The Approach to a Child with Dysmorphic Features: What the Pediatrician Should Know
Abstract
1. Introduction
2. Clinical Work-Up
2.1. Family and Medical History
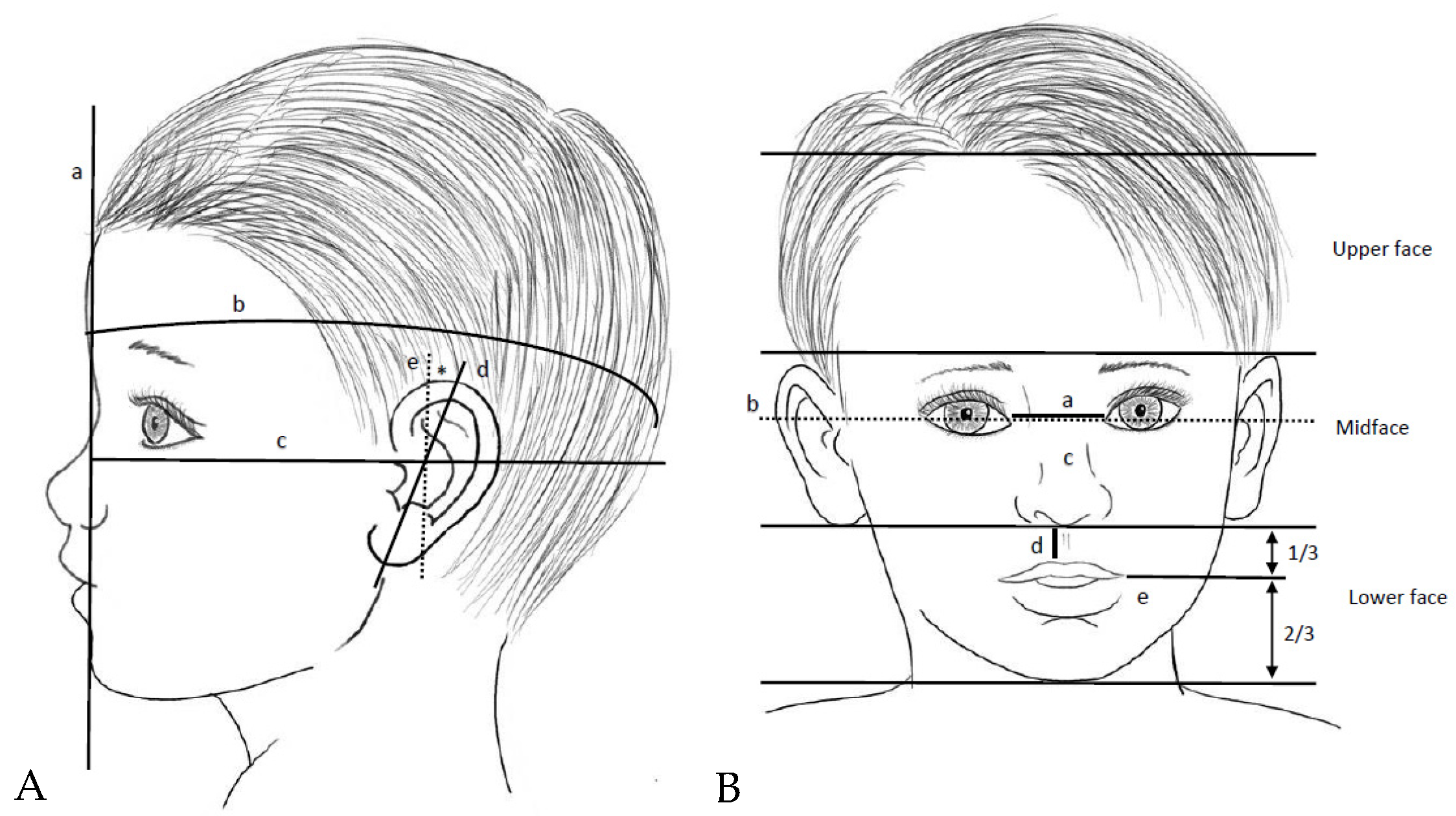
2.2. Physical Examination
3. Supportive Technological Tools
3.1. Human Malformation Terminology (National Human Genome Research Institute)
3.2. ACT Sheets and Algorithms
3.3. Online Mendelian Inheritance in Man
3.4. Risk Calculators
3.5. Face2Gene
3.6. DECIPHER
3.7. POSSUMweb
4. Genetic Testing
4.1. Karyotyping
4.2. Fluorescence In Situ Hybridization
4.3. Chromosomal Microarray
4.4. Multiplex Ligation-Dependent Probe Amplification
4.5. Next-Generation Genome Sequencing
5. Discussion
6. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Baird, P.A.; Anderson, T.W.; Newcombe, H.B.; Lowry, R.B. Genetic disorders in children and young adults: A population study. Am. J. Hum. Genet. 1988, 42, 677–693. [Google Scholar]
- Verma, I.C.; Puri, R.D. Global burden of genetic disease and the role of genetic screening. Semin. Fetal Neonatal Med. 2015, 20, 354–363. [Google Scholar] [CrossRef]
- CoCooper-Brown, L.; Copeland, S.; Dailey, S.; Downey, D.; Petersen, M.C.; Stimson, C.; Van Dyke, D.C. Feeding and swallowing dysfunction in genetic syndromes. Dev. Disabil. Res. Rev. 2008, 14, 147–157. [Google Scholar] [CrossRef]
- Prasad, A.N.; Prasad, C. Genetic evaluation of the floppy infant. Semin. Fetal Neonatal Med. 2011, 16, 99–108. [Google Scholar] [CrossRef]
- Oliveira, P.H.A.; Souza, B.S.; Pacheco, E.N.; Menegazzo, M.S.; Corrêa, I.S.; Zen, P.R.G.; Rosa, R.F.M.; Cesa, C.C.; Pellanda, L.C.; Vilela, M.A.P. Genetic syndromes associated with congenital cardiac defects and ophthalmologic changes—Systematization for diagnosis in the clinical practice. Arq. Bras. Cardiol. 2018, 110, 84–90. [Google Scholar] [CrossRef]
- Pauli, R.M. Achondroplasia: A comprehensive clinical review. Orphanet J. Rare Dis. 2019, 14, 1–49. [Google Scholar] [CrossRef]
- Girirajan, S.; Eichler, E.E. Phenotypic variability and genetic susceptibility to genomic disorders. Hum. Mol. Genet. 2010, 19, 176–187. [Google Scholar] [CrossRef]
- McDonald-McGinn, D.M.; Sullivan, K.E.; Marino, B.; Philip, N.; Swillen, A.; Vorstman, J.A.; Zackai, E.H.; Emanuel, B.S.; Vermeesch, J.R.; Morrow, B.E.; et al. 22q11.2 deletion syndrome. Nat. Rev. Dis. Primers. 2015, 1, 15071. [Google Scholar] [CrossRef]
- Léa Linglart, B.D.G. Congenital heart defects in Noonan syndrome: Diagnosis, management, and treatment. Am. J. Med Genet. Part C: Semin. Med Genet. 2020, 184, 73–80. [Google Scholar] [CrossRef]
- Villani, A.; Greer, M.-L.C.; Kalish, J.M.; Nakagawara, A.; Nathanson, K.L.; Pajtler, K.W.; Pfister, S.M.; Walsh, M.F.; Wasserman, J.D.; Zelley, K.; et al. Recommendations for cancer surveillance in individuals with RASopathies and other rare genetic conditions with increased cancer risk. Clin. Cancer Res. 2017, 23, e83–e90. [Google Scholar] [CrossRef]
- Zimmerman, R.; Schimenti, L.S.L. A catalog of genetic syndromes in childhood cancer. Pediatr. Blood Cancer 2015, 62, 2071–2075. [Google Scholar] [CrossRef] [PubMed]
- Spiker, A.M.; Troxell, T.; Ramsey, M.L. Gorlin Syndrome; 8 August 2023; StatPearls Publishing: Treasure Island, FL, USA, 2024. [Google Scholar] [PubMed]
