Further Evidence That Defects in Main Thyroid Dysgenesis-Related Genes Are an Uncommon Etiology for Primary Congenital Hypothyroidism in Mexican Patients: Report of Rare Variants in FOXE1, NKX2-5 and TSHR
Abstract
1. Introduction
2. Materials and Methods
2.1. Population Study
2.2. Molecular Analysis
2.2.1. Sanger Sequencing of NKX2-5, FOXE1, and TSHR
2.2.2. MLPA Analysis of TSHR, FOXE1, PAX8, and NKX2-1
2.3. Structural Modeling of the Human Forkhead Box E1 (FOXE1) and Thyrotropin Receptor Isoform 1 Precursor (TSHR) Proteins
3. Results
4. Discussion
5. Conclusions
- Although the Mexican population has one of the highest worldwide CH birth prevalences, our results along with the previously published findings confirm that small nucleotide and clinically relevant germline variants in the main TD-related genes of PAX8 [18], NKX2-1 [19], FOXE1, NKX2-5, and TSHR account for a minority (2.5%) of primary and permanent CH Mexican patients due to non-syndromic TD.
- Two previously unreported and clinically relevant genotypes, homozygous FOXE1 p.[Gly124Arg];[Gly124Arg] and compound heterozygous TSHR p.[Asp118Asn];[Trp422Arg], were found to account for 1.5% (N = 2/128) of the CH-TD patients, but further functional and segregation analyses are warranted.
- Unlike Polish CH patients [15], Mexican CH-TD patients did not harbor CNVs in PAX8, NKX2-1, FOXE1, or TSHR.
- Although we did not evaluate CNVs in NKX2-5, our data support the idea that it has negligible etiological participation in TD along with a benign character for its p.(Ala119Ser) allele [40].
- The protective effect on TD risk previously described for FOXE1 polyAla alleles larger than 16 alanines or their genotypes in French Caucasians [14] was not significantly supported in our Mexican CH-TD population.
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lorey, F.W.; Cunningham, G.C. Birth prevalence of primary congenital hypothyroidism by sex and ethnicity. Hum. Biol. 1992, 64, 531–538. [Google Scholar] [PubMed]
- Ford, G.; LaFranchi, S.H. Screening for congenital hypothyroidism: A worldwide view of strategies. Best Pract. Res. Clin. Endocrinol. Metab. 2014, 28, 175–187. [Google Scholar] [CrossRef] [PubMed]
- Mio, C.; Grani, G.; Durante, C.; Damante, G. Molecular defects in thyroid dysgenesis. Clin. Genet. 2020, 97, 222–231. [Google Scholar] [CrossRef] [PubMed]
- Castanet, M.; Polak, M.; Bonaïti-Pellié, C.; Lyonnet, S.; Czernichow, P.; Léger, J. Nineteen Years of National Screening for Congenital Hypothyroidism: Familial Cases with Thyroid Dysgenesis Suggest the Involvement of Genetic Factors. J. Clin. Endocrinol. Metab. 2001, 86, 2009–2014. [Google Scholar] [CrossRef] [PubMed]
- Stoppa-Vaucher, S.; Van Vliet, G.; Deladoëy, J. Variation by Ethnicity in the Prevalence of Congenital Hypothyroidism Due to Thyroid Dysgenesis. Thyroid 2011, 21, 13–18. [Google Scholar] [CrossRef]
- Yu, B.; Long, W.; Yang, Y.; Wang, Y.; Jiang, L.; Cai, Z.; Wang, H. Newborn Screening and Molecular Profile of Congenital Hypothyroidism in a Chinese Population. Front. Genet. 2018, 9, 509. [Google Scholar] [CrossRef]
