Functionalized Surfaces as a Tool for Virus Sensing: A Demonstration of Human mastadenovirus Detection in Environmental Waters
Abstract
1. Introduction
2. Materials and Methods
2.1. Samples and Sampling
2.1.1. Samples for Standardization of IMS Parameters
2.1.2. Validation in Naturally Contaminated Water Samples
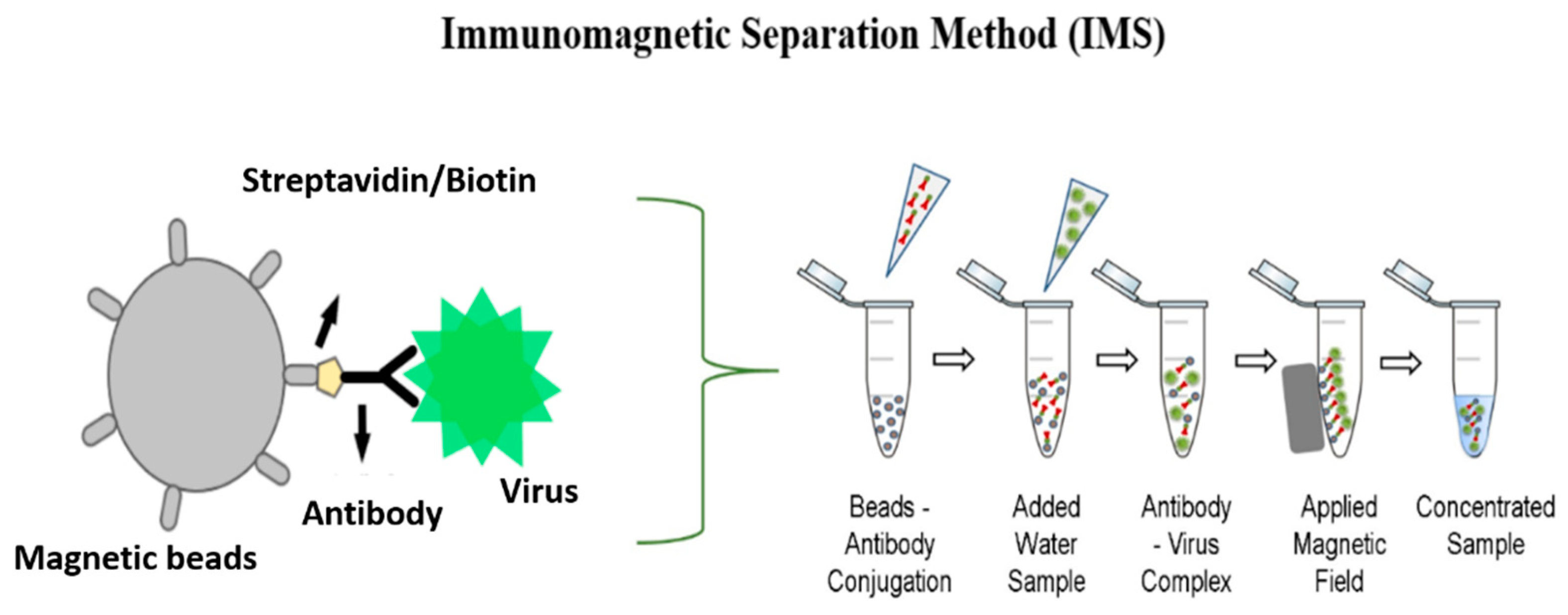
2.2. Magnetic Bead Functionalization
2.3. Concentration Steps
2.3.1. IMS Method
2.3.2. Ultracentrifugation Method
2.4. Viral Extraction
2.5. Quantitative Real-Time Polymerase Chain Reaction
2.6. Nested Polymerase Chain Reaction (Nested PCR)
2.7. Digital Polymerase Chain Reaction
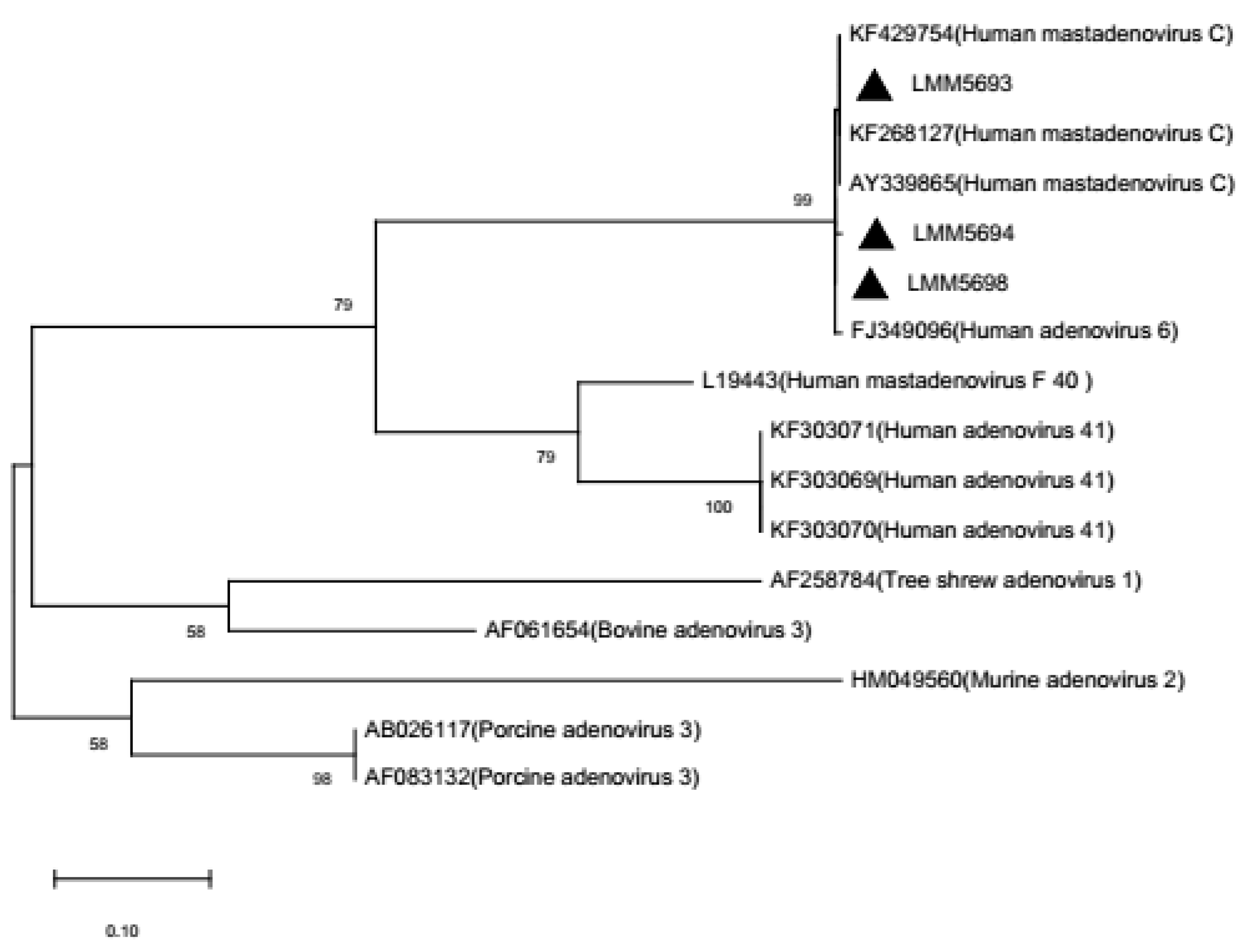
2.8. Phylogenetic Analysis
2.9. Viral Isolation
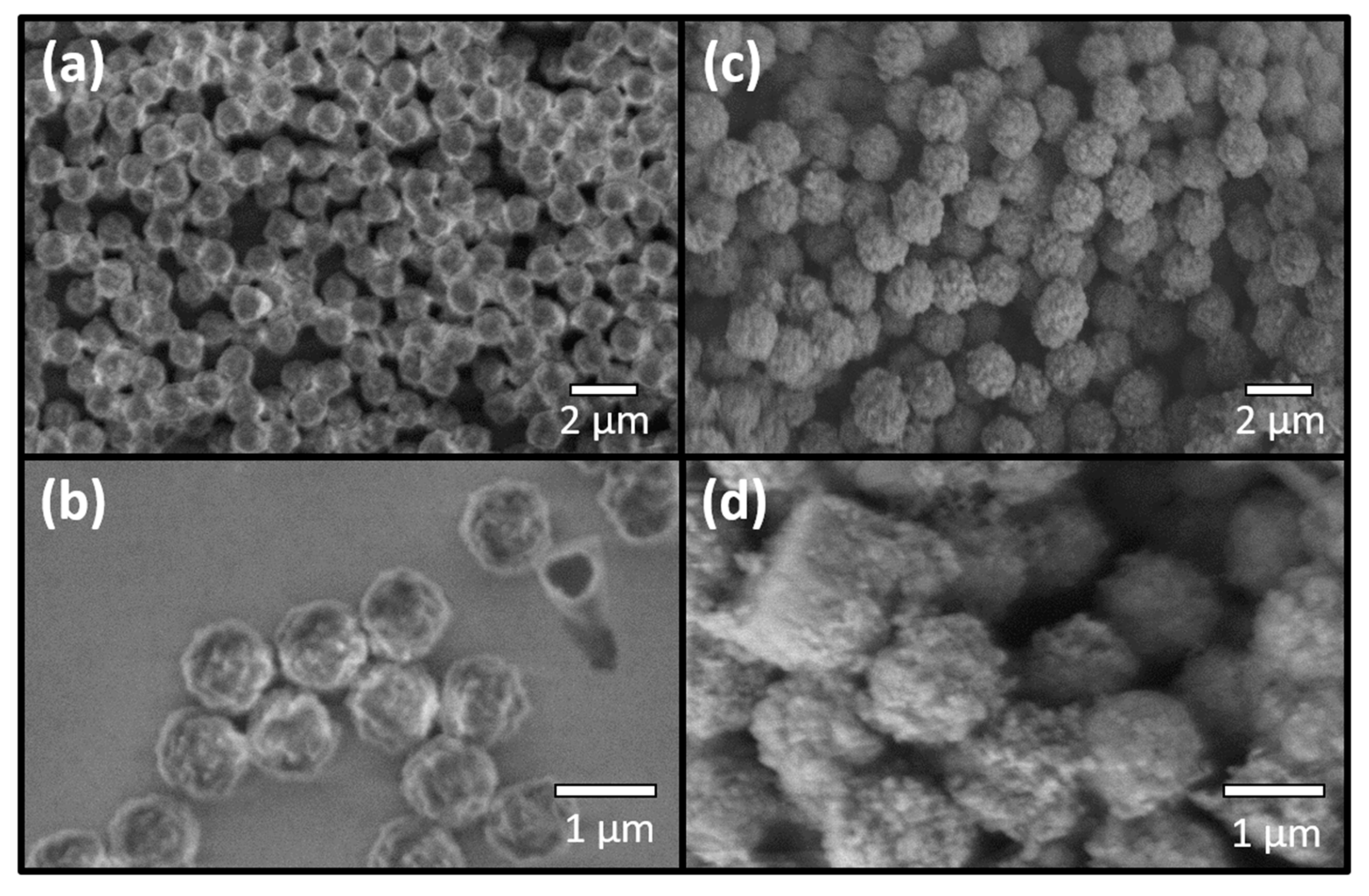
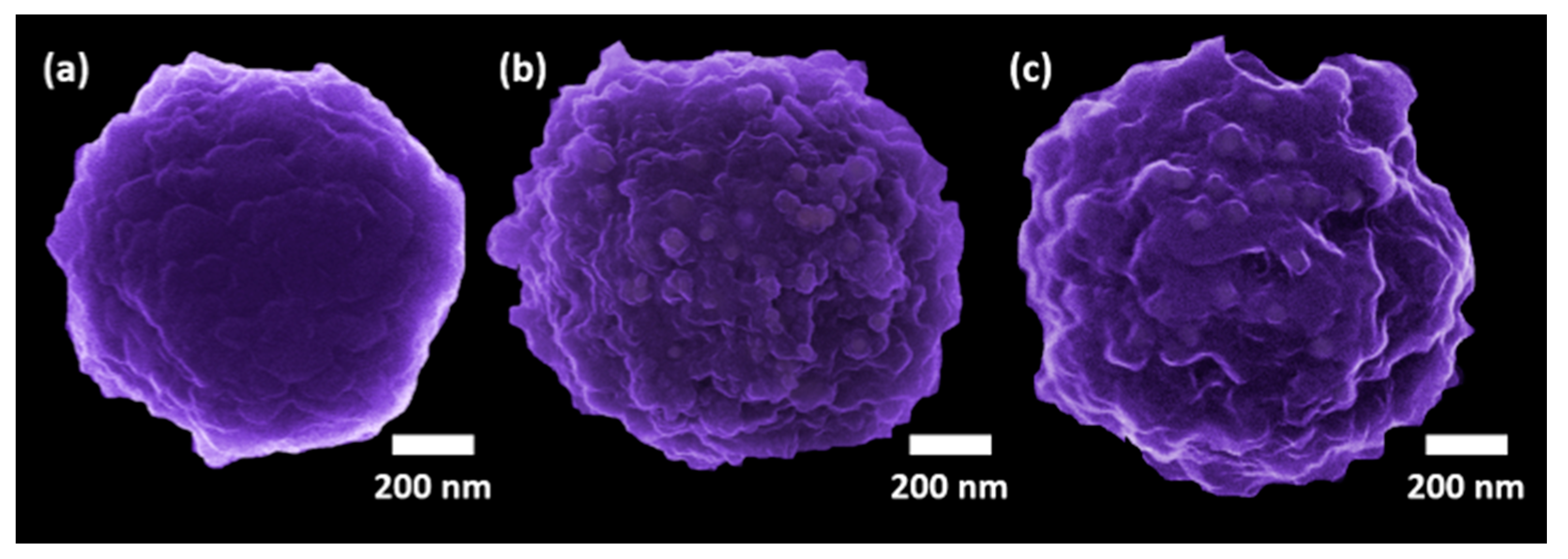
2.10. Microscopic Analysis
2.11. Viral Recovery
2.12. Statistical Analysis
3. Results
3.1. Standardization of IMS Parameters
3.2. Water Samples Analyses
4. Discussion
4.1. Standardization of IMS Parameters
4.2. Water Sample Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Val, A.L.; de Bicudo, C.E.M.; de Bicudo, D.C.; Florencio, D.G.; Spilki, F.R.; de Nogueira, I.S.; Hespanhol, I.; Cirilo, J.A.; Tundisi, J.G.; Val, P.; et al. Water quality in Brazil. In Water Quality in the Americas: Risks and Opportunities; Roldán, G., Tundisi, J., Jiménez, B., Vammen, K., Vaux, H., González, E., Doria, M., Eds.; Inter American Network of Academies of Sciences (IANAS): Trieste, Italy, 2019; pp. 104–126. ISBN 978-607-8379-33-0. [Google Scholar]
- Girones, R.; Ferrús, M.A.; Alonso, J.L.; Rodriguez-Manzano, J.; Calgua, B.; de Abreu Corrêa, A.; Hundesa, A.; Carratala, A.; Bofill-Mas, S. Molecular detection of pathogens in water—The pros and cons of molecular techniques. Water Res. 2010, 44, 4325–4339. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. The Top 10 Causes of Death. Available online: https://www.who.int/news-room/fact-sheets/detail/the-top-10-causes-of-death (accessed on 10 March 2020).
- Bosch, A.; Guix, S.; Sano, D.; Pintó, R.M. New tools for the study and direct surveillance of viral pathogens in water. Curr. Opin. Biotechnol. 2008, 19, 295–301. [Google Scholar] [CrossRef]