- De Onis, M.; Onyango, A.W.; Borghi, E.; Siyam, A.; Nishida, C.; Siekmann, J. Development of a WHO growth reference for school-aged children and adolescents. Bull. World Health Organ. 2007, 85, 660–667. [Google Scholar] [CrossRef] [PubMed]
- De Onis, M.; Garza, C.; Onyango, A.W.; Borghi, E. Comparison of the WHO child growth standards and the CDC 2000 growth charts. J. Nutr. 2007, 137, 144–148. [Google Scholar] [CrossRef] [PubMed]
- de Onis, M.; Onyango, A.; Borghi, E.; Siyam, A.; Blössner, M.; Lutter, C. Worldwide implementation of the WHO Child Growth Standards. Public. Health Nutr. 2012, 15, 1603–1610. [Google Scholar] [CrossRef] [PubMed]
- De Onis, M. Update on the implementation of the WHO child growth standards. World Rev. Nutr. Diet. 2013, 106, 75–82. [Google Scholar]
- Cacciari, E.; Milano, S.; Balsamo, A. Italian Cross Sectional Growth Charts for Height Weight and Italian Population From To Yr of Age Taken Are Based on a Sample of About Subjects Between and 4He Distribution of the Sample By Gender Age and Geographic Area Was Roughly Simil. J. Endocrinol. 2006, 29, 581–593. [Google Scholar]
- Khadilkar, A.; Khadikar, V.; Lohiya, N.N.; Karguppikar, M.B. Extended growth charts for Indian children. J. Pediatr. Endocrinol. Metab. 2021, 34, 357–362. [Google Scholar] [CrossRef] [PubMed]
- Hocquette, A.; Durox, M.; Wood, R.; Klungsøyr, K.; Szamotulska, K.; Berrut, S.; Rihs, T.; Kyprianou, T.; Sakkeus, L.; Lecomte, A.; et al. International versus national growth charts for identifying small and large-for-gestational age newborns: A population-based study in 15 European countries. Lancet Reg. Heal. Eur. 2021, 8, 100167. [Google Scholar] [CrossRef] [PubMed]
- Fredriks, A.M.; Van Buuren, S.; Van Heel, W.J.M.; Dijkman-Neerincx, R.H.M.; Verloove-Vanhorick, S.P.; Wit, J.M. Nationwide age references for sitting height, leg length, and sitting height/height ratio, and their diagnostic value for disproportionate growth disorders. Arch. Dis. Child. 2005, 90, 807–812. [Google Scholar] [CrossRef] [PubMed]
- Hawkes, C.P.; Mostoufi-Moab, S.; McCormack, S.E.; Grimberg, A.; Zemel, B.S. Sitting Height to Standing Height Ratio Reference Charts for Children in the United States. J. Pediatr. 2020, 226, 221–227.e15. [Google Scholar] [CrossRef]
- Grunauer, M.; Jorge, A.A.L. Genetic short stature. Growth Horm. IGF Res. 2018, 38, 29–33. [Google Scholar] [CrossRef] [PubMed]
- Wakeling, E.L.; Brioude, F.; Lokulo-Sodipe, O.; O’Connell, S.M.; Salem, J.; Bliek, J.; Canton, A.P.; Chrzanowska, K.H.; Davies, J.H.; Dias, R.P.; et al. Diagnosis and management of Silver-Russell syndrome: First international consensus statement. Nat. Rev. Endocrinol. 2017, 13, 105–124. [Google Scholar] [CrossRef] [PubMed]
- Zenker, M.; Edouard, T.; Blair, J.C.; Cappa, M. Noonan syndrome: Improving recognition and diagnosis. Arch. Dis. Child. 2022, 107, 1073–1078. [Google Scholar] [CrossRef] [PubMed]
- Angulo, M.A.; Butler, M.G.; Cataletto, M.E. Prader-Willi syndrome: A review of clinical, genetic, and endocrine findings. J. Endocrinol. Invest. 2015, 38, 1249–1263. [Google Scholar] [CrossRef] [PubMed]
