Contributions of Artificial Intelligence to Analysis of Gut Microbiota in Autism Spectrum Disorder: A Systematic Review
Abstract
1. Introduction
2. Materials and Methods
2.1. Protocol and Registration
2.2. Eligibility Criteria
2.3. Information Sources
2.4. Search
2.5. Study Selection
2.6. Data Collection and Data Items
3. Results
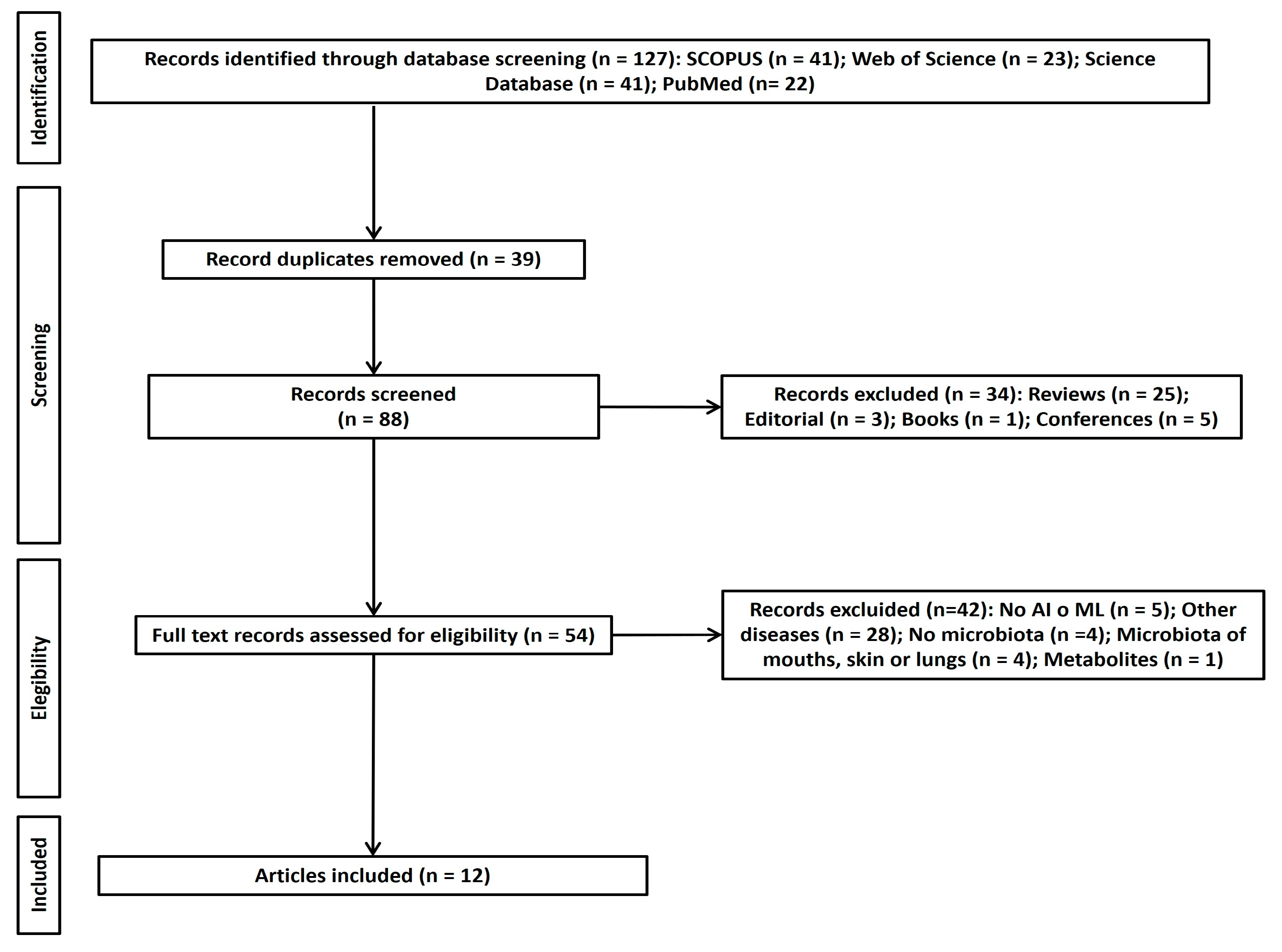
3.1. Study Selection
3.2. Individual Study Characteristics and Outcomes
3.3. Risk of Bias
3.4. Limitations
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- APA (American Psychiatric Association). Diagnostic and Statistical Manual of Mental Disorders, Fifth Edition Text Revision, DSM-5-TR; American Psychiatric Association: Washington, DC, USA, 2022. [Google Scholar]
- Maenner, M.J.; Shaw, K.A.; Baio, J.; Washington, A.; Patrick, M.; DiRienzo, M.; Christensen, D.L.; Wiggins, L.D.; Pettygrove, S.; Andrews, J.G.; et al. Prevalence of Autism Spectrum Disorder Among Children Aged 8 Years—Autism and Developmental Disabilities Monitoring Network, 11 Sites, United States, 2016. MMWR Surveill. Summ. 2020, 69, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Myers, S.M.; Voigt, R.G.; Colligan, R.C.; Weaver, A.L.; Storlie, C.B.; Stoeckel, R.E.; Port, J.D.; Katusic, S.K. Autism Spectrum Disorder: Incidence and Time Trends Over Two Decades in a Population-Based Birth Cohort. J. Autism Dev. Disord. 2019, 49, 1455–1474. [Google Scholar] [CrossRef] [PubMed]
- Russell, G.; Stapley, S.; Newlove-Delgado, T.; Salmon, A.; White, R.; Warren, F.; Pearson, A.; Ford, T. Time trends in autism diagnosis over 20 years: A UK population-based cohort study. J. Child Psychol. Psychiatry 2022, 63, 674–682. [Google Scholar] [CrossRef] [PubMed]
- Qiu, S.; Lu, Y.; Li, Y.; Shi, J.; Cui, H.; Gu, Y.; Li, Y.; Zhong, W.; Zhu, X.; Liu, Y.; et al. Prevalence of autism spectrum disorder in Asia: A systematic review and meta-analysis. Psychiatry Res. 2020, 284, 112679. [Google Scholar] [CrossRef] [PubMed]
- van ’t Hof, M.; Tisseur, C.; van Berckelear-Onnes, I.; van Nieuwenhuyzen, A.; Daniels, A.M.; Deen, M.; Hoek, H.W.; Ester, W.A. Age at autism spectrum disorder diagnosis: A systematic review and meta-analysis from 2012 to 2019. Autism 2021, 25, 862–873. [Google Scholar] [CrossRef] [PubMed]
- Chan, W.K.; Griffiths, R.; Price, D.J.; Mason, J.O. Cerebral organoids as tools to identify the developmental roots of autism. Mol. Autism 2020, 11, 58. [Google Scholar] [CrossRef] [PubMed]
- LaSalle, J.M. Epigenomic signatures reveal mechanistic clues and predictive markers for autism spectrum disorder. Mol. Psychiatry 2023, 28, 1890–1901. [Google Scholar] [CrossRef] [PubMed]
- Andreo-Martínez, P.; Rubio-Aparicio, M.; Sánchez-Meca, J.; Veas, A.; Martínez-González, A.E. A Meta-analysis of Gut Microbiota in Children with Autism. J. Autism Dev. Disord. 2022, 52, 1374–1387. [Google Scholar] [CrossRef]
- Martínez-González, A.E.; Andreo-Martínez, P. Implications of Gut Microbiota and Gastrointestinal Symptoms in Autism. In Advances in Health and Disease; Duncan, L.T., Ed.; NOVA Science Publishers: New York, NY, USA, 2020; Volume 29, pp. 16–21. [Google Scholar]
- Andreo-Martínez, P.; García-Martínez, N.; Sánchez-Samper, E.P.; Quesada-Medina, J.; MacFabe, D. Metabolites of the gut microbiota involved in the autism spectrum disorder. Rev. Dis. Clin. Neuro. 2018, 5, 39–48. [Google Scholar] [CrossRef]
- Andreo-Martínez, P.; García-Martínez, N.; Sánchez-Samper, E.P.; Martínez-González, A.E. An approach to gut microbiota profile in children with autism spectrum disorder. Environ. Microbiol. Rep. 2019, 12, 115–135. [Google Scholar] [CrossRef]