- Carter, M.J. Enterically infecting viruses: Pathogenicity, transmission and significance for food and waterborne infection. J. Appl. Microbiol. 2005, 98, 1354–1380. [Google Scholar] [CrossRef]
- ICTV—International Commitee on Taxonomy of Viruses. Adenoviridae. 2018. Available online: https://talk.ictvonline.org/ictv-reports/ictv_9th_report/dsdna-viruses-2011/w/dsdna_viruses/93/adenoviridae (accessed on 20 May 2020).
- Ryu, W.-S. Adenoviruses. In Molecular Virology of Human Pathogenic Viruses; Elsevier: London, UK, 2017; pp. 111–124. ISBN 9780128037089. [Google Scholar]
- Santos, N.S.O.; Soares, C.C. Viroses entéricas. In Virologia Humana; Santos, N.S.O., Romanos, M.T.V., Wigg, M.D., Eds.; Guanabara Koogan: Rio de Janeiro, Brazil, 2015; pp. 189–231. [Google Scholar]
- Haramoto, E.; Katayama, H.; Oguma, K.; Ohgaki, S. Quantitative analysis of human enteric adenoviruses in aquatic environments. J. Appl. Microbiol. 2007, 103, 2153–2159. [Google Scholar] [CrossRef]
- Mena, K.D.; Gerba, C.P. Waterborne adenovirus. Rev. Environ. Contam. Toxicol. 2009, 198, 133–167. [Google Scholar] [PubMed]
- Albinana-Gimenez, N.; Miagostovich, M.P.; Calgua, B.; Huguet, J.M.; Matia, L.; Girones, R. Analysis of adenoviruses and polyomaviruses quantified by qPCR as indicators of water quality in source and drinking-water treatment plants. Water Res. 2009, 43, 2011–2019. [Google Scholar] [CrossRef] [PubMed]
- Vecchia, A.D.; Rigotto, C.; Staggemeier, R.; Soliman, M.C.; de Souza, F.G.; Henzel, A.; Santos, E.L.; Do Nascimento, C.A.; De Quevedo, D.M.; Fleck, J.D.; et al. Surface water quality in the Sinos River basin, in Southern Brazil: Tracking microbiological contamination and correlation with physicochemical parameters. Environ. Sci. Pollut. Res. 2015, 22, 9899–9911. [Google Scholar] [CrossRef]
- Girardi, V.; Demoliner, M.; Rigotto, C.; Schneider, V.E.; Paesi, S.; Spilki, F.R. Assessment of diversity of adenovirus DNA polymerase gene in recreational waters facilitated by ultracentrifugal concentration. J. Water Health 2018, 1–10. [Google Scholar] [CrossRef]
- Verani, M.; Federigi, I.; Donzelli, G.; Cioni, L.; Carducci, A. Human adenoviruses as waterborne index pathogens and their use for Quantitative Microbial Risk Assessment. Sci. Total Environ. 2019, 651, 1469–1475. [Google Scholar] [CrossRef]