- Brioude, F.; Kalish, J.M.; Mussa, A.; Foster, A.C.; Bliek, J.; Ferrero, G.B.; Boonen, S.E.; Cole, T.; Baker, R.; Bertoletti, M.; et al. Clinical and molecular diagnosis, screening and management of Beckwith-Wiedemann syndrome: An international consensus statement M. made a substantial contribution to discussion of the content HHS Public Access. Nat. Rev. Endocrinol. 2018, 14, 229–249. Available online: www.imprinting-disorders.eu (accessed on 5 May 2024). [CrossRef]
- Adam, M.P.; Banka, S.; Bjornsson, H.T.; Bodamer, O.; E Chudley, A.; Harris, J.; Kawame, H.; Lanpher, B.C.; Lindsley, A.W.; Merla, G.; et al. Kabuki syndrome: International consensus diagnostic criteria. J. Med. Genet. 2019, 56, 89–95. [Google Scholar] [CrossRef] [PubMed]
- Amberger, J.S.; Bocchini, C.A.; Schiettecatte, F.; Scott, A.F.; Hamosh, A. OMIM.org: Online Mendelian Inheritance in Man (OMIM®), an Online catalog of human genes and genetic disorders. Nucleic Acids Res. 2015, 43, D789–D798. [Google Scholar] [CrossRef]
- Amberger, J.S.; Bocchini, C.A.; Scott, A.F.; Hamosh, A. OMIM.org: Leveraging knowledge across phenotype-gene relationships. Nucleic Acids Res. 2019, 47, D1038–D1043. [Google Scholar] [PubMed]
- Landrum, M.J.; Chitipiralla, S.; Brown, G.R.; Chen, C.; Gu, B.; Hart, J.; Hoffman, D.; Jang, W.; Kaur, K.; Liu, C.; et al. ClinVar: Improvements to accessing data. Nucleic Acids Res. 2020, 48, D835–D844. [Google Scholar] [CrossRef]
- Tan, M.-H.; Mester, J.; Peterson, C.; Yang, Y.; Chen, J.-L.; Rybicki, L.A.; Milas, K.; Pederson, H.; Remzi, B.; Orloff, M.S.; et al. A clinical scoring system for selection of patients for pten mutation testing is proposed on the basis of a prospective study of 3042 probands. Am. J. Hum. Genet. 2011, 88, 42–56. [Google Scholar] [CrossRef]
- Yehia, L.; Keel, E.; Eng, C. The Clinical Spectrum of PTEN Mutations. Annu. Rev. Med. 2020, 71, 103–116. [Google Scholar] [CrossRef]
- Loeys, B.L.; Dietz, H.C.; Braverman, A.C.; Callewaert, B.L.; De Backer, J.; Devereux, R.B.; Hilhorst-Hofstee, Y.; Jondeau, G.; Faivre, L.; Milewicz, D.M.; et al. The revised Ghent nosology for the Marfan syndrome. J. Med. Genet. 2010, 47, 476–485. [Google Scholar] [CrossRef]
- Gurovich, Y.; Hanani, Y.; Bar, O.; Nadav, G.; Fleischer, N.; Gelbman, D.; Basel-Salmon, L.; Krawitz, P.M.; Kamphausen, S.B.; Zenker, M.; et al. Identifying facial phenotypes of genetic disorders using deep learning. Nat. Med. 2019, 25, 60–64. [Google Scholar] [CrossRef] [PubMed]
- Latorre-Pellicer, A.; Ascaso, Á.; Trujillano, L.; Gil-Salvador, M.; Arnedo, M.; Lucia-Campos, C.; Antoñanzas-Pérez, R.; Marcos-Alcalde, I.; Parenti, I.; Bueno-Lozano, G.; et al. Evaluating face2gene as a tool to identify cornelia de lange syndrome by facial phenotypes. Int. J. Mol. Sci. 2020, 21, 1042. [Google Scholar] [CrossRef]
- Tekendo-Ngongang, C.; Owosela, B.; Fleischer, N.; Addissie, Y.A.; Malonga, B.; Badoe, E.; Gupta, N.; Moresco, A.; Huckstadt, V.; Ashaat, E.A.; et al. Rubinstein–Taybi syndrome in diverse populations. Am. J. Med. Genet. Part. A. 2020, 182, 2939–2950. [Google Scholar] [CrossRef]
- Tekendo-Ngongang, C.; Kruszka, P. Noonan syndrome on the African Continent. Birth Defects Res. 2020, 112, 718–724. [Google Scholar] [CrossRef] [PubMed]
- Liehr, T.; Acquarola, N.; Pyle, K.; St-Pierre, S.; Rinholm, M.; Bar, O.; Wilhelm, K.; Schreyer, I. Next generation phenotyping in Emanuel and Pallister-Killian syndrome using computer-aided facial dysmorphology analysis of 2D photos. Clin. Genet. 2018, 93, 378–381. [Google Scholar] [CrossRef]
