Colostrum Features of Active and Recovered COVID-19 Patients Revealed Using Next-Generation Proteomics Technique, SWATH-MS
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethics
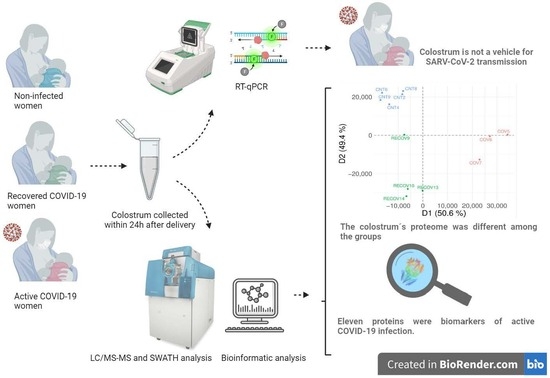
2.2. Research Design
2.3. Collection and Processing of Samples
2.4. RT-qPCR
2.5. Proteomic Analysis
2.5.1. Sample Processing
2.5.2. Protein Digestion and Sample Preparation
2.5.3. LC–MS/MS Analysis and Building a Spectral Library
2.5.4. SWATH–MS Analysis
2.6. Identification and Quantification of Proteins
2.7. Bioinformatic Analysis
3. Results
3.1. SARS-CoV-2 Detection in Colostrum
3.2. Proteomic Profile of the Colostrum Samples
3.3. Discriminant Analysis
3.4. Alteration of the Proteome Profile in Colostrum from COVID-19 Patients
3.5. The COVID-19-Recovered Group Differs from the Control and COVID-19 Groups
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mistry, P.; Barmania, F.; Mellet, J.; Peta, K.; Strydom, A.; Viljoen, I.M.; James, W.; Gordon, S.; Pepper, M.S. SARS-CoV-2 Variants, Vaccines, and Host Immunity. Front. Immunol. 2021, 12, 089244. [Google Scholar] [CrossRef]
- WHO Coronavirus (COVID-19) Dashboard|WHO Coronavirus (COVID-19) Dashboard with Vaccination Data. Available online: covid19.who.int (accessed on 27 April 2022).
- CDC COVID Data Tracker: Pregnant People: COVID-19 Illness and Outcomes. Available online: https://covid.cdc.gov/covid-data-tracker/#vaccinations-pregnant-women (accessed on 27 April 2022).
- Pacheco, F.; Sobral, M.; Guiomar, R.; de la Torre-Luque, A.; Caparros-Gonzalez, R.A.; Ganho-ávila, A. Breastfeeding during COVID-19: A Narrative Review of the Psychological Impact on Mothers. Behav. Sci. 2021, 11, 34. [Google Scholar] [CrossRef]
- Sims, C.R.; Lipsmeyer, M.E.; Turner, D.E.; Andres, A. Human Milk Composition Differs by Maternal BMI in the First 9 Months Postpartum. Am. J. Clin. Nutr. 2020, 112, 548–557. [Google Scholar] [CrossRef] [PubMed]
- Nuzzi, G.; Trambusti, I.; Di Cicco, M.E.; Peroni, D.G. Breast Milk: More than Just Nutrition! Minerva Pediatr. 2021, 73, 111–114. [Google Scholar] [CrossRef]
- Binns, C.; Lee, M.; Low, W.Y. The Long-Term Public Health Benefits of Breastfeeding. Asia Pac. J. Public Health 2016, 28, 7–14. [Google Scholar] [CrossRef]
- Eoh, K.J.; Park, E.Y.; Chang, Y.J.; Ha, H.I.; Hong, J.; Huang, D.; Nam, E.J.; Lim, M.C. The Preventive Effect of Breastfeeding against Ovarian Cancer in BRCA1 and BRCA2 Mutation Carriers: A Systematic Review and Meta-Analysis. Gynecol. Oncol. 2021, 163, 142–147. [Google Scholar] [CrossRef]
- Zhu, J.; Dingess, K.A. The Functional Power of the Human Milk Proteome. Nutrients 2019, 11, 1834. [Google Scholar] [CrossRef] [PubMed]
- Álvarez, D.; Muñoz, Y.; Ortiz, M.; Maliqueo, M.; Chouinard-Watkins, R.; Valenzuela, R. Impact of Maternal Obesity on the Metabolism and Bioavailability of Polyunsaturated Fatty Acids during Pregnancy and Breastfeeding. Nutrients 2021, 13, 19. [Google Scholar] [CrossRef] [PubMed]