- Gularte, J.S.; Girardi, V.; Demoliner, M.; de Souza, F.G.; Filippi, M.; Eisen, A.K.A.; Mena, K.D.; De Quevedo, D.M.; Rigotto, C.; de Barros, M.P.; et al. Human mastadenovirus in water, sediment, sea surface microlayer, and bivalve mollusk from southern Brazilian beaches. Mar. Pollut. Bull. 2019, 142, 335–349. [Google Scholar] [CrossRef]
- Fong, T.-T.; Lipp, E.K. Enteric viruses of humans and animals in aquatic environments: Health risks, detection, and potential water quality assessment tools. Microbiol. Mol. Biol. Rev. 2005, 69, 357–371. [Google Scholar] [CrossRef] [PubMed]
- Thurston-enriquez, J.A.; Haas, C.N.; Riley, K.; Gerba, C.P.; Jacangelo, J. Inactivation of Feline Calicivirus and Adenovirus Type 40 by UV Radiation Inactivation of Feline Calicivirus and Adenovirus Type 40 by UV Radiation. Appl. Environ. Mcrobiol. 2003, 69, 577–582. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez, R.A.; Pepper, I.L.; Gerba, C.P. Application of PCR-based methods to assess the infectivity of enteric viruses in environmental samples. Appl. Environ. Microbiol. 2009, 75, 297–307. [Google Scholar] [CrossRef] [PubMed]
- Lodder, W.J.; Van Den Berg, H.H.J.L.; Rutjes, S.A.; De Roda Husman, A.M. Presence of enteric viruses in source waters for drinking water production in the Netherlands. Appl. Environ. Microbiol. 2010, 76, 5965–5971. [Google Scholar] [CrossRef]
- Hamza, I.A.; Jurzik, L.; Überla, K.; Wilhelm, M. Methods to detect infectious human enteric viruses in environmental water samples. Int. J. Hyg. Environ. Health 2011, 214, 424–436. [Google Scholar] [CrossRef] [PubMed]
- Hamza, I.A.; Bibby, K. Critical issues in application of molecular methods to environmental virology. J. Virol. Methods 2019, 266, 11–24. [Google Scholar] [CrossRef] [PubMed]
- Prata, C.; Ribeiro, A.; Cunha, Â.; Gomes, N.C.M.; Almeida, A. Ultracentrifugation as a direct method to concentrate viruses in environmental waters: Virus-like particle enumeration as a new approach to determine the efficiency of recovery. J. Environ. Monit. 2012, 14, 64–70. [Google Scholar] [CrossRef]