- Narayanan, D.L.; Ranganath, P.; Aggarwal, S.; Dalal, A.; Phadke, S.R.; Mandal, K. Computer-aided Facial Analysis in Diagnosing Dysmorphic Syndromes in Indian Children. Indian. Pediatr. 2019, 56, 1017–1019. [Google Scholar] [CrossRef] [PubMed]
- Vorravanpreecha, N.; Lertboonnum, T.; Rodjanadit, R.; Sriplienchan, P.; Rojnueangnit, K. Studying Down syndrome recognition probabilities in Thai children with de-identified computer-aided facial analysis. Am. J. Med. Genet. Part. A 2018, 176, 1935–1940. [Google Scholar] [CrossRef]
- Ciancia, S.; Goedegebuure, W.J.; Grootjen, L.N.; Hokken-Koelega, A.C.S.; Kerkhof, G.F.; van der Kaay, D.C.M. Computer-aided facial analysis as a tool to identify patients with Silver-Russell syndrome and Prader-Willi syndrome. Eur. J. Pediatr. 2023, 182, 2607–2614. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Chatzimichali, E.A.; Brent, S.; Hutton, B.; Perrett, D.; Wright, C.F.; Bevan, A.P.; Hurles, M.E.; Firth, H.V.; Swaminathan, G.J. Facilitating Collaboration in Rare Genetic Disorders Through Effective Matchmaking in, D.E.C.I.P.H.E.R. Hum. Mutat. 2015, 36, 941–949. [Google Scholar] [CrossRef] [PubMed]
- Bragin, E.; Chatzimichali, E.A.; Wright, C.F.; Hurles, M.E.; Firth, H.V.; Bevan, A.P.; Swaminathan, G.J. DECIPHER: Database for the interpretation of phenotype-linked plausibly pathogenic sequence and copy-number variation. Nucleic Acids Res. 2014, 42, 993–1000. [Google Scholar] [CrossRef] [PubMed]
- Silva, M.; de Leeuw, N.; Mann, K.; Schuring-Blom, H.; Morgan, S.; Giardino, D.; Rack, K.; Hastings, R. European guidelines for constitutional cytogenomic analysis. Eur. J. Hum. Genet. 2019, 27, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Carroll, J.; Wigby, K.; Murray, S. Genetic testing strategies in the newborn. J. Perinatol. 2020, 40, 1007–1016. [Google Scholar] [CrossRef] [PubMed]
- Bick, D.; Jones, M.; Taylor, S.L.; Taft, R.J.; Belmont, J. Case for genome sequencing in infants and children with rare, undiagnosed or genetic diseases. J. Med. Genet. 2019, 56, 783–791. [Google Scholar] [CrossRef] [PubMed]
- Savatt, J.M.; Myers, S.M. Genetic Testing in Neurodevelopmental Disorders. Front. Pediatr. 2021, 9, 526779. [Google Scholar] [CrossRef] [PubMed]
- McGill, B.; Wakefield, C.; Vetsch, J.; Barlow-Stewart, K.; Kasparian, N.; Patenaude, A.; Young, M.; Cohn, R.; Tucker, K. Children and young people’s understanding of inherited conditions and their attitudes towards genetic testing: A systematic review. Clin. Genet. 2019, 95, 10–22. [Google Scholar] [CrossRef] [PubMed]
- Lim, Q.; McGill, B.; Quinn, V.; Tucker, K.; Mizrahi, D.; Patenaude, A.; Warby, M.; Cohn, R.; Wakefield, C. Parents’ attitudes toward genetic testing of children for health conditions: A systematic review. Clin. Genet. 2017, 92, 569–578. [Google Scholar] [CrossRef] [PubMed]
- American College of Medical Genetics and Genomics. Five Things Patients and Providers Should Question Is Uncertainty about the Validity of the Existing Test Result. 2015. Available online: http://www.choosingwisely.org/wp-content/uploads/2015/07/ACMG-Choosing-Wisely-List.pdf (accessed on 5 May 2024).