- Martínez-González, A.E.; Andreo-Martínez, P. The Role of Gut Microbiota in Gastrointestinal Symptoms of Children with ASD. Medicina 2019, 55, 408. [Google Scholar] [CrossRef] [PubMed]
- Berding, K.; Donovan, S.M. Diet Can Impact Microbiota Composition in Children With Autism Spectrum Disorder. Front. Neurosci. 2018, 12, 515. [Google Scholar] [CrossRef]
- Martínez-González, A.E.; Cervin, M.; Pérez-Sánchez, S. Assessing gastrointestinal symptoms in people with autism: Applying a new measure based on the Rome IV criteria. Dig. Liver Dis. 2024, in press. [CrossRef]
- Martínez-González, A.E.; Andreo-Martínez, P. Prebiotics, probiotics and fecal microbiota transplantation in autism: A systematic review. Rev. Psiquiatr. Salud Ment. 2020, 13, 150–164. [Google Scholar] [CrossRef] [PubMed]
- Lindström, E.S.; Langenheder, S. Local and regional factors influencing bacterial community assembly. Environ. Microbiol. Rep. 2012, 4, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T.A.; Worobey, M. Geographical variation of human gut microbial composition. Biol. Lett. 2014, 10, 20131037. [Google Scholar] [CrossRef] [PubMed]
- Gupta, V.K.; Paul, S.; Dutta, C. Geography, Ethnicity or Subsistence-Specific Variations in Human Microbiome Composition and Diversity. Front. Microbiol. 2017, 8, 1162. [Google Scholar] [CrossRef] [PubMed]
- Minsky, M. Steps toward Artificial Intelligence. Proc. IRE 1961, 49, 8–30. [Google Scholar] [CrossRef]
- Feigenbaum, E. Artificial intelligence research. IEEE Trans. Inf. Theory 1963, 9, 248–253. [Google Scholar] [CrossRef]
- Duan, Y.; Edwards, J.S.; Dwivedi, Y.K. Artificial intelligence for decision making in the era of Big Data—Evolution, challenges and research agenda. Int. J. Inf. Manag. 2019, 48, 63–71. [Google Scholar] [CrossRef]
- Stahl, D.; Pickles, A.; Elsabbagh, M.; Johnson, M.H.; The, B.T. Novel Machine Learning Methods for ERP Analysis: A Validation From Research on Infants at Risk for Autism. Dev. Neuropsychol. 2012, 37, 274–298. [Google Scholar] [CrossRef] [PubMed]
- Maenner, M.J.; Yeargin-Allsopp, M.; Van Naarden Braun, K.; Christensen, D.L.; Schieve, L.A. Development of a Machine Learning Algorithm for the Surveillance of Autism Spectrum Disorder. PLoS ONE 2016, 11, e0168224. [Google Scholar] [CrossRef] [PubMed]
- Rosenblatt, F. Principles of neurodynamics. perceptrons and the theory of brain mechanisms. Am. J. Psychol. 1963, 76, 705. [Google Scholar]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef] [PubMed]
- Krizhevsky, A.; Sutskever, I.; Hinton, G. ImageNet Classification with Deep Convolutional Neural Networks. Neural Inf. Process. Syst. 2012, 25, 84–90. [Google Scholar] [CrossRef]
- Graham, S.; Depp, C.; Lee, E.E.; Nebeker, C.; Tu, X.; Kim, H.C.; Jeste, D.V. Artificial Intelligence for Mental Health and Mental Illnesses: An Overview. Curr. Psychiatry Rep. 2019, 21, 116. [Google Scholar] [CrossRef] [PubMed]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. BMJ 2021, 372, n71. [Google Scholar] [CrossRef] [PubMed]
- Jung, Y.; Lee, T.; Oh, H.S.; Hyun, Y.; Song, S.; Chun, J.; Kim, H.W. Gut microbial and clinical characteristics of individuals with autism spectrum disorder differ depending on the ecological structure of the gut microbiome. Psychiatry Res. 2024, 335, 115775. [Google Scholar] [CrossRef] [PubMed]
- Olaguez-Gonzalez, J.M.; Chairez, I.; Breton-Deval, L.; Alfaro-Ponce, M. Machine Learning Algorithms Applied to Predict Autism Spectrum Disorder Based on Gut Microbiome Composition. Biomedicines 2023, 11, 2633. [Google Scholar] [CrossRef]
- Zou, R.; Xu, F.; Wang, Y.; Duan, M.; Guo, M.; Zhang, Q.; Zhao, H.; Zheng, H. Changes in the Gut Microbiota of Children with Autism Spectrum Disorder. Autism Res. 2020, 13, 1614–1625. [Google Scholar] [CrossRef]