- Ahmed, S.; Rubahn, H.-G.; Erdmann, H. Development of an Immunomagnetic Separation Method for Viable Salmonella Typhimurium Detected by Flow Cytometry. Online J. Biol. Sci. 2016, 16, 165–174. [Google Scholar] [CrossRef][Green Version]
- Haramoto, E.; Kitajima, M.; Katayama, H.; Ohgaki, S. Real-time PCR detection of adenoviruses, polyomaviruses, and torque teno viruses in river water in Japan. Water Res. 2010, 44, 1747–1752. [Google Scholar] [CrossRef]
- Kjelstrup-Hansen, J.; Tavares, L.; de Oliveira Hansen, R.; Liu, X.; Bordo, K.; Rubahn, H.-G. Optical properties of microstructured surface-grown and transferred organic nanofibers. J. Nanophotonics 2011, 5, 051701. [Google Scholar] [CrossRef]
- Wang, Y.; Costa, C.A.B.; Sobolewska, E.K.; Fiutowski, J.; Brehm, R.; Albers, J.; Nebling, E.; Lofink, F.; Wagner, B.; Benecke, W.; et al. Micro-cantilevers for optical sensing of biogenic amines. Microsyst. Technol. 2018, 24, 363–369. [Google Scholar] [CrossRef]
- Costa, C.A.B.; Grazhdan, D.; Fiutowski, J.; Nebling, E.; Blohm, L.; Lofink, F.; Rubahn, H.G.; de Oliveira Hansen, R. Meat and fish freshness evaluation by functionalized cantilever-based biosensors. Microsyst. Technol. 2020, 26, 867–871. [Google Scholar] [CrossRef]
- Korsa, M.T.; Maria Carmona Domingo, J.; Nsubuga, L.; Hvam, J.; Niekiel, F.; Lofink, F.; Rubahn, H.G.; Adam, J.; Hansen, R.D.O. Optimizing piezoelectric cantilever design for electronic nose applications. Chemosensors 2020, 8, 114. [Google Scholar] [CrossRef]
- Grinde, B.; Jonassen, T.; Ushijima, H. Sensitive detection of group A rotaviruses by immunomagnetic separation and reverse transcription-polymerase chain reaction. J. Virol. Methods 1995, 55, 327–338. [Google Scholar] [CrossRef]
- Yao, L.; Wu, Q.; Wang, D.; Kou, X.; Zhang, J. Development of monoclonal antibody-coated immunomagnetic beads for separation and detection of norovirus (genogroup II) in faecal extract samples. Lett. Appl. Microbiol. 2009, 49, 173–178. [Google Scholar] [CrossRef]
- Yang, W.; Gu, A.Z.; Zeng, S.Y.; Li, D.; He, M.; Shi, H.C. Development of a combined immunomagnetic separation and quantitative reverse transcription-PCR assay for sensitive detection of infectious rotavirus in water samples. J. Microbiol. Methods 2011, 84, 447–453. [Google Scholar] [CrossRef]