- Kalia, S.S.; Adelman, K.; Bale, S.J.; Chung, W.K.; Eng, C.; Evans, J.P.; Herman, G.E.; Hufnagel, S.B.; Klein, T.E.; Korf, B.R.; et al. Recommendations for reporting of secondary findings in clinical exome and genome sequencing, 2016 update (ACMG SF v2.0): A policy statement of the American College of Medical Genetics and Genomics. Genet. Med. 2017, 19, 249–255. [Google Scholar] [CrossRef]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.G.F.J. Standards and Guidelines for the Interpretation of Sequence Variants: A Joint Consensus Recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015, 17, 405–424. [Google Scholar] [CrossRef]
- Donald, C.G.; Sanders, A.K. The Genetic Information Nondiscrimination Act of 2008. J. Divers. Manag. 2008, 3, 33–46. [Google Scholar] [CrossRef][Green Version]
- Lalonde, E.; Rentas, S.; Lin, F.; Dulik, M.C.; Skraban, C.M.; Spinner, N.B. Genomic Diagnosis for Pediatric Disorders: Revolution and Evolution. Front. Pediatr. 2020, 8, 373. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhong, M.; Zheng, D. Chromosomal mosaicism detected by karyotyping and chromosomal microarray analysis in prenatal diagnosis. J. Cell Mol. Med. 2021, 25, 358–366. [Google Scholar] [CrossRef]
- Vermeesch, J.R.; Fiegler, H.; de Leeuw, N.; Szuhai, K.; Schoumans, J.; Ciccone, R.; Speleman, F.; Rauch, A.; Clayton-Smith, J.; Van Ravenswaaij, C.; et al. Guidelines for molecular karyotyping in constitutional genetic diagnosis. Eur. J. Hum. Genet. 2007, 15, 1105–1114. [Google Scholar] [CrossRef]
- Weise, A.; Mrasek, K.; Klein, E.; Mulatinho, M.; Llerena Jr, J.C.; Hardekopf, D.; Pekova, S.; Bhatt, S.; Kosyakova, N.; Liehr, T. Microdeletion and Microduplication Syndromes. J. Histochem. Cytochem. 2012, 60, 346–358. [Google Scholar] [CrossRef]
- Watson, C.T.; Marques-Bonet, T.; Sharp, A.J.M.H. The Genetics of Microdeletion and Microduplication Syndromes: An Update. Annu. Rev. Genom. Hum. Genet. 2014, 15, 215–244. [Google Scholar] [CrossRef]
- Carvill, G.L.; Mefford, H.C. Microdeletion syndromes. Curr. Opin. Genet. Dev. 2013, 23, 232–239. [Google Scholar] [CrossRef]
- Miller, D.T.; Adam, M.P.; Aradhya, S.; Biesecker, L.G.; Brothman, A.R.; Carter, N.P.; Church, D.M.; Crolla, J.A.; Eichler, E.E.; Epstein, C.J.; et al. Consensus Statement: Chromosomal Microarray Is a First-Tier Clinical Diagnostic Test for Individuals with Developmental Disabilities or Congenital Anomalies. Am. J. Hum. Genet. 2010, 86, 749–764. [Google Scholar] [CrossRef]
- Clark, M.M.; Stark, Z.; Farnaes, L.; Tan, T.Y.; White, S.M.; Dimmock, D.; Kingsmore, S.F. Meta-analysis of the diagnostic and clinical utility of genome and exome sequencing and chromosomal microarray in children with suspected genetic diseases. npj Genom. Med. 2018, 3, 16. [Google Scholar] [CrossRef]
- Ho, K.S.; Twede, H.; Vanzo, R.; Harward, E.; Hensel, C.H.; Martin, M.M.; Page, S.; Peiffer, A.; Mowery-Rushton, P.; Serrano, M.; et al. Clinical Performance of an Ultrahigh Resolution Chromosomal Microarray Optimized for Neurodevelopmental Disorders. BioMed Res. Int. 2016, 2016, 3284534. [Google Scholar] [CrossRef]
- Žilina, O.; Teek, R.; Tammur, P.; Kuuse, K.; Yakoreva, M.; Vaidla, E.; Mölter-Väär, T.; Reimand, T.; Kurg, A.; Õunap, K. Chromosomal microarray analysis as a first-tier clinical diagnostic test: Estonian experience. Mol. Genet. Genom. Med. 2014, 2, 166–175. [Google Scholar] [CrossRef]
