke, X.Y.; Vazac, S.; Biselli, S.; Niessner, R.; Knopp, D. Regenerable immuno-biochip for screening ochratoxin A in green coffee extract using an automated microarray chip reader with chemiluminescence detection. Anal. Chim. Acta 2011, 689, 234–242. [Google Scholar] [CrossRef]
- Oswald, S.; Karsunke, X.Y.; Dietrich, R.; Martlbauer, E.; Niessner, R.; Knopp, D. Automated regenerable microarray-based immunoassay for rapid parallel quantification of mycotoxins in cereals. Anal. Bioanal. Chem. 2013, 405, 6405–6415. [Google Scholar] [CrossRef]
- Oswald, S.; Dietrich, R.; Martlbauer, E.; Niessner, R.; Knopp, D. Microarray-Based Immunoassay for Parallel Quantification of Multiple Mycotoxins in Oat. Methods Mol. Biol. 2017, 1536, 143–156. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Chen, Q.; Xu, X.; Jin, Y.; Wang, Y.; Zhang, L.; Yang, W.; He, L.; Feng, X.; Chen, Y. Microarray surface enhanced Raman scattering based immunosensor for multiplexing detection of mycotoxin in foodstuff. Sens. Actuators B Chem. 2018, 266, 115–123. [Google Scholar] [CrossRef]
- Liu, R.; Li, W.; Cai, T.; Deng, Y.; Ding, Z.; Liu, Y.; Zhu, X.; Wang, X.; Liu, J.; Liang, B.; et al. TiO2 Nanolayer-Enhanced Fluorescence for Simultaneous Multiplex Mycotoxin Detection by Aptamer Microarrays on a Porous Silicon Surface. ACS Appl. Mater. Interfaces 2018, 10, 14447–14453. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Li, W.; Shen, P.; Liu, R.; Li, Y.; Xu, J.; Zheng, Q.; Zhang, Y.; Li, J.; Zheng, T. Aptamer fluorescence signal recovery screening for multiplex mycotoxins in cereal samples based on photonic crystal microsphere suspension array. Sens. Actuators B Chem. 2017, 248, 351–358. [Google Scholar] [CrossRef]
- Rasooly, A.; Herold, K.E. Food microbial pathogen detection and analysis using DNA microarray technologies. Foodborne Pathog. Dis. 2008, 5, 531–550. [Google Scholar] [CrossRef] [PubMed]
- Arora, P.; Sindhu, A.; Dilbaghi, N.; Chaudhury, A. Biosensors as innovative tools for the detection of food borne pathogens. Biosens. Bioelectron. 2011, 28, 1–12. [Google Scholar] [CrossRef] [PubMed]


| OTA Spiked-In (ng/g) | OTA Detected (ng/g) | Mean Recovery ACSB (%) | Coefficient of Variation, ACSB (%) | ||
|---|---|---|---|---|---|
| ELISA | ACSB | Pearson Correlation | |||
| 1 | 0.83 ± 0.07 | 0.91 ± 0.09 | r = 0.99 p < 0.0001 | 91 | 9.89 |
| 10 | 9.21 ± 0.41 | 9.03 ± 0.87 | 90 | 9.63 | |
| 100 | 88.56 ± 5.37 | 85.26 ± 7.31 | 85 | 8.57 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Y.; Peng, Z.; Li, Y.; Xiao, M.; Tan, G.; Wang, W.; Wang, Y.; Fang, M.; Zhang, S.; Tang, C.; et al. An Aptamer-Array-Based Sample-to-Answer Biosensor for Ochratoxin A Detection via Fluorescence Resonance Energy Transfer. Chemosensors 2021, 9, 309. https://doi.org/10.3390/chemosensors9110309
Li Y, Peng Z, Li Y, Xiao M, Tan G, Wang W, Wang Y, Fang M, Zhang S, Tang C, et al. An Aptamer-Array-Based Sample-to-Answer Biosensor for Ochratoxin A Detection via Fluorescence Resonance Energy Transfer. Chemosensors. 2021; 9(11):309. https://doi.org/10.3390/chemosensors9110309
Chicago/Turabian StyleLi, Yongning, Zhenfei Peng, Yaxi Li, Min Xiao, Gongjun Tan, Wenlian Wang, Yu Wang, Min Fang, Shu Zhang, Chenling Tang, and et al. 2021. "An Aptamer-Array-Based Sample-to-Answer Biosensor for Ochratoxin A Detection via Fluorescence Resonance Energy Transfer" Chemosensors 9, no. 11: 309. https://doi.org/10.3390/chemosensors9110309
APA StyleLi, Y., Peng, Z., Li, Y., Xiao, M., Tan, G., Wang, W., Wang, Y., Fang, M., Zhang, S., Tang, C., Yang, B., & Wu, T. (2021). An Aptamer-Array-Based Sample-to-Answer Biosensor for Ochratoxin A Detection via Fluorescence Resonance Energy Transfer. Chemosensors, 9(11), 309. https://doi.org/10.3390/chemosensors9110309