- Ding, X.; Xu, Y.; Zhang, X.; Zhang, L.; Duan, G.; Song, C.; Li, Z.; Yang, Y.; Wang, Y.; Wang, X.; et al. Gut microbiota changes in patients with autism spectrum disorders. J. Psychiatr. Res. 2020, 129, 149–159. [Google Scholar] [CrossRef] [PubMed]
- Olaguez-Gonzalez, J.M.; Schaeffer, S.E.; Breton-Deval, L.; Alfaro-Ponce, M.; Chairez, I. Assessment of machine learning strategies for simplified detection of autism spectrum disorder based on the gut microbiome composition. Neural Comput. Appl. 2024, 36, 8163–8180. [Google Scholar] [CrossRef]
- Dan, Z.; Mao, X.; Liu, Q.; Guo, M.; Zhuang, Y.; Liu, Z.; Chen, K.; Chen, J.; Xu, R.; Tang, J.; et al. Altered gut microbial profile is associated with abnormal metabolism activity of Autism Spectrum Disorder. Gut Microbes 2020, 11, 1246–1267. [Google Scholar] [CrossRef] [PubMed]
- Peralta-Marzal, L.N.; Rojas-Velazquez, D.; Rigters, D.; Prince, N.; Garssen, J.; Kraneveld, A.D.; Perez-Pardo, P.; Lopez-Rincon, A. A robust microbiome signature for autism spectrum disorder across different studies using machine learning. Sci. Rep. (Nat. Publ. Group) 2024, 14, 814. [Google Scholar] [CrossRef]
- Pietrucci, D.; Teofani, A.; Milanesi, M.; Fosso, B.; Putignani, L.; Messina, F.; Pesole, G.; Desideri, A.; Chillemi, G. Machine Learning Data Analysis Highlights the Role of Parasutterella and Alloprevotella in Autism Spectrum Disorders. Biomedicines 2022, 10, 2028. [Google Scholar] [CrossRef]
- Averina, O.V.; Kovtun, A.S.; Polyakova, S.I.; Savilova, A.M.; Rebrikov, D.V.; Danilenko, V.N. The bacterial neurometabolic signature of the gut microbiota of young children with autism spectrum disorders. J. Med. Microbiol. 2020, 69, 558–571. [Google Scholar] [CrossRef]
- Pulikkan, J.; Maji, A.; Dhakan, D.B.; Saxena, R.; Mohan, B.; Anto, M.M.; Agarwal, N.; Grace, T.; Sharma, V.K. Gut Microbial Dysbiosis in Indian Children with Autism Spectrum Disorders. Microb. Ecol. 2018, 76, 1102–1114. [Google Scholar] [CrossRef] [PubMed]
- Zurita, M.F.; Cárdenas, P.A.; Sandoval, M.E.; Peña, M.C.; Fornasini, M.; Flores, N.; Monaco, M.H.; Berding, K.; Donovan, S.M.; Kuntz, T.; et al. Analysis of gut microbiome, nutrition and immune status in autism spectrum disorder: A case-control study in Ecuador. Gut Microbes 2019, 11, 453–464. [Google Scholar] [CrossRef]
- Coretti, L.; Paparo, L.; Riccio, M.P.; Amato, F.; Cuomo, M.; Natale, A.; Borrelli, L.; Corrado, G.; Comegna, M.; Buommino, E.; et al. Gut Microbiota Features in Young Children With Autism Spectrum Disorders. Front. Microbiol. 2018, 9, 3146. [Google Scholar] [CrossRef]
- Son, J.S.; Zheng, L.J.; Rowehl, L.M.; Tian, X.; Zhang, Y.; Zhu, W.; Litcher-Kelly, L.; Gadow, K.D.; Gathungu, G.; Robertson, C.E.; et al. Comparison of Fecal Microbiota in Children with Autism Spectrum Disorders and Neurotypical Siblings in the Simons Simplex Collection. PLoS ONE 2015, 10, e0137725. [Google Scholar] [CrossRef]
- Rojas-Velazquez, D.; Kidwai, S.; Kraneveld, A.D.; Tonda, A.; Oberski, D.; Garssen, J.; Lopez-Rincon, A. Methodology for biomarker discovery with reproducibility in microbiome data using machine learning. BMC Bioinform. 2024, 25, 26. [Google Scholar] [CrossRef] [PubMed]
- David, M.M.; Tataru, C.; Daniels, J.; Schwartz, J.; Keating, J.; Hampton-Marcell, J.; Gottel, N.; Gilbert, J.A.; Wall, D.P. Children with Autism and Their Typically Developing Siblings Differ in Amplicon Sequence Variants and Predicted Functions of Stool-Associated Microbes. mSystems 2021, 6, e00193-20. [Google Scholar] [CrossRef] [PubMed]