- Tao, V.Q.; Chan, K.Y.K.; Chu, Y.W.Y.; Mok, G.T.K.; Tan, T.Y.; Yang, W.; Lee, S.L.; Tang, W.F.; Tso, W.W.Y.; Lau, E.T.; et al. The clinical impact of chromosomal microarray on paediatric care in Hong Kong. PLoS ONE 2014, 9, e109629. [Google Scholar] [CrossRef]
- Battaglia, A.; Doccini, V.; Bernardini, L.; Novelli, A.; Loddo, S.; Capalbo, A.; Filippi, T.; Carey, J.C. Confirmation of chromosomal microarray as a first-tier clinical diagnostic test for individuals with developmental delay, intellectual disability, autism spectrum disorders and dysmorphic features. Eur. J. Paediatr. Neurol. 2013, 17, 589–599. [Google Scholar] [CrossRef]
- Levy, B.; Burnside, R.D. Are all chromosome microarrays the same? What clinicians need to know. Prenat. Diagn. 2019, 39, 157–164. [Google Scholar] [CrossRef]
- Keren, B. The advantages of SNP arrays over CGH arrays. Mol. Cytogenet. 2014, 7 (Suppl. S1), I31. Available online: http://www.molecularcytogenetics.org/content/7/S1/I31 (accessed on 5 May 2024).
- Wiszniewska, J.; Bi, W.; Shaw, C.; Stankiewicz, P.; Kang, S.-H.L.; Pursley, A.N.; Lalani, S.; Hixson, P.; Gambin, T.; Tsai, C.-H.; et al. Combined array CGH plus SNP genome analyses in a single assay for optimized clinical testing. Eur. J. Hum. Genet. 2014, 22, 79–87. [Google Scholar] [CrossRef]
- Schouten, J.; Vught PVan Galjaard, R. For Prenatal Diagnosis of Common. Aneuploidies 2019, 1885, 161–170. [Google Scholar]
- Stuppia, L.; Antonucci, I.; Palka, G.; Gatta, V. Use of the MLPA assay in the molecular diagnosis of gene copy number alterations in human genetic diseases. Int. J. Mol. Sci. 2012, 13, 3245–3276. [Google Scholar] [CrossRef]
- Ji, X.; Zhang, J.; Xu, Y.; Long, F.; Sun, W.; Liu, X.; Chen, Y.; Jiang, W. MLPA Application in Clinical Diagnosis of DMD/BMD in Shanghai. J. Clin. Lab. Anal. 2015, 29, 405–411. [Google Scholar] [CrossRef]
- Moelans, C.B.; Atanesyan, L.; Savola, S.P.; Van Diest, P.J. Methylation-Specific Multiplex Ligation-Dependent Probe Amplification (MS-MLPA), DNA Methylation Protocols, Methods in Molecular Biology; Humana Press: New York, NY, USA, 2018; Volume 1708, pp. 537–549. [Google Scholar]
- Bean, L.J.H.; Funke, B.; Carlston, C.M.; Gannon, J.L.; Kantarci, S.; Krock, B.L.; Zhang, S.; Bayrak-Toydemir, P. Diagnostic gene sequencing panels: From design to report—A technical standard of the American College of Medical Genetics and Genomics (ACMG). Genet. Med. 2020, 22, 453–461. [Google Scholar] [CrossRef]
- Petersen, B.S.; Fredrich, B.; Hoeppner, M.P.; Ellinghaus, D.; Franke, A. Opportunities and challenges of whole-genome and -exome sequencing. BMC Genet. 2017, 18, 1749–1751. [Google Scholar] [CrossRef]
- Barbosa-Gouveia, S.; Vázquez-Mosquera, M.E.; González-Vioque, E.; Álvarez, J.V.; Chans, R.; Laranjeira, F.; Martins, E.; Ferreira, A.C.; Avila-Alvarez, A.C.M. Utility of Gene Panels for the Diagnosis of Inborn Errors of Metabolism in a Metabolic Reference Center. Genes 2021, 12, 1262. [Google Scholar] [CrossRef] [PubMed]
- Santani, A.; Murrell, J.; Funke, B.; Yu, Z.; Hegde, M.; Mao, R.; Ferreira-Gonzalez, A.; Voelkerding, K.V.; Weck, K.E. Development and validation of targeted next-generation sequencing panels for detection of germline variants in inherited diseases. Arch. Pathol. Lab. Med. 2017, 141, 787–797. [Google Scholar] [CrossRef] [PubMed]