- Andreas, N.J.; Kampmann, B.; Mehring Le-Doare, K. Human Breast Milk: A Review on Its Composition and Bioactivity. Early Hum. Dev. 2015, 91, 629–635. [Google Scholar] [CrossRef]
- Vukavić, T. Timing of the Gut Closure. J. Pediatr. Gastroenterol. Nutr. 1984, 3, 700–703. [Google Scholar] [CrossRef]
- Jansen, S.; Wasityastuti, W.; Astarini, F.D.; Hartini, S. Mothers’ Knowledge of Breastfeeding and Infant Feeding Types Affect Acute Respiratory Infections. J. Prev. Med. Hyg. 2020, 61, E401–E408. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Shang, Y.; Ren, Y.; Bie, Y.; Qiu, Y.; Yuan, Y.; Zhao, Y.; Zou, L.; Lin, S.-H.; Zhou, X. Omics Study Reveals Abnormal Alterations of Breastmilk Proteins and Metabolites in Puerperant Women With COVID-19. Signal Transduct. Target. Ther. 2020, 5, 247. [Google Scholar] [CrossRef] [PubMed]
- Kimberlin, D.W.; Puopolo, K.M.; Kimberlin, D.W. Breast Milk and COVID-19: What Do We Know? Clin. Infect. Dis. 2021, 72, 131–132. [Google Scholar] [CrossRef]
- Fouda, G.G.; Martinez, D.R.; Swamy, G.K.; Permar, S.R. The Impact of IgG Transplacental Transfer on Early Life Immunity. Immunohorizons 2018, 2, 14. [Google Scholar] [CrossRef]
- Morniroli, D.; Consales, A.; Crippa, B.L.; Vizzari, G.; Ceroni, F.; Cerasani, J.; Colombo, L.; Mosca, F.; Giannì, M.L. The Antiviral Properties of Human Milk: A Multitude of Defence Tools from Mother Nature. Nutrients 2021, 13, 694. [Google Scholar] [CrossRef]
- Hanson, L.Å.; Korotkova, M. The Role of Breastfeeding in Prevention of Neonatal Infection. Semin. Neonatol. 2002, 7, 275–281. [Google Scholar] [CrossRef]
- Lubbe, W.; Botha, E.; Niela-Vilen, H.; Reimers, P. Breastfeeding during the COVID-19 Pandemic—A Literature Review for Clinical Practice. Int. Breastfeed J. 2020, 15, 82. [Google Scholar] [CrossRef] [PubMed]
- Krasny, L.; Bland, P.; Kogata, N.; Wai, P.; Howard, B.A.; Natrajan, R.C.; Huang, P.H. SWATH Mass Spectrometry as a Tool for Quantitative Profiling of the Matrisome. J. Proteom. 2018, 189, 11–22. [Google Scholar] [CrossRef] [PubMed]
- Ludwig, C.; Gillet, L.; Rosenberger, G.; Amon, S.; Collins, B.C.; Aebersold, R. Data-independent Acquisition-based SWATH-MS for Quantitative Proteomics: A Tutorial. Mol. Syst. Biol. 2018, 14, e8126. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING Database in 2021: Customizable Protein–Protein Networks, and Functional Characterization of User-Uploaded Gene/Measurement Sets. Nucleic Acids Res. 2021, 49, D605. [Google Scholar] [CrossRef]
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2009. [Google Scholar] [CrossRef]
- Gu, Z.; Eils, R.; Schlesner, M. Complex Heatmaps Reveal Patterns and Correlations in Multidimensional Genomic Data. Bioinformatics 2016, 32, 2847–2849. [Google Scholar] [CrossRef]