- IBGE Seberi, RS, BR. Available online: https://cidades.ibge.gov.br/brasil/rs/seberi/panorama (accessed on 27 October 2019).
- Wolf, S.; Hewitt, J.; Greening, G.E. Viral multiplex quantitative PCR assays for tracking sources of fecal contamination. Appl. Environ. Microbiol. 2010, 76, 1388–1394. [Google Scholar] [CrossRef]
- Li, Y.; Ge, X.; Zhang, H.; Zhou, P.; Zhu, Y.; Zhang, Y.; Yuan, J.; Wang, L.F.; Shi, Z. Host range, prevalence, and genetic diversity of adenoviruses in bats. J. Virol. 2010, 84, 3889–3897. [Google Scholar] [CrossRef]
- Majumdar, N.; Wessel, T.; Marks, J. Digital PCR Modeling for Maximal Sensitivity, Dynamic Range and Measurement Precision. PLoS ONE 2015, 10, e0118833. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The Neighbor-joining Method: A New Method for Reconstructing Phylogenetic Trees’. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Novikov, S.M.; Popok, V.N.; Evlyukhin, A.B.; Hanif, M.; Morgen, P.; Fiutowski, J.; Beermann, J.; Rubahn, H.-G.; Bozhevolnyi, S.I. Highly Stable Monocrystalline Silver Clusters for Plasmonic Applications. Langmuir 2017, 33, 6062–6070. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Li, Y.; Tao, T.; Bie, X.; Lu, F.; Lu, Z. Development and application of a sensitive, rapid, and reliable immunomagnetic separation-PCR detection method for Cronobacter spp. J. Dairy Sci. 2016, 100, 961–969. [Google Scholar] [CrossRef] [PubMed]
- Ha, J.-H.; Kim, M.L.; Choi, C.; Choi, I.-S.; Myoung, J.; Ha, S.-D. Recovery of structurally intact norovirus from food-contact surfaces. Food Control. 2015, 47, 564–568. [Google Scholar] [CrossRef]
- Lee, H.-M.; Yang, J.-S.; Yoon, S.-R.; Lee, J.Y.; Kim, S.-J.; Lee, H.-W.; Kim, S.H.; Ha, J.-H. Immunomagnetic separation combined with RT-qPCR for evaluating the effect of disinfectant treatments against norovirus on food contact surfaces. LWT 2018, 97, 83–86. [Google Scholar] [CrossRef]
- Haramoto, E.; Kitajima, M.; Hata, A.; Torrey, J.R.; Masago, Y.; Sano, D.; Katayama, H. A review on recent progress in the detection methods and prevalence of human enteric viruses in water. Water Res. 2018, 135, 168–186. [Google Scholar] [CrossRef]