- Makretskaya, N.; Bezlepkina, O.; Kolodkina, A.; Kiyaev, A.; Vasilyev, E.V.; Petrov, V.; Kalinenkova, S.; Malievsky, O.; Dedov, I.I.; Tiulpakov, A. High frequency of mutations in ’dyshormonogenesis genes’ in severe congenital hypothyroidism. PLoS ONE 2018, 13, e0204323. [Google Scholar] [CrossRef]
- Hinojosa-Trejo, M.A.; Vela-Amieva, M.; Ibarra-González, I.; Cosío-Farias, A.P.; Herrera-Pérez, L.A.; Caamal-Parra, G.; Bolaños-Córdova, L.E.; García-Flores, E.P. Prevalencia al nacimiento de hipotiroidismo congénito. Acta Pediátr. Mexico 2018, 39, 5–13. [Google Scholar] [CrossRef]
- Kreisner, E.; Neto, E.; Gross, J. High Prevalence of Extrathyroid Malformations in a Cohort of Brazilian Patients with Permanent Primary Congenital Hypothyroidism. Thyroid 2005, 15, 165–169. [Google Scholar] [CrossRef]
- Monroy-Santoyo, S.; Ibarra-González, I.; Fernández-Lainez, C.; Greenawalt-Rodríguez, S.; Chacón-Rey, J.; Calzada-León, R.; Vela-Amieva, M. Higher incidence of thyroid agenesis in Mexican newborns with congenital hypothyroidism associated with birth defects. Early Hum. Dev. 2012, 88, 61–64. [Google Scholar] [CrossRef]
- Kumorowicz-Czoch, M.; Tylek-Lemanska, D.; Wyrobek, L.; Grodzicka, T.; Starzyk, J. Thyroid developmental anomalies among first-degree relatives of children with thyroid dysgenesis and congenital hypothyroidism. J. Pediatr. Endocrinol. Metab. 2012, 25, 413–418. [Google Scholar] [CrossRef]
- Sindhuja, L.; Dayal, D.; Sodhi, K.S.; Sachdeva, N.; Bhattacharya, A. Thyroid dysfunction and developmental anomalies in first degree relatives of children with thyroid dysgenesis. World J. Pediatr. 2016, 12, 215–218. [Google Scholar] [CrossRef]
- Sun, F.; Zhang, J.-X.; Yang, C.-Y.; Gao, G.-Q.; Zhu, W.-B.; Han, B.; Zhang, L.-L.; Wan, Y.-Y.; Ye, X.-P.; Ma, Y.-R.; et al. The genetic characteristics of congenital hypothyroidism in China by comprehensive screening of 21 candidate genes. Eur. J. Endocrinol. 2018, 178, 623–633. [Google Scholar] [CrossRef] [PubMed]
- Carré, A.; Castanet, M.; Sura-Trueba, S.; Szinnai, G.; Van Vliet, G.; Trochet, D.; Amiel, J.; Léger, J.; Czernichow, P.; Scotet, V.; et al. Polymorphic length of FOXE1 alanine stretch: Evidence for genetic susceptibility to thyroid dysgenesis. Hum. Genet. 2007, 122, 467–476. [Google Scholar] [CrossRef]
- Kumorowicz-Czoch, M.; Madetko-Talowska, A.; Tylek-Lemanska, D.; Pietrzyk, J.J.; Starzyk, J. Identification of deletions in children with congenital hypothyroidism and thyroid dysgenesis with the use of multiplex ligation-dependent probe amplification. J. Pediatr. Endocrinol. Metab. 2015, 28, 171–176. [Google Scholar] [CrossRef] [PubMed]
- Narumi, S.; Muroya, K.; Asakura, Y.; Adachi, M.; Hasegawa, T. Transcription Factor Mutations and Congenital Hypothyroidism: Systematic Genetic Screening of a Population-Based Cohort of Japanese Patients. J. Clin. Endocrinol. Metab. 2010, 95, 1981–1985. [Google Scholar] [CrossRef]
- Vela-Amieva, M.; Gamboa-Cardiel, S.; Pérez-Andrade, E.M.; Ortiz-Cortés, J.; González-Contreras, C.R.; Ortega-Velázquez, V. Epidemiología del hipotiroidismo congénito en Mexico. Salud Pública Mexico 2004, 46, 141–148. [Google Scholar] [CrossRef][Green Version]