- Vacanti, N.M. The Fundamentals of Constructing and Interpreting Heat Maps. Methods Mol. Biol. 2019, 1862, 279–291. [Google Scholar] [CrossRef] [PubMed]
- Jassal, B.; Matthews, L.; Viteri, G.; Gong, C.; Lorente, P.; Fabregat, A.; Sidiropoulos, K.; Cook, J.; Gillespie, M.; Haw, R.; et al. The Reactome Pathway Knowledgebase. Nucleic Acids Res. 2020, 48, D498–D503. [Google Scholar] [CrossRef]
- Supek, F.; Bošnjak, M.; Škunca, N.; Šmuc, T. REVIGO Summarizes and Visualizes Long Lists of Gene Ontology Terms. PLoS ONE 2011, 6, e21800. [Google Scholar] [CrossRef] [PubMed]
- Salonia, A.; Pontillo, M.; Capogrosso, P.; Gregori, S.; Carenzi, C.; Ferrara, A.M.; Rowe, I.; Boeri, L.; Larcher, A.; Ramirez, G.A.; et al. Testosterone in Males with COVID-19: A 7-Month Cohort Study. Andrology 2022, 10, 34–41. [Google Scholar] [CrossRef]
- Singh, K.S.; Singh, B.P.; Rokana, N.; Singh, N.; Kaur, J.; Singh, A.; Panwar, H. Bio-Therapeutics from Human Milk: Prospects and Perspectives. J. Appl. Microbiol. 2021, 131, 2669–2687. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; McMahon, R.J.; Woo, J.G.; Davidson, B.S.; Morrow, A.L.; Zhang, Q. Temporal Changes in Milk Proteomes Reveal Developing Milk Functions. J. Proteome Res. 2012, 11, 3897–3907. [Google Scholar] [CrossRef]
- Zhang, L.; de Waard, M.; Verheijen, H.; Boeren, S.; Hageman, J.A.; van Hooijdonk, T.; Vervoort, J.; van Goudoever, J.B.; Hettinga, K. Changes over Lactation in Breast Milk Serum Proteins Involved in the Maturation of Immune and Digestive System of the Infant. J. Proteom. 2016, 147, 40–47. [Google Scholar] [CrossRef]
- Raheem, R.A.; Binns, C.W.; Chih, H.J. Protective Effects of Breastfeeding against Acute Respiratory Tract Infections and Diarrhoea: Findings of a Cohort Study. J. Paediatr. Child Health 2017, 53, 271–276. [Google Scholar] [CrossRef]
- Centeno-Tablante, E.; Medina-Rivera, M.; Finkelstein, J.L.; Rayco-Solon, P.; Garcia-Casal, M.N.; Rogers, L.; Ghezzi-Kopel, K.; Ridwan, P.; Peña-Rosas, J.P.; Mehta, S. Transmission of SARS-CoV-2 through Breast Milk and Breastfeeding: A Living Systematic Review. Ann. N. Y. Acad. Sci. 2021, 1484, 32–54. [Google Scholar] [CrossRef]
- Groß, R.; Conzelmann, C.; Müller, J.A.; Stenger, S.; Steinhart, K.; Kirchhoff, F.; Münch, J. Detection of SARS-CoV-2 in Human Breastmilk. Lancet 2020, 395, 1757–1758. [Google Scholar] [CrossRef]
- Guo, J.; Tan, M.; Zhu, J.; Tian, Y.; Liu, H.; Luo, F.; Wang, J.; Huang, Y.; Zhang, Y.; Yang, Y.; et al. Proteomic Analysis of Human Milk Reveals Nutritional and Immune Benefits in the Colostrum from Mothers with COVID-19. Nutrients 2022, 14, 2513. [Google Scholar] [CrossRef]