- Singh, A.; Chaudhary, S.; Agarwal, A.; Verma, A.S. Antibodies: Monoclonal and polyclonal. In Animal Biotechnology: Models in Discovery and Translation; Verma, A.S., Singh, A., Eds.; Elsevier: Amsterdam, The Netherlands, 2014; pp. 265–287. ISBN 978-0-12-416002-6. [Google Scholar]
- Hwang, Y.; Leong, O.M.; Chen, W.; Yates, M. V Comparison of a Reporter Assay and Immunomagnetic Separation Real-Time Reverse Transcription-PCR for the Detection of Enteroviruses in Seeded Environmental Water Samples. Appl. Environ. Microbiol. 2007, 73, 2338–2340. [Google Scholar] [CrossRef]
- Ko, G.; Cromeans, T.L.; Sobsey, M.D. Detection of infectious Adenovirus by mRNA RT-PCR in cell culture. Appl. Environ. Microbiol. 2003, 69, 7377–7384. [Google Scholar] [CrossRef][Green Version]
- Bibby, K.; Peccia, J. Prevalence of respiratory adenovirus species B and C in sewage sludge. Environ. Sci. Process. Impacts 2013, 15, 336–338. [Google Scholar] [CrossRef]
- Staggemeier, R.; Heck, T.M.S.; Demoliner, M.; Ritzel, R.G.F.; Röhnelt, N.M.S.; Girardi, V.; Venker, C.A.; Spilki, F.R. Enteric viruses and adenovirus diversity in waters from 2016 Olympic venues. Sci. Total Environ. 2017, 586, 304–312. [Google Scholar] [CrossRef]
- Moresco, V.; Viancelli, A.; Nascimento, M.A.; Souza, D.S.M.; Ramos, A.P.D.; Garcia, L.A.T.; Simões, C.M.O.; Barardi, C.R.M. Microbiological and physicochemical analysis of the coastal waters of southern Brazil. Mar. Pollut. Bull. 2012, 64, 40–48. [Google Scholar] [CrossRef]
- Couceiro, N.S.O. Viroses respiratórias. In Virologia Humana; Santos, N.S.O., Romanos, M.T.V., Wigg, M.D., Eds.; Guanabara Koogan: Rio de Janeiro, Brazil, 2015; pp. 101–140. [Google Scholar]
- Rames, E.; Roiko, A.; Stratton, H.; Macdonald, J. Technical aspects of using human adenovirus as a viral water quality indicator. Water Res. 2016, 96, 308–326. [Google Scholar] [CrossRef]
- Zhao, M.; Guo, Y.; Qiu, F.; Wang, L.; Yang, S.; Feng, Z.; Li, G. Molecular and clinical characterization of human adenovirus associated with acute respiratory tract infection in hospitalized children. J. Clin. Virol. 2020, 123, 104254. [Google Scholar] [CrossRef]
