Cost-Effective Real-Time Metabolic Profiling of Cancer Cell Lines for Plate-Based Assays
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plates and Plate Readers
2.2. Fluorescent Dyes and Drugs
2.3. Media
2.4. Cells
3. Results
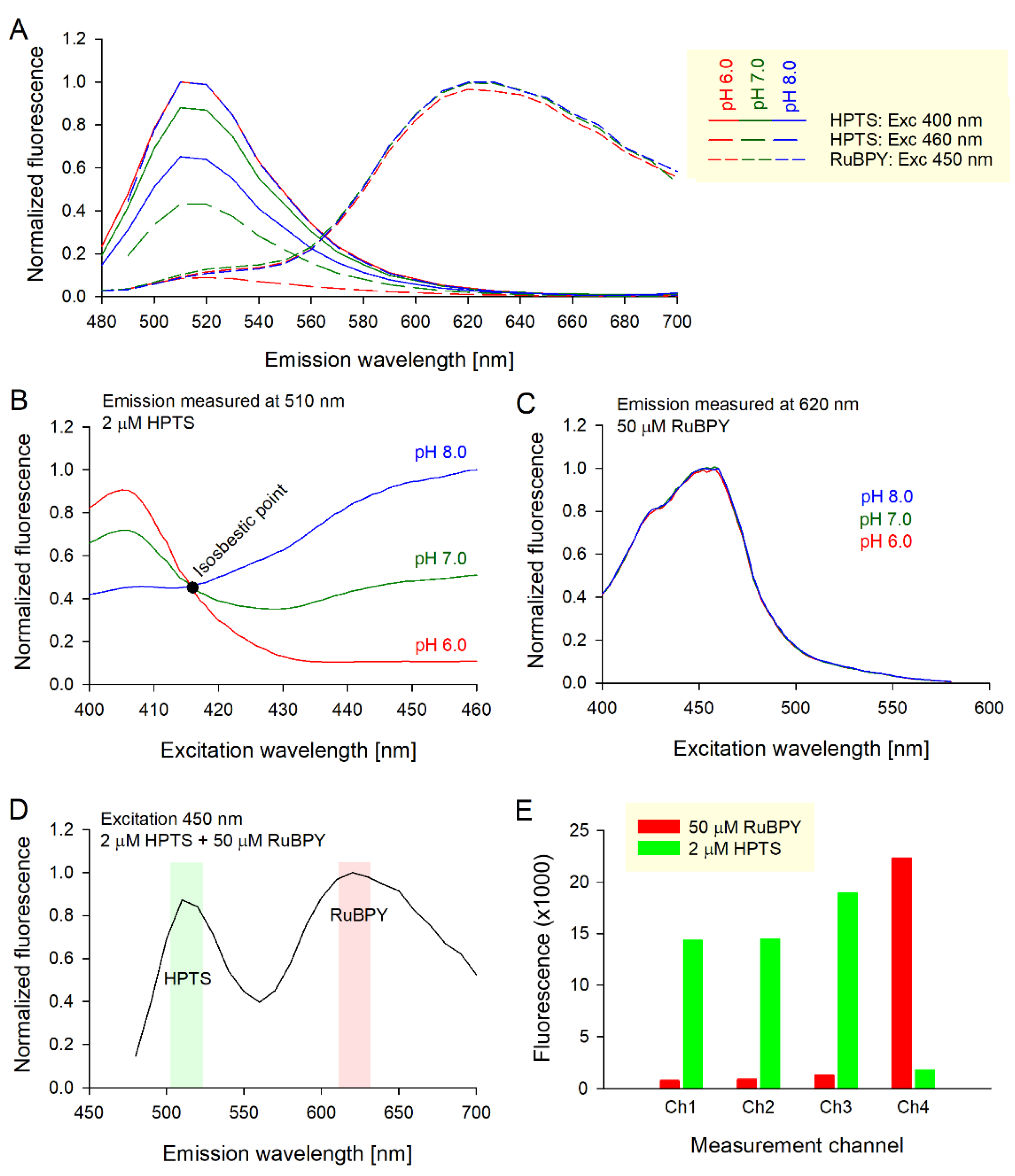
3.1. Selecting Cost-Effective pH and Oxygen Probes for Assaying Glycolysis and Respiration
- Ch1: Excitation 400 nm/emission 510 nm, optimized for HPTS fluorescence at low pH;
- Ch2: Excitation 460 nm/emission 510 nm, optimized for HPTS fluorescence at high pH;
- Ch3: Excitation 416 nm/emission 510 nm, optimized for pH-insensitive HPTS fluorescence as an O2-insensitive reference to O2-sensitive RuBPY;
- Ch4: Excitation 450 nm/emission 620 nm, optimized for RuBPY fluorescence, which is quenched by oxygen, i.e., inversely related to oxygen tension.