- Mandelker, D.; Schmidt, R.J.; Ankala, A.; Gibson, K.M.; Bowser, M.; Sharma, H.; Duffy, E.; Hegde, M.; Santani, A.; Lebo, M.; et al. Navigating highly homologous genes in a molecular diagnostic setting: A resource for clinical next-generation sequencing. Genet. Med. 2016, 18, 1282–1289. [Google Scholar] [CrossRef] [PubMed]
- Retterer, K.; Juusola, J.; Cho, M.T.; Vitazka, P.; Millan, F.; Gibellini, F.; Vertino-Bell, A.; Smaoui, N.; Neidich, J.; Monaghan, K.G.; et al. Clinical application of whole-exome sequencing across clinical indications. Genet. Med. 2016, 18, 696–704. [Google Scholar] [CrossRef] [PubMed]
- Mahfouz, N.A.; Kizhakkedath, P.; Ibrahim, A.; El Naofal, M.; Ramaswamy, S.; Harilal, D.; Qutub, Y.; Uddin, M.; Taylor, A.; Alloub, Z.; et al. Utility of clinical exome sequencing in a complex Emirati pediatric cohort. Comput. Struct. Biotechnol. J. 2020, 18, 1020–1027. [Google Scholar] [CrossRef]
- Lee, H.; Deignan, J.L.; Dorrani, N.; Strom, S.P.; Kantarci, S.; Quintero-Rivera, F.; Das, K.; Toy, T.; Harry, B.; Yourshaw, M.; et al. Clinical Exome Sequencing for Genetic Identification of Rare Mendelian Disorders. JAMA 2014, 312, 1880–1887. [Google Scholar] [CrossRef]
- Monroe, G.R.; Frederix, G.W.; Savelberg, S.M.C.; de Vries, T.I.; Duran, K.J.; van der Smagt, J.J.; Terhal, P.A.; van Hasselt, P.M.; Kroes, H.Y.; Verhoeven-Duif, N.M.; et al. Effectiveness of whole-exome sequencing and costs of the traditional diagnostic trajectory in children with intellectual disability. Genet. Med. 2016, 18, 949–956. [Google Scholar] [CrossRef]
- Srivastava, S.; Love-Nichols, J.A.; Dies, K.A.; Ledbetter, D.H.; Martin, C.L.; Chung, W.K.; Firth, H.V.; Frazier, T.; Hansen, R.L.; Prock, L.; et al. Meta-analysis and multidisciplinary consensus statement: Exome sequencing is a first-tier clinical diagnostic test for individuals with neurodevelopmental disorders. Genet. Med. 2019, 21, 2413–2421. [Google Scholar] [CrossRef]
- Burdick, K.J.; Cogan, J.D.; Rives, L.C.; Robertson, A.K.; Koziura, M.E.; Brokamp, E.; Duncan, L.; Hannig, V.; Pfotenhauer, J.; Vanzo, R.; et al. Limitations of exome sequencing in detecting rare and undiagnosed diseases. Am. J. Med Genet. Part A 2020, 182, 1400–1406. [Google Scholar] [CrossRef]
- Gross, A.M.; Ajay, S.S.; Rajan, V.; Brown, C.; Bluske, K.; Burns, N.J.; Chawla, A.; Coffey, A.J.; Malhotra, A.; Scocchia, A.; et al. Copy-number variants in clinical genome sequencing: Deployment and interpretation for rare and undiagnosed disease. Genet. Med. 2019, 21, 1121–1130. [Google Scholar] [CrossRef]
- Zhang, F.; Lupski, J.R. Non-coding genetic variants in human disease. Hum. Mol. Genet. 2015, 24, R102–R110. [Google Scholar] [CrossRef] [PubMed]
- Belkadi, A.; Bolze, A.; Itan, Y.; Cobat, A.; Vincent, Q.B.; Antipenko, A.; Shang, L.; Boisson, B.; Casanova, J.-L.; Abel, L. Whole-genome sequencing is more powerful than whole-exome sequencing for detecting exome variants. Proc. Natl. Acad. Sci. USA 2015, 112, 5473–5478. [Google Scholar] [CrossRef] [PubMed]
- Lionel, A.C.; Costain, G.; Monfared, N.; Walker, S.; Reuter, M.S.; Hosseini, S.M.; Thiruvahindrapuram, B.; Merico, D.; Jobling, R.; Nalpathamkalam, T.; et al. Improved diagnostic yield compared with targeted gene sequencing panels suggests a role for whole-genome sequencing as a first-tier genetic test. Genet. Med. 2018, 20, 435–443. [Google Scholar] [CrossRef] [PubMed]