- Vetter, M.R.; Staggemeier, R.; Vecchia, A.D.; Henzel, A.; Rigotto, C.; Spilki, F.R. Seasonal variation on the presence of adenoviruses in stools from non-diarrheic patients. Braz. J. Microbiol. 2015, 46, 749–752. [Google Scholar] [CrossRef][Green Version]
- Kosulin, K.; Geiger, E.; Vécsei, A.; Huber, W.D.; Rauch, M.; Brenner, E.; Wrba, F.; Hammer, K.; Innerhofer, A.; Pötschger, U.; et al. Persistence and reactivation of human adenoviruses in the gastrointestinal tract. Clin. Microbiol. Infect. 2016, 22, 381.e1–381.e8. [Google Scholar] [CrossRef]
- La Rosa, G.; Della Libera, S.; Petricca, S.; Iaconelli, M.; Donia, D.; Saccucci, P.; Cenko, F.; Xhelilaj, G.; Divizia, M. Genetic diversity of human adenovirus in children with acute gastroenteritis, Albania, 2013-2015. Biomed. Res. Int. 2015, 1–7. [Google Scholar] [CrossRef]
- Girardi, V.; Mena, K.D.; Albino, S.M.; Demoliner, M.; Gularte, J.S.; de Souza, F.G.; Rigotto, C.; Quevedo, D.M.; Schneider, V.E.; Paesi, S.O.; et al. Microbial risk assessment in recreational freshwaters from southern Brazil. Sci. Total Environ. 2019, 651. [Google Scholar] [CrossRef]
- Summa, M.; von Bonsdorff, C.H.; Maunula, L. Evaluation of four virus recovery methods for detecting noroviruses on fresh lettuce, sliced ham, and frozen raspberries. J. Virol. Methods 2012, 183, 154–160. [Google Scholar] [CrossRef]
- Chaturvedi, A.; Gorthi, S.S. Automated blood sample preparation unit (ABSPU) for portable microfluidic flow cytometry. SLAS Technol. 2017, 22, 73–80. [Google Scholar] [CrossRef]
- Chen, S.L.; Chang, W.H.; Wang, C.H.; You, H.L.; Wu, J.J.; Liu, T.H.; Lee, M.S.; Lee, G. Bin An integrated microfluidic system for live bacteria detection from human joint fluid samples by using ethidium monoazide and loop-mediated isothermal amplification. Microfluid. Nanofluidics 2017, 21, 1–12. [Google Scholar] [CrossRef]
- Ahmed, S.; Noh, J.W.; Hoyland, J.; de Oliveira Hansen, R.; Erdmann, H.; Rubahn, H.-G. On-chip immunomagnetic separation of bacteria by in-flow dynamic manipulation of paramagnetic beads. Appl. Phys. A 2016, 122, 955. [Google Scholar] [CrossRef]



| Virus | Target Gene | Name | Sequence (5′–3′) | Sense | Product Length | References |
|---|---|---|---|---|---|---|