3.2. Calibration of Signals to Units of pH and Oxygen Tension
3.3. Converting pH and Oxygen Time Courses into Fluxes
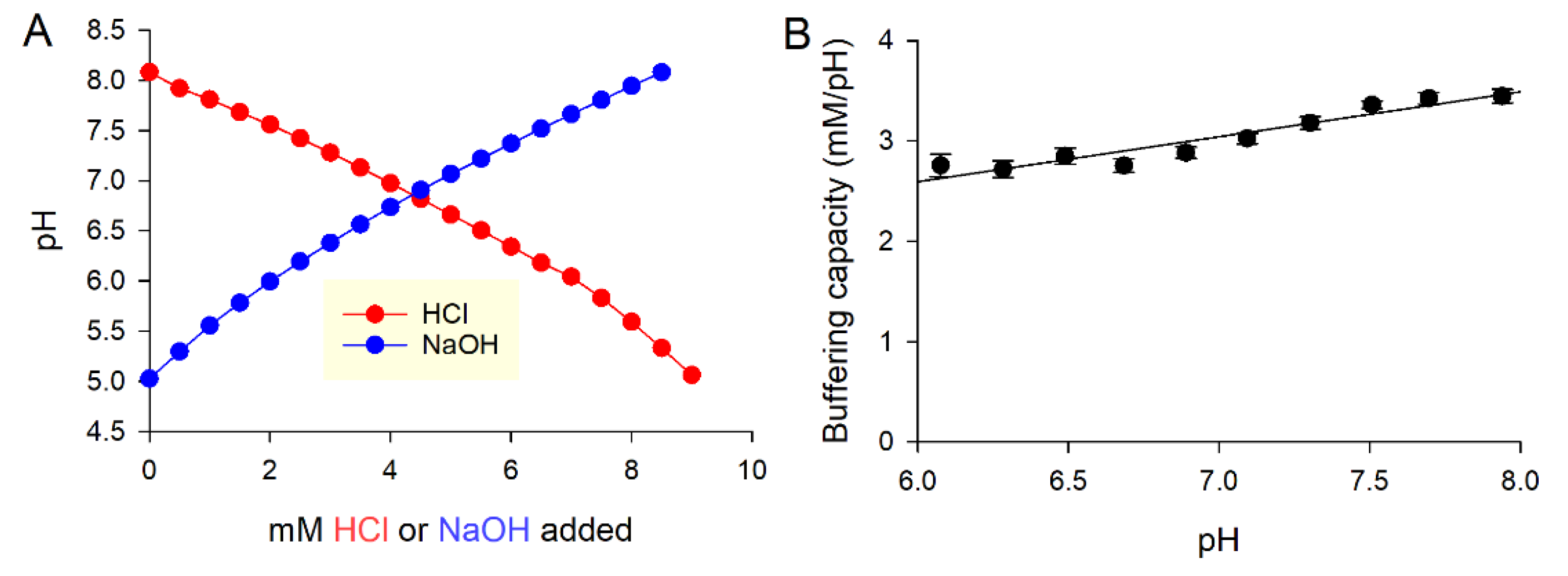
3.3.1. Buffering
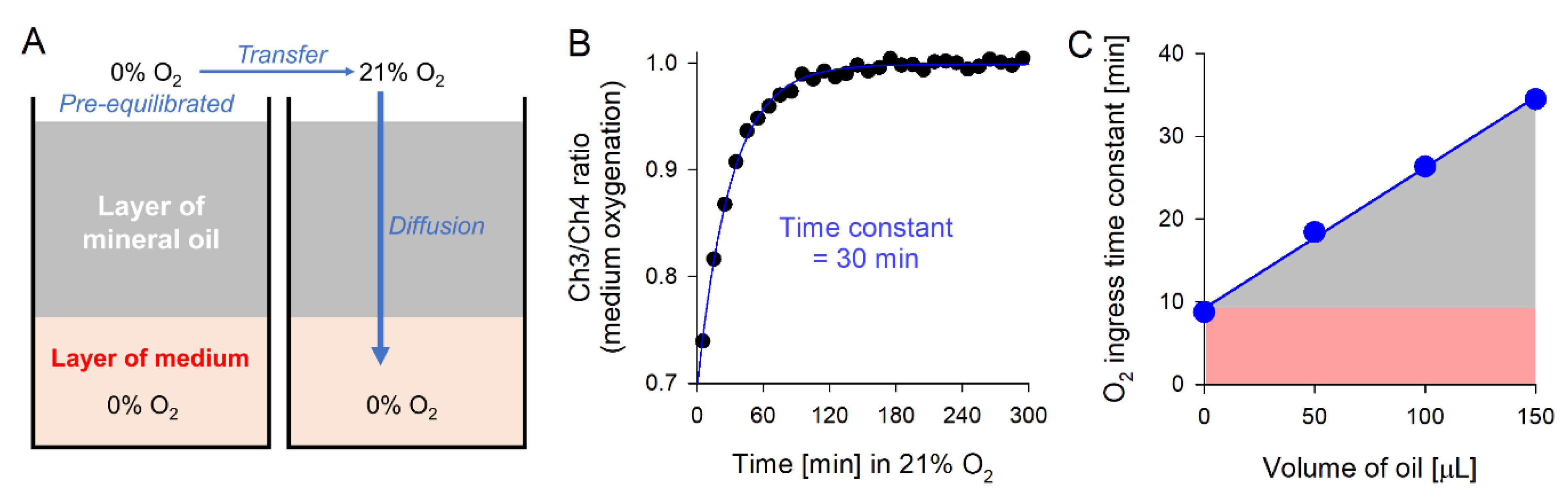
3.3.2. Open and Closed Systems
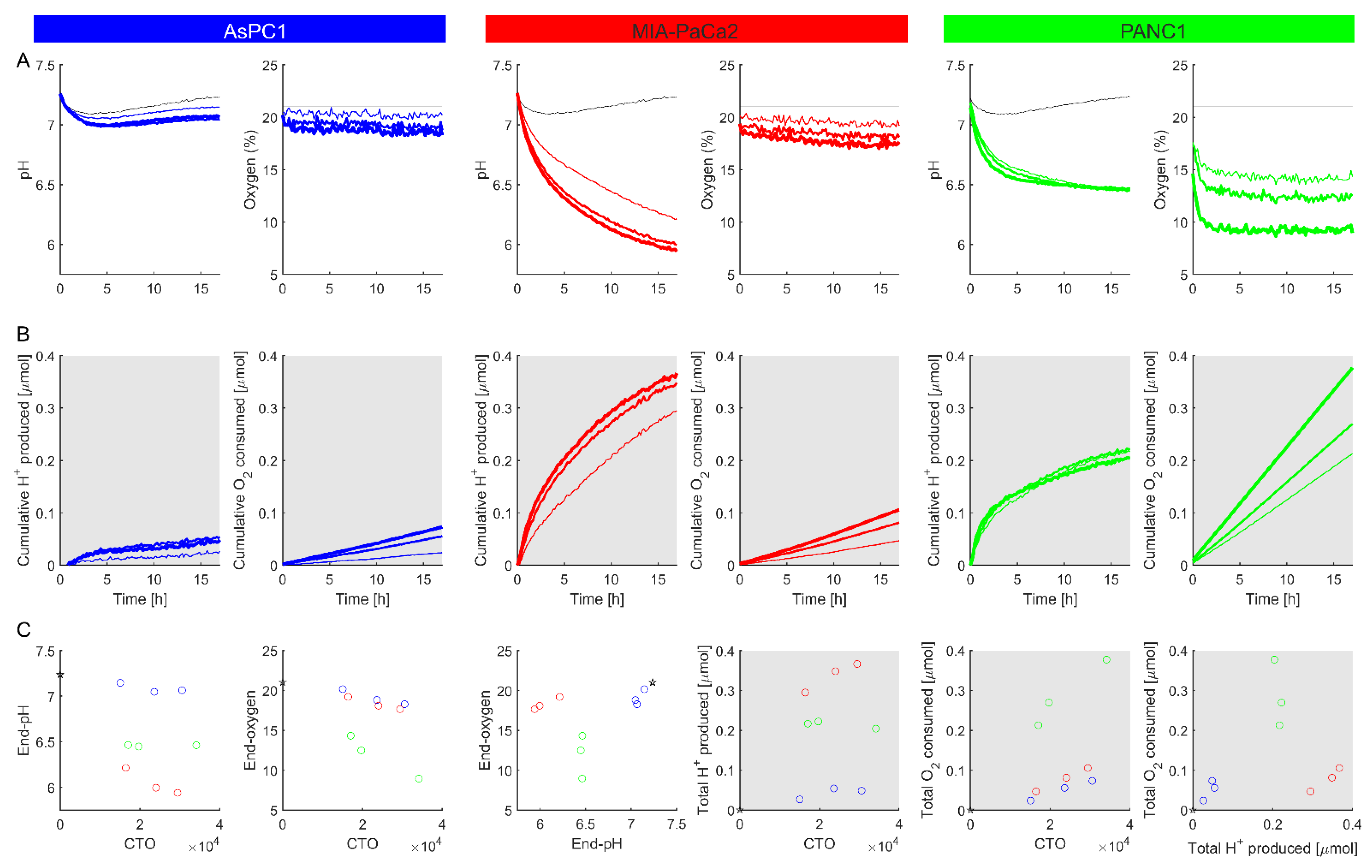
3.4. Implementation of Assay to Metabolically Phenotype Cancer Cells
3.4.1. Normalizing for Cell Number
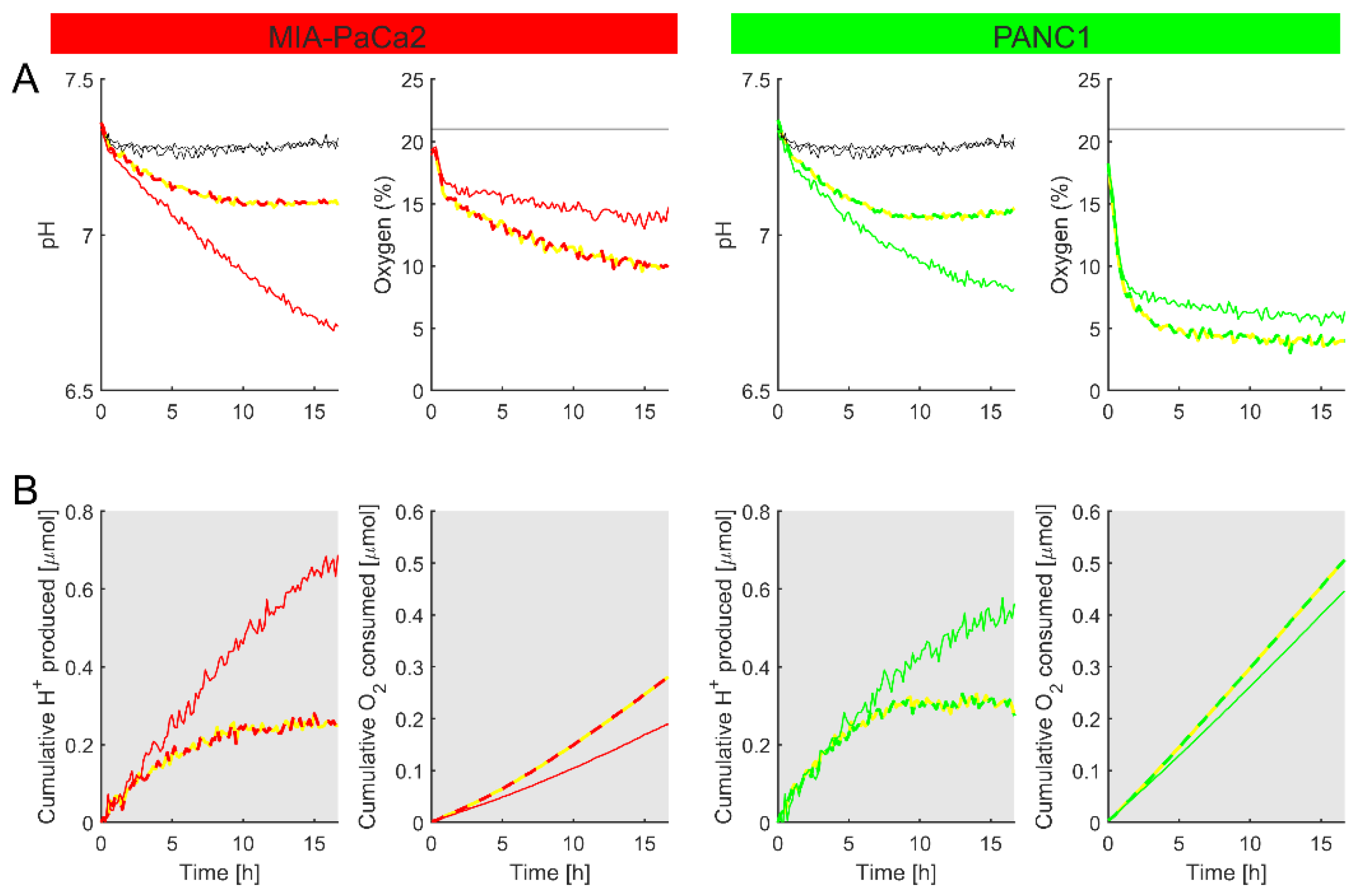
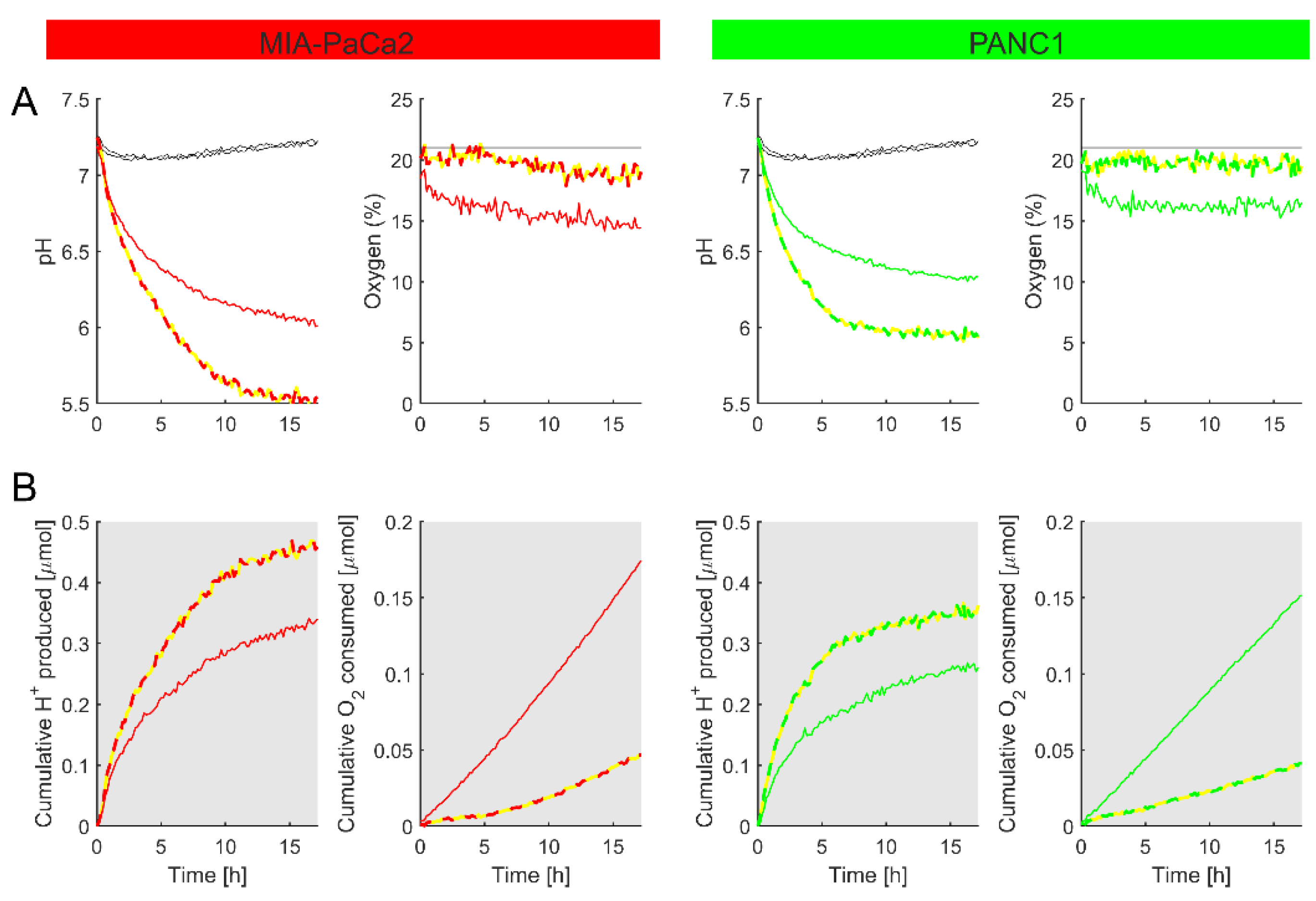
3.4.2. Real-Time Monitoring of Metabolic Fluxes
3.4.3. Measuring Metabolic Responses
3.5. Proof-of-Principle Measurements on Non-Adherent Cells
4. Discussion
5. Appendix: Step-by-Step Protocol
- (1)
- Dissolve HPTS and RuBPY in sterile, deionized water to obtain stocks of 4 and 100 mM, respectively. Mix both dyes in a 1:1 ratio, divide them into aliquots, and store them at −20 °C. Avoid multiple thaw-freeze cycles.
- (2)
- Seed cells onto a black, fluorescence-compatible, flat-bottom 96-well plate at the desired density and leave to attach overnight. Higher densities will produce larger and more resolvable fluxes. When planning the plate, ensure that some wells are cell-free (blanks) to serve as reference points for pH and O2. Recommendation: Add PBS to the outermost wells to help maintain humidity and prevent evaporation in the remainder of the plate.
- (3)
- Thaw and vortex the dye mixture. Dissolve (1:1000 v/v) in Phenol Red-free medium of choice. Allow aliquots of 100 µL per well.
- (4)
- Replace media that had bathed cells during the settling period with the dye-containing medium of the desired composition (e.g., pH, buffering, inhibitors etc). Whilst tilting the plate slightly, gently add 150 µL of mineral oil to each well to cover the medium and introduce a controlled diffusion barrier to gas exchange. Volumes of medium and mineral oil should be optimized for the given oxygen consumption and acid production rate.
- (5)
- Place the plate into the plate-reader, keeping the lid on. Start collecting the data immediately to capture the initial state.