- Alcántara-Ortigoza, M.A.; Angel, A.G.-D.; Martínez-Cruz, V.; Vela-Amieva, M.; Sánchez-Pérez, C.; Moreno-Rojas, R.; Estandía-Ortega, B.; Hernández-Martínez, N. Molecular analysis of the PAX8 gene in a sample of Mexican patients with primary congenital hypothyroidism: Identification of the recurrent p.Arg31His mutation. Clin. Endocrinol. 2012, 76, 148–150. [Google Scholar] [CrossRef]
- González-del Angel, A.; Fernández-Hernández, L.; Sánchez-Verdiguel, I.; González-Núñez, A.; Martínez-Cruz, V.; Sánchez, C.; Moreno-Rojas, R.; Alcántara-Ortigoza, M.A. Gene Variants in NKX2-1 Do Not Represent a Major Etiological Factor of Primary Congenital Hypothyroidism in Mexican Population. J. Pediatr. Genet. 2019, 8, 41–46. [Google Scholar] [CrossRef]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015, 17, 405–423. [Google Scholar] [CrossRef] [PubMed]
- Kleinberger, J.; Maloney, K.; Pollin, T.I.; Jeng, L.J.B. An openly available online tool for implementing the ACMG/AMP standards and guidelines for the interpretation of sequence variants. Genet. Med. 2016, 18, 1165. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Yan, R.; Roy, A.; Xu, D.; Poisson, J.; Zhang, Y. The I-TASSER Suite: Protein structure and function prediction. Nat. Methods 2015, 12, 7–8. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Zhang, Y. LOMETS: A local meta-threading-server for protein structure prediction. Nucleic Acids Res. 2007, 35, 3375–3382. [Google Scholar] [CrossRef]
- Tsai, K.-L.; Huang, C.-Y.; Chang, C.-H.; Sun, Y.-J.; Hsiao, C.-D. Crystal Structure of the Human FOXK1a-DNA Complex and Its Implications on the Diverse Binding Specificity of Winged Helix/Forkhead Proteins. J. Biol. Chem. 2006, 281, 17400–17409. [Google Scholar] [CrossRef] [PubMed]
- Sanders, P.; Young, S.; Sanders, J.; Kabelis, K.; Baker, S.; Sullivan, A.; Evans, M.; Clark, J.; Wilmot, J.; Hu, X.; et al. Crystal structure of the TSH receptor bound to a blocking type TSHR autoantibody. J. Mol. Endocrinol. 2011, 46, 81–99. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Williams, C.J.; Headd, J.J.; Moriarty, N.W.; Prisant, M.G.; Videau, L.L.; Deis, L.N.; Verma, V.; Keedy, D.A.; Hintze, B.; Chen, V.B.; et al. MolProbity: More and better reference data for improved all-atom structure validation. Protein Sci. 2018, 27, 293–315. [Google Scholar] [CrossRef]
- Jo, S.; Kim, T.; Iyer, V.G.; Im, W. CHARMM-GUI: A web-based graphical user interface for CHARMM. J. Comput. Chem. 2008, 29, 1859–1865. [Google Scholar] [CrossRef]
- Omasits, U.; Ahrens, C.; Müller, S.; Wollscheid, B. Protter: Interactive protein feature visualization and integration with experimental proteomic data. Bioinformatics 2014, 30, 884–886. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Cerqueira, T.L.D.O.; Ramos, Y.R.; Strappa, G.B.; De Jesus, M.S.; Santos, J.G.; Sousa, C.; Carvalho, G.; Fernandes, V.; Boa-Sorte, N.; Amorim, T.; et al. Mutation screening in the genes PAX-8, NKX2-5, TSH-R, HES-1 in cohort of 63 Brazilian children with thyroid dysgenesis. Arch. Endocrinol. Metab. 2018, 62, 466–471. [Google Scholar] [CrossRef]