- Vernocchi, P.; Marangelo, C.; Guerrera, S.; Del Chierico, F.; Guarrasi, V.; Gardini, S.; Conte, F.; Paci, P.; Ianiro, G.; Gasbarrini, A.; et al. Gut microbiota functional profiling in autism spectrum disorders: Bacterial VOCs and related metabolic pathways acting as disease biomarkers and predictors. Front. Microbiol. 2023, 14, 1287350. [Google Scholar] [CrossRef] [PubMed]
- Vernocchi, P.; Ristori, M.V.; Guerrera, S.; Guarrasi, V.; Conte, F.; Russo, A.; Lupi, E.; Albitar-Nehme, S.; Gardini, S.; Paci, P.; et al. Gut Microbiota Ecology and Inferred Functions in Children With ASD Compared to Neurotypical Subjects. Front. Microbiol. 2022, 13, 871086. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.B.; Doenyas, C.; Wan, J.; Zeng, S.J.; Cai, C.Q.; Zhou, J.X.; Liu, Y.Q.; Yin, Z.Q.; Zhou, W.H. Virulence factor-related gut microbiota genes and immunoglobulin A levels as novel markers for machine learning-based classification of autism spectrum disorder. Comp. Struct. Biotechnol. J. 2021, 19, 545–554. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Wan, J.; Rong, H.; He, F.; Wang, H.; Zhou, J.; Cai, C.; Wang, Y.; Xu, R.; Yin, Z.; et al. Alterations in Gut Glutamate Metabolism Associated with Changes in Gut Microbiota Composition in Children with Autism Spectrum Disorder. mSystems 2019, 4, e00321-18. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; He, F.; Yang, F.; Yang, Z.; Xie, Y.; Zhou, S.; Liang, J.; Xu, R.; Wang, Y.; Guo, H.; et al. Increased stool immunoglobulin A level in children with autism spectrum disorders. Res. Dev. Disabil. 2018, 82, 90–94. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.J.; Fu, P.C. Gut Microbiota Analysis and In Silico Biomarker Detection of Children with Autism Spectrum Disorder across Cohorts. Microorganisms 2023, 11, 19. [Google Scholar] [CrossRef]
- Kovtun, A.S.; Averina, O.V.; Alekseeva, M.G.; Danilenko, V.N. Antibiotic Resistance Genes in the Gut Microbiota of Children with Autistic Spectrum Disorder as Possible Predictors of the Disease. Microb. Drug Resist. 2020, 26, 1307–1320. [Google Scholar] [CrossRef]
- Wu, T.; Wang, H.C.; Lu, W.W.; Zhai, Q.X.; Zhang, Q.X.; Yuan, W.W.; Gu, Z.N.; Zhao, J.X.; Zhang, H.; Chen, W. Potential of gut microbiome for detection of autism spectrum disorder. Microb. Pathog. 2020, 149, 10. [Google Scholar] [CrossRef]
- Strati, F.; Cavalieri, D.; Albanese, D.; De Felice, C.; Donati, C.; Hayek, J.; Jousson, O.; Leoncini, S.; Renzi, D.; Calabro, A.; et al. New evidences on the altered gut microbiota in autism spectrum disorders. Microbiome 2017, 5, 24. [Google Scholar] [CrossRef] [PubMed]
- Kang, D.W.; Park, J.G.; Ilhan, Z.E.; Wallstrom, G.; Labaer, J.; Adams, J.B.; Krajmalnik-Brown, R. Reduced incidence of Prevotella and other fermenters in intestinal microflora of autistic children. PLoS ONE 2013, 8, e68322. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Liu, X.; Xiong, X.-Q.; Yang, T.; Cui, T.; Hou, N.-L.; Lai, X.; Liu, S.; Guo, M.; Liang, X.-H.; et al. Effect of vitamin A supplementation on gut microbiota in children with autism spectrum disorders—A pilot study. BMC Microbiol. 2017, 17, 204. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.S.; Wang, Y.H.; Xu, J.S.; Song, Y.; Liu, B.Q.; Xiong, Z.F. Leveraging Existing 16SrRNA Microbial Data to Define a Composite Biomarker for Autism Spectrum Disorder. Microbiol. Spectr. 2022, 10, 12. [Google Scholar] [CrossRef] [PubMed]