- (6)
- Perform calculations according to the equations described herein. Note: Media prepared for the assay must be characterized in separate experiments in terms of buffering capacity and oxygen diffusivity.
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Birsoy, K.; Possemato, R.; Lorbeer, F.K.; Bayraktar, E.C.; Thiru, P.; Yucel, B.; Wang, T.; Chen, W.W.; Clish, C.B.; Sabatini, D.M. Metabolic determinants of cancer cell sensitivity to glucose limitation and biguanides. Nature 2014, 508, 108–112. [Google Scholar] [CrossRef] [Green Version]
- Nowell, P.C. The clonal evolution of tumor cell populations. Science 1976, 194, 23–28. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.; Wang, Z.; Tang, Y.; Lu, X.; Chen, J.; Dong, Y.; Wu, B.; Wang, C.; Yang, L.; Guo, Z.; et al. Liquid biopsy-based single-cell metabolic phenotyping of lung cancer patients for informative diagnostics. Nat. Commun. 2019, 10, 3856. [Google Scholar] [CrossRef] [Green Version]
- Ke, C.; Li, A.; Hou, Y.; Sun, M.; Yang, K.; Cheng, J.; Wang, J.; Ge, T.; Zhang, F.; Li, Q.; et al. Metabolic phenotyping for monitoring ovarian cancer patients. Sci. Rep. 2016, 6, 23334. [Google Scholar] [CrossRef] [Green Version]
- Jain, M.; Nilsson, R.; Sharma, S.; Madhusudhan, N.; Kitami, T.; Souza, A.L.; Kafri, R.; Kirschner, M.W.; Clish, C.B.; Mootha, V.K. Metabolite profiling identifies a key role for glycine in rapid cancer cell proliferation. Science 2012, 336, 1040–1044. [Google Scholar] [CrossRef] [Green Version]
- San-Millan, I.; Brooks, G.A. Reexamining cancer metabolism: Lactate production for carcinogenesis could be the purpose and explanation of the Warburg Effect. Carcinogenesis 2017, 38, 119–133. [Google Scholar] [CrossRef] [PubMed]
- Mayer, A.; Vaupel, P. Hypoxia, lactate accumulation, and acidosis: Siblings or accomplices driving tumor progression and resistance to therapy? Adv. Exp. Med. Biol. 2013, 789, 203–209. [Google Scholar] [CrossRef]