- Liu, W.; Wang, J.; Li, W.; Zhou, Z.; Liu, S.; Rong, Z. Clinical Characteristics of 19 Neonates Born to Mothers with COVID-19. Front. Med. 2020, 14, 193–198. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Liu, W.; Su, H.; Li, S.; Shereen, M.A.; Lv, Z.; Niu, Z.; Li, D.; Liu, F.; Luo, Z.; et al. Breastfeeding Risk from Detectable Severe Acute Respiratory Syndrome Coronavirus 2 in Breastmilk. J. Infect. 2020, 81, 452. [Google Scholar] [CrossRef] [PubMed]
- Pace, R.M.; Williams, J.E.; Järvinen, K.M.; Belfort, M.B.; Pace, C.D.W.; Lackey, K.A.; Gogel, A.C.; Nguyen-Contant, P.; Kanagaiah, P.; Fitzgerald, T.; et al. Characterization of SARS-CoV-2 RNA, Antibodies, and Neutralizing Capacity in Milk Produced by Women with COVID-19. mBio 2021, 12, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Cao, X.; Wu, R.; Liu, B.; Ye, W.; Yue, X.; Wu, J. Comparative Proteomic Exploration of Whey Proteins in Human and Bovine Colostrum and Mature Milk Using ITRAQ-Coupled LC-MS/MS. Int. J. Food Sci. Nutr. 2017, 68, 671–681. [Google Scholar] [CrossRef]
- Bardanzellu, F.; Fanos, V.; Reali, A. Omics in Human Colostrum and Mature Milk: Looking to Old Data with New Eyes. Nutrients 2017, 9, 843. [Google Scholar] [CrossRef]
- Shen, B.; Yi, X.; Sun, Y.; Bi, X.; Du, J.; Zhang, C.; Quan, S.; Zhang, F.; Sun, R.; Qian, L.; et al. Proteomic and Metabolomic Characterization of COVID-19 Patient Sera. Cell 2020, 182, 59. [Google Scholar] [CrossRef] [PubMed]
- Connors, J.M.; Levy, J.H. COVID-19 and Its Implications for Thrombosis and Anticoagulation. Blood 2020, 135, 2033–2040. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Mesa, J.E.; Galindo-Coral, S.; Montes, M.C.; Muñoz Martin, A.J. Thrombosis and Coagulopathy in COVID-19. Curr. Probl. Cardiol. 2021, 46, 100742. [Google Scholar] [CrossRef]
- Levi, M.; Thachil, J.; Iba, T.; Levy, J.H. Coagulation Abnormalities and Thrombosis in Patients with COVID-19. Lancet Haematol. 2020, 7, e438–e440. [Google Scholar] [CrossRef] [PubMed]
- Vandooren, J.; Itoh, Y. Alpha-2-Macroglobulin in Inflammation, Immunity and Infections. Front. Immunol. 2021, 12, 803244. [Google Scholar] [CrossRef] [PubMed]
- Speeckaert, M.M.; Delanghe, J.R. A Key Role for Vitamin D Binding Protein in COVID-19? Eur. J. Nutr. 2021, 60, 2259. [Google Scholar] [CrossRef]
- Yağcı, S.; Serin, E.; Acicbe, Ö.; Zeren, M.İ.; Odabaşı, M.S. The Relationship between Serum Erythropoietin, Hepcidin, and Haptoglobin Levels with Disease Severity and Other Biochemical Values in Patients with COVID-19. Int. J. Lab. Hematol. 2021, 43 (Suppl. 1), 142–151. [Google Scholar] [CrossRef]
- Glogauer, M.; Marchal, C.C.; Zhu, F.; Worku, A.; Clausen, B.E.; Foerster, I.; Marks, P.; Downey, G.P.; Dinauer, M.; Kwiatkowski, D.J. Rac1 Deletion in Mouse Neutrophils Has Selective Effects on Neutrophil Functions. J. Immunol. 2003, 170, 5652–5657. [Google Scholar] [CrossRef]