- Brust, E.S.; Beltrao, C.B.; Chammas, M.C.; Watanabe, T.; Sapienza, M.T.; Marui, S. Absence of mutations in PAX8, NKX2.5, and TSH receptor genes in patients with thyroid dysgenesis. Arq. Bras. Endocrinol. Metabol. 2012, 56, 173–177. [Google Scholar] [CrossRef] [PubMed]
- Léger, J.; Olivieri, A.; Donaldson, M.; Torresani, T.; Krude, H.; Van Vliet, G.; Polak, M.; Butler, G. European Society for Paediatric Endocrinology Consensus Guidelines on Screening, Diagnosis, and Management of Congenital Hypothyroidism. Horm. Res. Paediatr. 2014, 81, 80–103. [Google Scholar] [CrossRef]
- Santos-Silva, R.; Rosário, M.; Grangeia, A.; Costa, C.; Castro-Correia, C.; Alonso, I.; Leão, M.; Fontoura, M. Genetic analyses in a cohort of Portuguese pediatric patients with congenital hypothyroidism. J. Pediatr. Endocrinol. Metab. 2019, 32, 1265–1273. [Google Scholar] [CrossRef] [PubMed]
- Löf, C.; Patyra, K.; Kuulasmaa, T.; Vangipurapu, J.; Undeutsch, H.; Jaeschke, H.; Pajunen, T.; Kero, A.; Krude, H.; Biebermann, H.; et al. Detection of Novel Gene Variants Associated with Congenital Hypothyroidism in a Finnish Patient Cohort. Thyroid 2016, 26, 1215–1224. [Google Scholar] [CrossRef] [PubMed]
- Long, W.; Zhou, L.; Wang, Y.; Liu, J.; Wang, H.; Yu, B. Complicated Relationship between Genetic Mutations and Phenotypic Characteristics in Transient and Permanent Congenital Hypothyroidism: Analysis of Pooled Literature Data. Int. J. Endocrinol. 2020, 2020, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Castanet, M.; Polak, M. Spectrum of Human Foxe1/TTF2 Mutations. Horm. Res. Paediatr. 2010, 73, 423–429. [Google Scholar] [CrossRef]
- Dentice, M.; Cordeddu, V.; Rosica, A.; Ferrara, A.M.; Santarpia, L.; Salvatore, L.C.D.; Chiovato, L.; Perri, A.; Moschini, L.; Fazzini, C.; et al. Missense Mutation in the Transcription Factor NKX2–5: A Novel Molecular Event in the Pathogenesis of Thyroid Dysgenesis. J. Clin. Endocrinol. Metab. 2006, 91, 1428–1433. [Google Scholar] [CrossRef]
- Al Taji, E.; Biebermann, H.; Límanová, Z.; Hníková, O.; Zikmund, J.; Dame, C.; Grüters, A.; Lebl, J.; Krude, H. Screening for mutations in transcription factors in a Czech cohort of 170 patients with congenital and early-onset hypothyroidism: Identification of a novel PAX8 mutation in dominantly inherited early-onset non-autoimmune hypothyroidism. Eur. J. Endocrinol. 2007, 156, 521–529. [Google Scholar] [CrossRef]
- Van Engelen, K.; Mommersteeg, M.T.M.; Baars, M.J.H.; Lam, J.; Ilgun, A.; Van Trotsenburg, A.S.P.; Smets, A.M.J.B.; Christoffels, V.M.; Mulder, B.J.M.; Postma, A.V. The Ambiguous Role of NKX2-5 Mutations in Thyroid Dysgenesis. PLoS ONE 2012, 7, e52685. [Google Scholar] [CrossRef]
- Khatami, M.; Heidari, M.M.; Tabesh, F.; Ordooei, M.; Salehifar, Z. Mutation analysis of the NKX2.5 gene in Iranian pediatric patients with congenital hypothyroidism. J. Pediatr. Endocrinol. Metab. 2017, 30, 857–862. [Google Scholar] [CrossRef] [PubMed]