- Chiappori, F.; Cupaioli, F.A.; Consiglio, A.; Di Nanni, N.; Mosca, E.; Licciulli, V.F.; Mezzelani, A. Analysis of Faecal Microbiota and Small ncRNAs in Autism: Detection of miRNAs and piRNAs with Possible Implications in Host-Gut Microbiota Cross-Talk. Nutrients 2022, 14, 1340. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Shi, K.; Liu, X.; Dai, Y.; Liu, Y.; Zhang, L.; Du, X.; Zhu, T.; Yu, J.; Fang, S.; et al. Gut microbial profile is associated with the severity of social impairment and IQ performance in children with autism spectrum disorder. Front. Psychiatry 2021, 12, 789864. [Google Scholar] [CrossRef] [PubMed]
- Ha, S.; Oh, D.; Lee, S.; Park, J.; Ahn, J.; Choi, S.; Cheon, K.A. Altered Gut Microbiota in Korean Children with Autism Spectrum Disorders. Nutrients 2021, 13, 3300. [Google Scholar] [CrossRef]
- Huang, M.; Liu, K.; Wei, Z.; Feng, Z.; Chen, J.; Yang, J.; Zhong, Q.; Wan, G.; Kong, X.J. Serum Oxytocin Level Correlates With Gut Microbiome Dysbiosis in Children With Autism Spectrum Disorder. Front. Neurosci. 2021, 15, 721884. [Google Scholar] [CrossRef] [PubMed]
- Zhao, R.H.; Zheng, P.Y.; Liu, S.M.; Tang, Y.C.; Li, E.Y.; Sun, Z.Y.; Jiang, M.M. Correlation between gut microbiota and behavior symptoms in children with autism spectrum disorder. Zhongguo Dang Dai Er Ke Za Zhi 2019, 21, 663–669. [Google Scholar] [CrossRef]
- Shi, K.; Zhang, L.; Yu, J.; Chen, Z.; Lai, S.; Zhao, X.; Li, W.G.; Luo, Q.; Lin, W.; Feng, J.; et al. A 12-genus bacterial signature identifies a group of severe autistic children with differential sensory behavior and brain structures. Clin. Transl. Med. 2021, 11, e314. [Google Scholar] [CrossRef]
- Liu, T.; Pan, X.; Wang, X.; Feenstra, K.A.; Heringa, J.; Huang, Z. Predicting the relationships between gut microbiota and mental disorders with knowledge graphs. Health Inf. Sci. Syst. 2021, 9, 3. [Google Scholar] [CrossRef] [PubMed]
- Viswanathan, M.; Patnode, C.D.; Berkman, N.D.; Bass, E.B.; Chang, S.; Hartling, L.; Murad, M.H.; Treadwell, J.R.; Kane, R.L. Assessing the risk of bias in systematic reviews of health care interventions. In Methods Guide for Effectiveness and Comparative Effectiveness Reviews [Internet]; Agency for Healthcare Research and Quality (US): Rockville, MD, USA, 2017. [Google Scholar]
- Andreo-Martínez, P.; Ortiz-Martínez, V.M.; Salar-García, M.J.; Veiga-del-Baño, J.M.; Chica, A.; Quesada-Medina, J. Waste animal fats as feedstock for biodiesel production using non-catalytic supercritical alcohol transesterification: A perspective by the PRISMA methodology. Energy Sustain. Dev. 2022, 69, 150–163. [Google Scholar] [CrossRef]
- Guillamón, E.; Andreo-Martínez, P.; Mut-Salud, N.; Fonollá, J.; Baños, A. Beneficial Effects of Organosulfur Compounds from Allium cepa on Gut Health: A Systematic Review. Foods 2021, 10, 1680. [Google Scholar] [CrossRef] [PubMed]
- Ortiz-Martínez, V.M.; Andreo-Martínez, P.; García-Martínez, N.; Pérez de los Ríos, A.; Hernández-Fernández, F.J.; Quesada-Medina, J. Approach to biodiesel production from microalgae under supercritical conditions by the PRISMA method. Fuel Process. Technol. 2019, 191, 211–222. [Google Scholar] [CrossRef]
- Castro, K.; Faccioli, L.S.; Baronio, D.; Gottfried, C.; Perry, I.S.; dos Santos Riesgo, R. Effect of a ketogenic diet on autism spectrum disorder: A systematic review. Res. Autism. Spectr. Disord. 2015, 20, 31–38. [Google Scholar] [CrossRef]
- Martínez-González, A.E.; Piqueras, J.A. Differences in the severity of Autistic Spectrum Disorder symptoms according to the educational context. Eur. J. Educ. Psychol. 2019, 12, 153–164. [Google Scholar] [CrossRef]
| Ref | Participant Characteristic | Country | Data Source | Study Type | Main Finding | ML/DL Method Used | Model Predicting Microbiome | |
|---|---|---|---|---|---|---|---|---|
| Experimental Group | Control Group | |||||||