- Gordón-Alonso, M.; Rocha-Perugini, V.; Álvarez, S.; Moreno-Gonzalo, O.; Ursa, Á.; López-Martín, S.; Izquierdo-Useros, N.; Martínez-Picado, J.; Muñoz-Fernández, M.Á.; Yáñez-Mó, M.; et al. The PDZ-Adaptor Protein Syntenin-1 Regulates HIV-1 Entry. Mol. Biol. Cell 2012, 23, 2253–2263. [Google Scholar] [CrossRef] [PubMed]
- Ma, D.; Liu, S.; Hu, L.; He, Q.; Shi, W.; Yan, D.; Cao, Y.; Zhang, G.; Wang, Z.; Wu, J.; et al. Single-Cell RNA Sequencing Identify SDCBP in ACE2-Positive Bronchial Epithelial Cells Negatively Correlates with COVID-19 Severity. J. Cell Mol. Med. 2021, 25, 7001–7012. [Google Scholar] [CrossRef]
- Fraser, D.D.; Cepinskas, G.; Patterson, E.K.; Slessarev, M.; Martin, C.; Daley, M.; Patel, M.A.; Miller, M.R.; O’Gorman, D.B.; Gill, S.E.; et al. Novel Outcome Biomarkers Identified With Targeted Proteomic Analyses of Plasma From Critically Ill Coronavirus Disease 2019 Patients. Crit. Care Explor. 2020, 2, e0189. [Google Scholar] [CrossRef]
- Zhang, Q.; Cundiff, J.K.; Maria, S.D.; McMahon, R.J.; Woo, J.G.; Davidson, B.S.; Morrow, A.L. Quantitative Analysis of the Human Milk Whey Proteome Reveals Developing Milk and Mammary-Gland Functions across the First Year of Lactation. Proteomes 2013, 1, 128. [Google Scholar] [CrossRef]
- D’Alessandro, A.; Scaloni, A.; Zolla, L. Human Milk Proteins: An Interactomics and Updated Functional Overview. J. Proteome Res. 2010, 9, 3339–3373. [Google Scholar] [CrossRef]
- Yasar, S.; Kucukakcali, Z.; Doganer, A. Ensemble Learning-Based Prediction of COVID-19 Positive Patient Groups Determined by IL-6 Levels and Control Individuals Based on the Proteomics Data. Med. Sci. 2021, 10, 1516–1523. [Google Scholar] [CrossRef]
- Shekar, P.C.; Goel, S.; Rani, S.D.S.; Sarathi, D.P.; Alex, J.L.; Singh, S.; Kumar, S. Kappa-Casein-Deficient Mice Fail to Lactate. Proc. Natl. Acad. Sci. USA 2006, 103, 8000–8005. [Google Scholar] [CrossRef] [PubMed]
- Knoll, R.; Schultze, J.L.; Schulte-Schrepping, J. Monocytes and Macrophages in COVID-19. Front. Immunol. 2021, 12, 720109. [Google Scholar] [CrossRef]
- Hellerud, B.C.; Orrem, H.L.; Dybwik, K.; Pischke, S.E.; Baratt-Due, A.; Castellheim, A.; Fure, H.; Bergseth, G.; Christiansen, D.; Nunn, M.A.; et al. Combined Inhibition of C5 and CD14 Efficiently Attenuated the Inflammatory Response in a Porcine Model of Meningococcal Sepsis. J. Intensive Care 2017, 5, 21. [Google Scholar] [CrossRef] [PubMed]
- Medjeral-Thomas, N.R.; Troldborg, A.; Hansen, A.G.; Pihl, R.; Clarke, C.L.; Peters, J.E.; Thomas, D.C.; Willicombe, M.; Palarasah, Y.; Botto, M.; et al. Protease Inhibitor Plasma Concentrations Associate with COVID-19 Infection. Oxf. Open Immunol. 2021, 2, iqab014. [Google Scholar] [CrossRef]
- Geyer, P.E.; Arend, F.M.; Doll, S.; Louiset, M.; Virreira Winter, S.; Müller-Reif, J.B.; Torun, F.M.; Weigand, M.; Eichhorn, P.; Bruegel, M.; et al. High-Resolution Serum Proteome Trajectories in COVID-19 Reveal Patient-Specific Seroconversion. EMBO Mol. Med. 2021, 13, e14167. [Google Scholar] [CrossRef] [PubMed]