- Kato, Y.; Ozawa, S.; Miyamoto, C.; Maehata, Y.; Suzuki, A.; Maeda, T.; Baba, Y. Acidic extracellular microenvironment and cancer. Cancer Cell Int. 2013, 13, 89. [Google Scholar] [CrossRef] [Green Version]
- Sun, C.; Liu, X.; Wang, B.; Wang, Z.; Liu, Y.; Di, C.; Si, J.; Li, H.; Wu, Q.; Xu, D.; et al. Endocytosis-mediated mitochondrial transplantation: Transferring normal human astrocytic mitochondria into glioma cells rescues aerobic respiration and enhances radiosensitivity. Theranostics 2019, 9, 3595–3607. [Google Scholar] [CrossRef]
- O’Neill, S.; Porter, R.K.; McNamee, N.; Martinez, V.G.; O’Driscoll, L. 2-Deoxy-D-Glucose inhibits aggressive triple-negative breast cancer cells by targeting glycolysis and the cancer stem cell phenotype. Sci. Rep. 2019, 9, 3788. [Google Scholar] [CrossRef] [Green Version]
- Guo, J.; Su, Y.; Zhang, M. Circ_0000140 restrains the proliferation, metastasis and glycolysis metabolism of oral squamous cell carcinoma through upregulating CDC73 via sponging miR-182-5p. Cancer Cell Int. 2020, 20, 407. [Google Scholar] [CrossRef]
- John, G.T.; Goelling, D.; Klimant, I.; Schneider, H.; Heinzle, E. PH-sensing 96-well microtitre plates for the characterization of acid production by dairy starter cultures. J. Dairy Res. 2003, 70, 327–333. [Google Scholar] [CrossRef] [PubMed]
- Heux, S.; Philippe, B.; Portais, J.C. High-throughput workflow for monitoring and mining bioprocess data and its application to inferring the physiological response of Escherichia coli to perturbations. Appl. Environ. Microbiol. 2011, 77, 7040–7049. [Google Scholar] [CrossRef] [Green Version]
- Wu, S.; Wu, S.; Yi, Z.; Zeng, F.; Wu, W.; Qiao, Y.; Zhao, X.; Cheng, X.; Tian, Y. Hydrogel-Based Fluorescent Dual pH and Oxygen Sensors Loaded in 96-Well Plates for High-Throughput Cell Metabolism Studies. Sensors 2018, 18, 564. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kieninger, J.; Aravindalochanan, K.; Sandvik, J.A.; Pettersen, E.O.; Urban, G.A. Pericellular oxygen monitoring with integrated sensor chips for reproducible cell culture experiments. Cell Prolif. 2014, 47, 180–188. [Google Scholar] [CrossRef] [PubMed]