| [30] | ASD (n = 249) | NT siblings (n = 106) NT control (n = 101) | South Korea | Original | PCR, 16S rRNA (V3–V4 regions) | Negative association between Bifidobacterium longum and Childhood Autism Rating Scale outcomes, as well as a negative association between Streptococcus salivarus and Social Responsiveness Scale (SRS) outcomes in ASD. | ML: XGB regression of high/low SRS/VABS values Prediction of microbial age | Bacteroides vulgatus, Roseburia cecicola group, Lachnospiraceae and Agathobaculum butyriproducience showed significantly different abundances between high and low SRS groups in the E1 model (p ≤ 0.01), but not in the E2-SRS classification model. Streptococcus salivarus significantly differed between high and low SRS groups in model E2 (p ≤ 0.01), but not in E1. |
| [31] | ASD (n = 48) (from 2 to 7 years old, average 5, average BMI = 17.4, 10 females and 38 males) ASD (n = 77) | NT (n = 48) (all at 48 months, no allergies, 24 females and 24 males, average BMI = 16.3). NT (n = 50) | Mexico | [32,33] | 16S rRNA (V3–V4 regions) (V4 region) | See [32,33] | ML: SVM, RF DL: ANNs Classification ASD v. HC | Lachnospira (primary predictor in the RF- and ANN-based models, ranking second in the SVM) Of the five main predictors in SVM and ANN models: Bacteroides (p = 2.4 × 10−3), Escherichia–Shigella (p = 2.39 × 10−2), Akkermansia (p = 2.51 × 10−2) and Dialister (p = 3.67 × 10−2) are statistically different. SVM [32]: Bacteroides, Lachnospira, Blautia, Lachnoclostridium and Subdoligranulum ANN [32]: Lachnospira, Bacteroides, Lachnoclostridium, Blautia and Subdoligranulum RF [32]: Lachnospira, Escherichia–Shigella, Bacteroides, Blautia and Roseburia ANN performed better than SVM on training and validation partitions, with 97.01% for training and 82.21% for validation. SVM [33]: Ruminococcus torques, Anaerobutyricum Dorea, Subdoligranulum and Bacteroides ANN [33]: Anaerobutyricum, Bacteroides, Ruminococcus torques, Dorea and Subdoligranulum RF [33]: Anaerobutyricum, Faecalibacterium, Clostridium sensu stricto, Ruminococcus torques and Agathobacter |
| [34] | ASD (n = 111) | NT (n = 143) | Mexico | [35] | 16S rRNA (V4 region) | See [35] | ML: RF, SVM, kNN, NB DL: ANNs Classification ASD v. HC | Main predictor: Prevotella_2. Other significant predictors: Ruminiclostridium_6 and the Alloprevotella. The ANN model demonstrates a 6% increase in sensitivity compared to kNN and RF models |
| [36] | ASD (n = 60) ASD (n = 77) ASD (n = 48) | Siblings (n = 57) HC (n = 50) HC (n = 48) | The Netherlands | Original [32,33] | 16S rRNA (V3–V4 regions) (V4 region) | See [32,33] | ML: REFS Feature selection through REFS | ASVs: 26 ASVs for differential abundances. ↓Actinobacteria phylum, Bifidobacterium and Collinsella in ASD ↑Bacteroidota phylum, Prevotellaceae and Parabacteroides in ASD ↑bacterial taxa in ASD phenotype: Clostridia, Sarcina and Parabacteroides |
| [37] | ASD (n = 540) | HC (n = 419) | Italy | [35,38,39,40,41,42] | 16S rRNA (Different regions) | See cited articles | ML: RF, SVM, GB Classification ASD v. HC | Main bacterial generates for all three algorithms: Alloprevotella, Sutterella, Haemophilus, Faecalibacterium and an unclassified Clostridia ‘UCG 014’. RF and the GBM algorithms: [Eubacterium] siraeum_group, Tyzzerella, Negativibacillus, Muribaculaceae, Gastranaerophilales, Megamonas and Rombustia. GBM and SVM algorithms: Bacteroides and Subdoligranulum identified as important. ↓Alloprevotella genus in ASD sample (abundance in ASD samples = 0.34 ± 0.20, abundance in HC samples = 0.12 ± 0.14). ↑Parasutterella (ASD samples = 0.57 ± 0.16, HC samples = 0.38 ± 0.17), Haemophilus (ASD samples = 0.57 ± 0.19, HC samples = 0.33 ± 0.17), Faecalibacterium (ASD samples = 0.86 ± 0.14, HC samples = 0.70 ± 0.21) and Clostridiales UCG 14 (ASD samples = 0.60 ± 0.21, HC samples = 0.34 ± 0.17) in ASD. |