- Papp, E.; Nardai, G.; Soti, C.; Csermely, P. Molecular Chaperones, Stress Proteins and Redox Homeostasis. Biofactors 2003, 17, 249–257. [Google Scholar] [CrossRef]
- Greulich, W.; Wagner, M.; Gaidt, M.M.; Stafford, C.; Cheng, Y.; Linder, A.; Carell, T.; Hornung, V. TLR8 Is a Sensor of RNase T2 Degradation Products. Cell 2019, 179, 1264–1275.e13. [Google Scholar] [CrossRef] [PubMed]
- Beltrán-Camacho, L.; Eslava-Alcón, S.; Rojas-Torres, M.; Sánchez-Morillo, D.; Martinez-Nicolás, M.P.; Martín-Bermejo, V.; de la Torre, I.G.; Berrocoso, E.; Moreno, J.A.; Moreno-Luna, R.; et al. The Serum of COVID-19 Asymptomatic Patients up-Regulates Proteins Related to Endothelial Dysfunction and Viral Response in Circulating Angiogenic Cells Ex-Vivo. Mol. Med. 2022, 28, 40. [Google Scholar] [CrossRef] [PubMed]
- Köhler, J.; Maletzki, C.; Koczan, D.; Frank, M.; Springer, A.; Steffen, C.; Revenko, A.S.; MacLeod, A.R.; Mikkat, S.; Kreikemeyer, B.; et al. Kininogen Supports Inflammation and Bacterial Spreading during Streptococccus pyogenes Sepsis. EBioMedicine 2020, 58, 102908. [Google Scholar] [CrossRef]











| Criteria | Characteristics |
|---|---|
| Inclusion criteria | Full-term pregnancy |
| Absence of infection during gestation period (except SARS-CoV-2 infection in recovered group) | |
| Absence of infection at the moment of delivery (except SARS-CoV-2 infection in COVID-19 group) | |
| Exclusion criteria | Preterm pregnancy |
| Infectious disease during gestation (except SARS-CoV-2 infection in recovered group) | |
| Infectious disease at the moment of the delivery (except SARS-CoV-2 infection in COVID-19 group) | |
| Immunocompromised mothers | |
| Mothers whose babies are in Neonatal Intensive Care Unit | |
| Caesarea |
| Characteristics | Noninfected (n = 5) | Recovered COVID-19 (n = 4) | COVID-19 (n = 3) |
|---|---|---|---|
| Maternal age: mean years (SD) | 27.37 (7.86) | 33.53 (1.89) | 31.85 (9.06) |
| Ethnicity | Caucasian | Caucasian | Caucasian |
| Infectious diseases (apart from COVID-19) | No | No | No |
| Other complications occurred during pregnancy (diabetes, preeclampsia, anemia, etc.) | ICP * (donor CNT6) | No | No |
| Women vaccinated against SARS-CoV-2 | - | 2 Donors (RECOV10 and 13) | - |
| Number of vaccine doses | - | 2 | - |
| Vaccine type (adenovirus-based/mRNA) | - | mRNA | - |
| COVID-19 detection: mean days before delivery (SD) | - | 123.25 (34.29) | 1 |
| PCR by nasal swabs at the delivery day (negative/positive) | Negative | Negative | Positive |
| Gravidity: mean (SD) | 2 (1.2) | 2.75 (1.5) | 2.66 (2.08) |
| Type of delivery (vaginal/caesarean) | Vaginal | Vaginal | Vaginal |
| Birth week: mean (SD) | 38.88 (1.54) | 39.61 (0.7) | 37.85 (0.66) |
| Collection of colostrum postdelivery: mean hours (SD) | 19.2 (6.6) | 24 | 16 (6.9) |