- Steinegger, A.; Wolfbeis, O.S.; Borisov, S.M. Optical Sensing and Imaging of pH Values: Spectroscopies, Materials, and Applications. Chem. Rev. 2020, 120, 12357–12489. [Google Scholar] [CrossRef]
- Wang, X.D.; Wolfbeis, O.S. Optical methods for sensing and imaging oxygen: Materials, spectroscopies and applications. Chem. Soc. Rev. 2014, 43, 3666–3761. [Google Scholar] [CrossRef] [Green Version]
- Nonnenmacher, Y.; Palorini, R.; Hiller, K. Determining Compartment-Specific Metabolic Fluxes. Methods Mol. Biol. 2019, 1862, 137–149. [Google Scholar] [CrossRef]
- Zhang, J.; Zhang, Q. Using Seahorse Machine to Measure OCR and ECAR in Cancer Cells. Methods Mol. Biol. 2019, 1928, 353–363. [Google Scholar] [CrossRef]
- Hynes, J.; Swiss, R.L.; Will, Y. High-Throughput Analysis of Mitochondrial Oxygen Consumption. Methods Mol. Biol. 2018, 1782, 71–87. [Google Scholar] [CrossRef] [PubMed]
- Naciri, M.; Kuystermans, D.; Al-Rubeai, M. Monitoring pH and dissolved oxygen in mammalian cell culture using optical sensors. Cytotechnology 2008, 57, 245–250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arguello, R.J.; Combes, A.J.; Char, R.; Gigan, J.P.; Baaziz, A.I.; Bousiquot, E.; Camosseto, V.; Samad, B.; Tsui, J.; Yan, P.; et al. SCENITH: A Flow Cytometry-Based Method to Functionally Profile Energy Metabolism with Single-Cell Resolution. Cell Metab. 2020, 32, 1063–1075.e1067. [Google Scholar] [CrossRef] [PubMed]
- Tanumihardja, E.; Slaats, R.H.; van der Meer, A.D.; Passier, R.; Olthuis, W.; van den Berg, A. Measuring Both pH and O2 with a Single On-Chip Sensor in Cultures of Human Pluripotent Stem Cell-Derived Cardiomyocytes to Track Induced Changes in Cellular Metabolism. ACS Sens. 2021, 6, 267–274. [Google Scholar] [CrossRef] [PubMed]
- Diepart, C.; Verrax, J.; Calderon, P.B.; Feron, O.; Jordan, B.F.; Gallez, B. Comparison of methods for measuring oxygen consumption in tumor cells in vitro. Anal. Biochem. 2010, 396, 250–256. [Google Scholar] [CrossRef] [PubMed]