| [43] | ASD (n = 60) ASD (n = 77) ASD (n = 48) | HC (n = 57) HC (n = 50) HC (n = 48) | The Netherlands | [32,33,44] | 16S rRNA (V3–V4 regions) (V4 region) | See cited articles | ML: REFS Feature selection: biomarker identification | Better performance in AUC and MCC compared to K-Best and 10-time random selection methods. ↓Bifidobacterium, Enterobacteriaceae, Lachnospira, Lachnospiraceae and Clostridium in ASD |
| [45] | ASD (n = 41) | NT (n = 35) | Italy | Original | PCR 16S rRNA (V3–V4 regions) | Bifidobacterium was negatively correlated with indole and skatole Positive correlations between Carnobacteriaceae, Actinobacillus, Pepetostreptococcaceae, pentanoic acid, 2.6-dimethyl-pyrazine, nonadecane and 3-methyl-butanoic acid. | ML: PCA, PLS-DA Feature selection: biomarker identification | Hist Gradient Boosting Classifier was the best performing model with 89% accuracy. VOCs associated with ASDs: methyl isobutyl ketone, benzeneacetaldehyde, phenyl ethyl alcohol, ethanol, butanoic acid, octadecane, acetic acid, skatole and tetradecanal (myristyl aldehyde) Positive correlations with OTUs-VOCs couples: -Bifidobacteriaceae/2-dodecanol -Serratia/benzyl alcohol -Roseburia/1-butanol -Firmicutes/butanoic acid -Pasteurellaceae/3-methyl 1-butanol. |
| [46] | ASD (n = 41) | NT (n = 35) | Italy | Original | PCR 16S rRNA (V3–V4 regions) | 30 ASD with GI symptoms: 93% with a high level of severity Phylum level: ↓Actinobacteria, Cyanobacteria and TM7 in ASD ↑Proteobacteria and Bacteroidetes in ASD (Bacteroidetes was observed in the ASD without GI symptoms group) Family level: ↓Coriobacteriaceae, Bifidobacteriaceae, Actynomicetaceae and (Tissierellaceae) in ASD. ↑Alcaligenaceae, Lactobacillaceae, Prevotellaceaeae and Bacteroidaceae in ASD. ↑Bacteroidaceae and Lactobacillaceae ASD without GI symptoms. Genus level: ↑Bacteroides and Klebsiella in ASD. Klebsiella and Lactobacillus were higher in ASD without GI symptoms. ↓Bifidobacterium and Actinomyces in ASD. ASD-related microbial biomarkers (p-value < 0.05): -Bacteroidetes/Proteobacteria. -Bacteroidaceae/Rikenellaceae/ Lactobacillaceae/Prevotellaceae/Pasteurellaceae/Alcaligenaceae. -Bacteroides/Lactobacillus/ Prevotella/Klebsiella/Roseburia/Haemophilus/Sutterella. | ML: LR, SGD, RF, ET, GB, XGB, etc. DL: MLP Classification ASD v. HC | Contextually, model classification analysis based on ML identified both KOs and ko pathways able to classify 73% of patients with ASD versus CTRLs (p-value < 0.05). Specific selected OTUs for ASD and CTRLs revealed the main bacteria: ↓Bacteroides, Lactobacillus, Prevotella, Staphylococcus and Sutturella in ASD ↓Ruminococcus, Blautia, Coprococcus, Bifodobacterium and Streptococcus in ASD. |
| [47] | ASD (n = 43) | TD (n = 31) | China | [48,49] | IgA detection via ELISA [49] StoolGen fecal DNA extraction kit (CWBiotech Co., Beijing, China) and NanoDrop 2000 (Thermo Scientific, Foster City, CA, USA). A total of 5 µg (or more) of DNA was required for library construction using the TruSeq DNA sample preparation kit (Illumina, San Diego, CA, USA) [48]. | VFGM genes related to ASD: cpsH, cpsJ and cpsO genes related to high levels of Streptococcus agalactiae 2603 V/R in the gut of ASD children with/without GI symptoms | ML: RF Classification ASD v. HC | The main genes involved according to machine learning via the random forest method were mtrE, kfiC, pvdM and hasA. |