| Severity of the COVID-19 infection (mild/severe) | - | Mild | Mild |
| Mild symptoms (% of mother with at least one mild symptom: myalgia, headache, anosmia, low-grade fever) | - | 75% | 66% |
| Severe symptoms (% of mother with severe symptoms: trouble breathing, persistent pressure or pain in the chest, confusion, pale, grey or blue-colored skin | - | 0% | 0% |
| Name | Description | Oligonucleotide Sequence (5′ > 3′) |
|---|---|---|
| 2019-nCoV_N1-F | 2019-nCoV_N1 forward primer | GAC CCC AAA ATC AGC GAA AT |
| 2019-nCoV_N1-R | 2019-nCoV_N1 reverse primer | TCT GGT TAC TGC CAG TTG AAT CTG |
| 2019-nCoV_N1-P | 2019-nCoV_N1 probe | FAM-ACC CCG CAT TAG GTT TGG TGG ACC-BHQ1 |
| RP-F | RNase P forward primer | AGA TTT GGA CCT GCG AGC G |
| RP-R | RNase P reverse primer | GAG CGG CTG TCT CCA CAA GT |
| RP-P | RNase P probe | FAM-TTC TGA CCT GAA GGC TCT GCG CG-BHQ-1 |
| E_Sarbeco_F1 | E_Sarbeco forward primer | ACAGGTACGTTAATAGTTAATAGCGT |
| E_Sarbeco_R2 | E_Sarbeco reverse primer | ATATTGCAGCAGTACGCACACA |
| E_Sarbeco_P1 | E_Sarbeco probe | FAM-ACACTAGCCATCCTTACTGCGCTTCGBBQ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hernández-Caravaca, I.; Moros-Nicolás, C.; González-Brusi, L.; Romero de Ávila, M.J.; De Paco Matallana, C.; Pelegrín, P.; Castaño-Molina, M.Á.; Díaz-Meca, L.; Sánchez-Romero, J.; Martínez-Alarcón, L.; et al. Colostrum Features of Active and Recovered COVID-19 Patients Revealed Using Next-Generation Proteomics Technique, SWATH-MS. Children 2023, 10, 1423. https://doi.org/10.3390/children10081423
Hernández-Caravaca I, Moros-Nicolás C, González-Brusi L, Romero de Ávila MJ, De Paco Matallana C, Pelegrín P, Castaño-Molina MÁ, Díaz-Meca L, Sánchez-Romero J, Martínez-Alarcón L, et al. Colostrum Features of Active and Recovered COVID-19 Patients Revealed Using Next-Generation Proteomics Technique, SWATH-MS. Children. 2023; 10(8):1423. https://doi.org/10.3390/children10081423
Chicago/Turabian StyleHernández-Caravaca, Iván, Carla Moros-Nicolás, Leopoldo González-Brusi, Mª José Romero de Ávila, Catalina De Paco Matallana, Pablo Pelegrín, María Ángeles Castaño-Molina, Lucía Díaz-Meca, Javier Sánchez-Romero, Laura Martínez-Alarcón, and et al. 2023. "Colostrum Features of Active and Recovered COVID-19 Patients Revealed Using Next-Generation Proteomics Technique, SWATH-MS" Children 10, no. 8: 1423. https://doi.org/10.3390/children10081423
APA StyleHernández-Caravaca, I., Moros-Nicolás, C., González-Brusi, L., Romero de Ávila, M. J., De Paco Matallana, C., Pelegrín, P., Castaño-Molina, M. Á., Díaz-Meca, L., Sánchez-Romero, J., Martínez-Alarcón, L., Avilés, M., & Izquierdo-Rico, M. J. (2023). Colostrum Features of Active and Recovered COVID-19 Patients Revealed Using Next-Generation Proteomics Technique, SWATH-MS. Children, 10(8), 1423. https://doi.org/10.3390/children10081423