- Ge, X.; Hanson, M.; Shen, H.; Kostov, Y.; Brorson, K.A.; Frey, D.D.; Moreira, A.R.; Rao, G. Validation of an optical sensor-based high-throughput bioreactor system for mammalian cell culture. J. Biotechnol. 2006, 122, 293–306. [Google Scholar] [CrossRef]
- Alim Uysal, B.A.; Kotan, G.; Guneser, M.B.; Dincer, A.N.; Senturk, H.; Rafiqi, A.M. Investigation of the effect of different chelation solutions on penetration of resin-based and bioceramic sealers with a novel method. Microsc. Res. Tech. 2021. [Google Scholar] [CrossRef]
- Michl, J.; Park, K.C.; Swietach, P. Evidence-based guidelines for controlling pH in mammalian live-cell culture systems. Commun. Biol. 2019, 2, 144. [Google Scholar] [CrossRef]
- Yao, J.; Czaplinska, D.; Ialchina, R.; Schnipper, J.; Liu, B.; Sandelin, A.; Pedersen, S.F. Cancer Cell Acid Adaptation Gene Expression Response Is Correlated to Tumor-Specific Tissue Expression Profiles and Patient Survival. Cancers 2020, 12, 2183. [Google Scholar] [CrossRef]
- Forciniti, S.; Dalla Pozza, E.; Greco, M.R.; Amaral Carvalho, T.M.; Rolando, B.; Ambrosini, G.; Carmona-Carmona, C.A.; Pacchiana, R.; Di Molfetta, D.; Donadelli, M.; et al. Extracellular Matrix Composition Modulates the Responsiveness of Differentiated and Stem Pancreatic Cancer Cells to Lipophilic Derivate of Gemcitabine. Int. J. Mol. Sci. 2020, 22, 29. [Google Scholar] [CrossRef]
- Ford, K.L.; Moorhouse, E.L.; Bortolozzi, M.; Richards, M.A.; Swietach, P.; Vaughan-Jones, R.D. Regional acidosis locally inhibits but remotely stimulates Ca2+ waves in ventricular myocytes. Cardiovasc. Res. 2017, 113, 984–995. [Google Scholar] [CrossRef]
- Chung, Y.J.; Luo, A.; Park, K.C.; Loonat, A.A.; Lakhal-Littleton, S.; Robbins, P.A.; Swietach, P. Iron-deficiency anemia reduces cardiac contraction by downregulating RyR2 channels and suppressing SERCA pump activity. JCI Insight 2019, 4, e125618. [Google Scholar] [CrossRef]