| [50] | ASD (n = 73) | TD (n = 71) | China | [48,51] | Illumina NovaSeq 6000 Illumina HiSeq 4000, Illumina Inc. San Diego, CA, USA | See cited articles | ML: RF Classification ASD v. HC | Predicted performance was evaluated according to AUROC. In the China cohort, a high AUROC value of 0.984 and 97% accuracy were achieved with only one round of a 100-iteration run. The Moscow cohort produced a poor average AUROC outcome of 0.81 and only 67% accuracy following six rounds of the 100-iteration run. Overall, average values for AUROC and accuracy were 0.86 and 80%, respectively, with an average feature set of 67 species. Eubacterium_sp_CAG_248 and Prevotella copri were the most likely biomarkers involved in ASD. |
| [52] | ASD (n = 169) | NT (n = 128) | China | [39,42,53,54,55] | PCR 16S rRNA (Different regions) | See cited articles | ML: LDA (LEfSe)+ RF, kSVM + RBF, DT DL: MLP Feature selection + Classification | Dominant major genera: ↓Prevotella, ↓Ruminococcus and Roseburia as potential biomarkers of ASD. Prevotella, Roseburia, Ruminococcus, Megasphaera and Catenibacterium as potential biomarkers in ASD patients. However, only Prevotella significantly differed between the two groups. |
| [56] | ASD (n = 569) | HC (n = 450) | China | [32,33,35,38,40,41,46,57,58,59,60,61] | PCR 16S rRNA (V3–V4, V4, V4–V5 regions) Illumina MiSeq | See cited articles | ML: RF Classification of ASD v. HC (after feature selection) | AUC of the training set and verification set was 0.688 and 0.706. Dominant genera of the ASD group included Lachnospiracea_incertae_sedis, Clostridium_XVIII, Eubacterium, Anaerostipes, Clostridium_sensu_stricto, Coprococcus, Dorea and Faecalibacterium. Subgroup analysis followed different sequencing platforms to examine dominant genera in ASD. Dominant genera in the ASD group included Eubacterium, Bifidobacterium, Blautia, Dialister, Coprococcus and Lachnospiracea_ incertae_sedis. |
| Item | [31] | [34] | [36] | [37] | [43] | [45] | [46] | [47] | [50] | [52] | [56] | [30] |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 1 | 2 | 2 | 2 |
| 1 | 2 | 2 | 2 | 2 | 1 | 1 | 1 | 2 | 2 | 2 | 2 |
| 1 | 1 | 2 | 2 | 1 | 1 | 2 | 1 | 1 | 1 | 2 | 2 |
| 1 | 1 | 2 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 2 | 2 |
| 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 2 |
| 2 | 2 | 2 | 2 | 2 | 2 | 2 | 1 | 1 | 2 | 2 | 2 |
| TOTAL | 8 | 9 | 11 | 10 | 9 | 8 | 11 | 8 | 6 | 9 | 11 | 12 |
| Risk of bias | 4 | 3 | 1 | 2 | 3 | 4 | 1 | 4 | 6 | 3 | 1 | 0 |
| Risk of bias classification | M | M | L | L | M | M | L | M | M | M | L | L |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Climent-Pérez, P.; Martínez-González, A.E.; Andreo-Martínez, P. Contributions of Artificial Intelligence to Analysis of Gut Microbiota in Autism Spectrum Disorder: A Systematic Review. Children 2024, 11, 931. https://doi.org/10.3390/children11080931
Climent-Pérez P, Martínez-González AE, Andreo-Martínez P. Contributions of Artificial Intelligence to Analysis of Gut Microbiota in Autism Spectrum Disorder: A Systematic Review. Children. 2024; 11(8):931. https://doi.org/10.3390/children11080931
Chicago/Turabian StyleCliment-Pérez, Pau, Agustín Ernesto Martínez-González, and Pedro Andreo-Martínez. 2024. "Contributions of Artificial Intelligence to Analysis of Gut Microbiota in Autism Spectrum Disorder: A Systematic Review" Children 11, no. 8: 931. https://doi.org/10.3390/children11080931
APA StyleCliment-Pérez, P., Martínez-González, A. E., & Andreo-Martínez, P. (2024). Contributions of Artificial Intelligence to Analysis of Gut Microbiota in Autism Spectrum Disorder: A Systematic Review. Children, 11(8), 931. https://doi.org/10.3390/children11080931