| HAdV-C (1,2,5,6) | Hexon | VTB2-HAdVCf | GAGACGTACTTCAGCCTGAAT | + | 101 bp | [33] |
| VTB2-HAdVCr | GATGAACCGCAGCGTCAA | - | ||||
| VTB2-HAdVCprobe | CCTACGCACGACGTGACCACAGA | + | ||||
| HAdV-F (40,41) | Hexon | VTB1-HAdVFf | GCCTGGGGAACAAGTTCAGA | + | 137 bp | [33] |
| VTB1-HAdVFr | GCGTAAAGCGCACTTTGTAAG | - | ||||
| VTB1-HAdVFprobe | CAGTCGCTGYGACCTGTCTGTGGTT | - | ||||
| AdV | DNA polymerase | Pol-F | CAGCCKCKGTTRTGYAGGGT | + | 261 bp | [34] |
| Pol-R | GCHACCATYAGCTCCAACTC | - | ||||
| Pol-nF | GGGCTCRTTRGTCCAGCA | + | ||||
| Pol-nR | TAYGACATCTGYGGCATGTA | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gularte, J.S.; de Oliveira Hansen, R.; Demoliner, M.; Fiutowski, J.; Eisen, A.K.A.; Heldt, F.H.; Rodrigues de Almeida, P.; Müller de Quevedo, D.; Rubahn, H.-G.; Rosado Spilki, F. Functionalized Surfaces as a Tool for Virus Sensing: A Demonstration of Human mastadenovirus Detection in Environmental Waters. Chemosensors 2021, 9, 19. https://doi.org/10.3390/chemosensors9020019
Gularte JS, de Oliveira Hansen R, Demoliner M, Fiutowski J, Eisen AKA, Heldt FH, Rodrigues de Almeida P, Müller de Quevedo D, Rubahn H-G, Rosado Spilki F. Functionalized Surfaces as a Tool for Virus Sensing: A Demonstration of Human mastadenovirus Detection in Environmental Waters. Chemosensors. 2021; 9(2):19. https://doi.org/10.3390/chemosensors9020019
Chicago/Turabian StyleGularte, Juliana Schons, Roana de Oliveira Hansen, Meriane Demoliner, Jacek Fiutowski, Ana Karolina Antunes Eisen, Fagner Henrique Heldt, Paula Rodrigues de Almeida, Daniela Müller de Quevedo, Horst-Günter Rubahn, and Fernando Rosado Spilki. 2021. "Functionalized Surfaces as a Tool for Virus Sensing: A Demonstration of Human mastadenovirus Detection in Environmental Waters" Chemosensors 9, no. 2: 19. https://doi.org/10.3390/chemosensors9020019
APA StyleGularte, J. S., de Oliveira Hansen, R., Demoliner, M., Fiutowski, J., Eisen, A. K. A., Heldt, F. H., Rodrigues de Almeida, P., Müller de Quevedo, D., Rubahn, H.-G., & Rosado Spilki, F. (2021). Functionalized Surfaces as a Tool for Virus Sensing: A Demonstration of Human mastadenovirus Detection in Environmental Waters. Chemosensors, 9(2), 19. https://doi.org/10.3390/chemosensors9020019