- Hulikova, A.; Harris, A.L.; Vaughan-Jones, R.D.; Swietach, P. Regulation of intracellular pH in cancer cell lines under normoxia and hypoxia. J. Cell Physiol. 2013, 228, 743–752. [Google Scholar] [CrossRef]
- Jo, J.; Lee, C.H.; Kopelman, R.; Wang, X. In vivo quantitative imaging of tumor pH by nanosonophore assisted multispectral photoacoustic imaging. Nat. Commun. 2017, 8, 471. [Google Scholar] [CrossRef] [Green Version]
- Contreras-Baeza, Y.; Sandoval, P.Y.; Alarcon, R.; Galaz, A.; Cortes-Molina, F.; Alegria, K.; Baeza-Lehnert, F.; Arce-Molina, R.; Guequen, A.; Flores, C.A.; et al. Monocarboxylate transporter 4 (MCT4) is a high affinity transporter capable of exporting lactate in high-lactate microenvironments. J. Biol. Chem. 2019, 294, 20135–20147. [Google Scholar] [CrossRef] [PubMed]
- Bachmeier, C.J.; Trickler, W.J.; Miller, D.W. Drug efflux transport properties of 2′,7′-bis(2-carboxyethyl)-5(6)-carboxyfluorescein acetoxymethyl ester (BCECF-AM) and its fluorescent free acid, BCECF. J. Pharm. Sci. 2004, 93, 932–942. [Google Scholar] [CrossRef] [PubMed]
- Lucien, F.; Harper, K.; Pelletier, P.P.; Volkov, L.; Dubois, C.M. Simultaneous pH measurement in endocytic and cytosolic compartments in living cells using confocal microscopy. J. Vis. Exp. 2014, 86, 51395. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hinton, A.; Sennoune, S.R.; Bond, S.; Fang, M.; Reuveni, M.; Sahagian, G.G.; Jay, D.; Martinez-Zaguilan, R.; Forgac, M. Function of a subunit isoforms of the V-ATPase in pH homeostasis and in vitro invasion of MDA-MB231 human breast cancer cells. J. Biol. Chem. 2009, 284, 16400–16408. [Google Scholar] [CrossRef] [Green Version]
- Ray, A.; Koo Lee, Y.E.; Epstein, T.; Kim, G.; Kopelman, R. Two-photon nano-PEBBLE sensors: Subcellular pH measurements. Analyst 2011, 136, 3616–3622. [Google Scholar] [CrossRef]
- Willoughby, D.; Schwiening, C.J. Electrically evoked dendritic pH transients in rat cerebellar Purkinje cells. J. Physiol. 2002, 544, 487–499. [Google Scholar] [CrossRef] [PubMed]