- Lelieveld, S.H.; Spielmann, M.; Mundlos, S.; Veltman, J.A.; Gilissen, C. Comparison of Exome and Genome Sequencing Technologies for the Complete Capture of Protein-Coding Regions. Hum. Mutat. 2015, 36, 815–822. [Google Scholar] [CrossRef] [PubMed]
- Nambot, S.; Thevenon, J.; Kuentz, P.; Duffourd, Y.; Tisserant, E.; Bruel, A.-L.; Mosca-Boidron, A.-L.; Masurel-Paulet, A.; Lehalle, D.; Jean-Marçais, N.; et al. Clinical whole-exome sequencing for the diagnosis of rare disorders with congenital anomalies and/or intellectual disability: Substantial interest of prospective annual reanalysis. Genet. Med. 2018, 20, 645–654. [Google Scholar] [CrossRef]
- Pena, L.D.; Jiang, Y.-H.; Schoch, K.; Spillmann, R.C.; Stong, N.; Horn, S.R.; Sullivan, J.A.; McConkie-Rosell, A.; Kansagra, S.; Smith, E.C.; et al. Looking beyond the exome: A phenotype-first approach to molecular diagnostic resolution in rare and undiagnosed diseases. Genet. Med. 2018, 20, 464–469. [Google Scholar] [CrossRef]

| Prenatal Diagnosis | Postnatal Diagnosis |
|---|---|
| Abnormal fetal ultrasound | Abnormal clinical phenotype |
| Maternal serum screening or non-invasive prenatal testing (NIPT) indicating an increased risk of a fetus carrying a chromosomal abnormality | Multiple congenital abnormalities |
| Intellectual disability | |
| Parental chromosome rearrangement, mosaicism, or previous aneuploidy | Epilepsy |
| Autism | |
| Previous livebirth/stillbirth with a chromosome abnormality | Early onset obesity |
| Familial monogenic disorder | Early onset hearing loss |
| Early onset visual problem | |
| Clinically significant abnormal growth (short stature, excessive growth, microcephaly, macrocephaly) | |
| Ambiguous genitalia | |
| Family history of chromosome rearrangements in a symptomatic child |
| Method | Main Indications in Children | Point of Strength | Limitations |
|---|---|---|---|
| Karyotyping |
|
|
|
| Fluorescence in situ hybridization (FISH) |
|
|
|
| Chromosomal microarray (CMA): array-CGH SNP array |
|
|
|
| Multiplex Ligation-Dependent Probe Amplification (MLPA) |
|
|
|
| Gene panel |
|
|
|
| Whole Exome sequencing (WES) |
|
|
|
| Whole Genome sequencing (WGS) |
|
|
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ciancia, S.; Madeo, S.F.; Calabrese, O.; Iughetti, L. The Approach to a Child with Dysmorphic Features: What the Pediatrician Should Know. Children 2024, 11, 578. https://doi.org/10.3390/children11050578
Ciancia S, Madeo SF, Calabrese O, Iughetti L. The Approach to a Child with Dysmorphic Features: What the Pediatrician Should Know. Children. 2024; 11(5):578. https://doi.org/10.3390/children11050578
Chicago/Turabian StyleCiancia, Silvia, Simona Filomena Madeo, Olga Calabrese, and Lorenzo Iughetti. 2024. "The Approach to a Child with Dysmorphic Features: What the Pediatrician Should Know" Children 11, no. 5: 578. https://doi.org/10.3390/children11050578
APA StyleCiancia, S., Madeo, S. F., Calabrese, O., & Iughetti, L. (2024). The Approach to a Child with Dysmorphic Features: What the Pediatrician Should Know. Children, 11(5), 578. https://doi.org/10.3390/children11050578