- Lima, S.; Bonifacio, R.L.; Azzellini, G.C.; Coichev, N. Ruthenium(II) tris(bipyridyl) ion as a luminescent probe for oxygen uptake on the catalyzed oxidation of HSO3-. Talanta 2002, 56, 547–556. [Google Scholar] [CrossRef]
- Zhou, C.; Zhao, W.X.; You, F.T.; Geng, Z.X.; Peng, H.S. Highly Stable and Luminescent Oxygen Nanosensor Based on Ruthenium-Containing Metallopolymer for Real-Time Imaging of Intracellular Oxygenation. ACS Sens. 2019, 4, 984–991. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Z.; Yu, X.; Zhai, S.; Hao, Y. Ratiometric Dissolved Oxygen Sensors Based on Ruthenium Complex Doped with Silver Nanoparticles. Sensors 2017, 17, 548. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scholler-Mann, A.; Matt, K.; Hochecker, B.; Bergemann, J. Ex vivo Assessment of Mitochondrial Function in Human Peripheral Blood Mononuclear Cells Using XF Analyzer. Bio. Protoc. 2021, 11, e3980. [Google Scholar] [CrossRef]
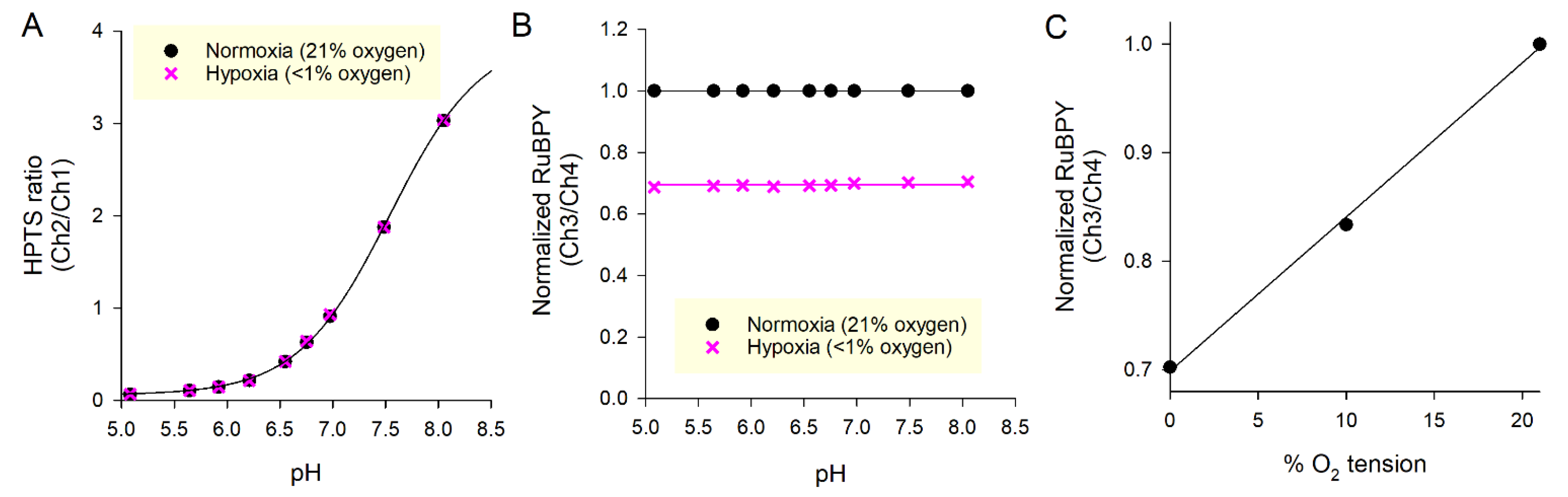


| Calibration Variable | Best-Fit |
|---|---|
| pKa | 7.5383 |
| rmax | 3.9503 |
| rmin | 0.0603 |
| ranoxia | 0.6945 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Blaszczak, W.; Tan, Z.; Swietach, P. Cost-Effective Real-Time Metabolic Profiling of Cancer Cell Lines for Plate-Based Assays. Chemosensors 2021, 9, 139. https://doi.org/10.3390/chemosensors9060139
Blaszczak W, Tan Z, Swietach P. Cost-Effective Real-Time Metabolic Profiling of Cancer Cell Lines for Plate-Based Assays. Chemosensors. 2021; 9(6):139. https://doi.org/10.3390/chemosensors9060139
Chicago/Turabian StyleBlaszczak, Wiktoria, Zhengchu Tan, and Pawel Swietach. 2021. "Cost-Effective Real-Time Metabolic Profiling of Cancer Cell Lines for Plate-Based Assays" Chemosensors 9, no. 6: 139. https://doi.org/10.3390/chemosensors9060139
APA StyleBlaszczak, W., Tan, Z., & Swietach, P. (2021). Cost-Effective Real-Time Metabolic Profiling of Cancer Cell Lines for Plate-Based Assays. Chemosensors, 9(6), 139. https://doi.org/10.3390/chemosensors9060139