- Hermanns, P.; Grasberger, H.; Refetoff, S.; Pohlenz, J. Mutations in the NKX2.5 Gene and the PAX8 Promoter in a Girl with Thyroid Dysgenesis. J. Clin. Endocrinol. Metab. 2011, 96, E977–E981. [Google Scholar] [CrossRef] [PubMed]
- Persani, L.; Calebiro, D.; Cordella, D.; Weber, G.; Gelmini, G.; Libri, M.; De Filippis, T.; Bonomi, M. Genetics and phenomics of hypothyroidism due to TSH resistance. Mol. Cell. Endocrinol. 2010, 322, 72–82. [Google Scholar] [CrossRef] [PubMed]
- Szidonya, L.; Cserző, M.; Hunyady, L. Dimerization and oligomerization of G-protein-coupled receptors: Debated structures with established and emerging functions. J. Endocrinol. 2008, 196, 435–453. [Google Scholar] [CrossRef]
- Latif, R.; Michalek, K.; Morshed, S.A.; Davies, T.F. A Tyrosine Residue on the TSH Receptor Stabilizes Multimer Formation. PLoS ONE 2010, 5, e9449. [Google Scholar] [CrossRef]
- Smits, G.; Campillo, M.; Govaerts, C.; Janssens, V.; Richter, C.; Vassart, G.; Pardo, L.; Costagliola, S. Glycoprotein hormone receptors: Determinants in leucine-rich repeats responsible for ligand specificity. EMBO J. 2003, 22, 2692–2703. [Google Scholar] [CrossRef]
- Lábadi, Á.; Grassi, E.S.; Gellén, B.; Kleinau, G.; Biebermann, H.; Ruzsa, B.; Gelmini, G.; Rideg, O.; Miseta, A.; Kovács, G.L.; et al. Loss-of-Function Variants in a Hungarian Cohort Reveal Structural Insights on TSH Receptor Maturation and Signaling. J. Clin. Endocrinol. Metab. 2015, 100, E1039–E1045. [Google Scholar] [CrossRef][Green Version]
- Ruchała, M.; Szczepanek, E.; Sowiński, J. Diagnostic value of radionuclide scanning and ultrasonography in thyroid developmental anomaly imaging. Nucl. Med. Rev. Cent. East Eur. 2011, 14, 21–28. [Google Scholar] [CrossRef]
- Camilot, M.; Teofoli, F.; Gandini, A.; Franceschi, R.; Rapa, A.; Corrias, A.; Bona, G.; Radetti, G.; Tatò, L. Thyrotropin receptor gene mutations and TSH resistance: Variable expressivity in the heterozygotes. Clin. Endocrinol. 2005, 63, 146–151. [Google Scholar] [CrossRef]
- Zdraveska, N.; Kocova, M.; Nicholas, A.K.; Anastasovska, V.; Schoenmakers, N. Genetics of Gland-in-situ or Hypoplastic Congenital Hypothyroidism in Macedonia. Front. Endocrinol. 2020, 11, 413. [Google Scholar] [CrossRef]
- Nicholas, A.K.; Serra, E.G.; Cangul, H.; Alyaarubi, S.; Ullah, I.; Schoenmakers, E.; Deeb, A.; Habeb, A.M.; Almaghamsi, M.; Peters, C.; et al. Comprehensive Screening of Eight Known Causative Genes in Congenital Hypothyroidism with Gland-in-Situ. J. Clin. Endocrinol. Metab. 2016, 101, 4521–4531. [Google Scholar] [CrossRef] [PubMed]
- Santarpia, L.; Valenzise, M.; Di Pasquale, G.; Arrigo, T.; Martino, G.S.; Cicciò, M.P.; Trimarchi, F.; De Luca, F.; Benvenga, S. TTF-2/FOXE1 gene polymorphisms in Sicilian patients with permanent primary congenital hypothyroidism. J. Endocrinol. Investig. 2007, 30, 13–19. [Google Scholar] [CrossRef] [PubMed]
- Szczepanek, E.; Ruchala, M.; Szaflarski, W.; Budny, B.; Kilinska, L.; Jaroniec, M.; Niedziela, M.; Zabel, M.; Sowiński, J. FOXE1 Polyalanine Tract Length Polymorphism in Patients with Thyroid Hemiagenesis and Subjects with Normal Thyroid. Horm. Res. Paediatr. 2011, 75, 329–334. [Google Scholar] [CrossRef] [PubMed]
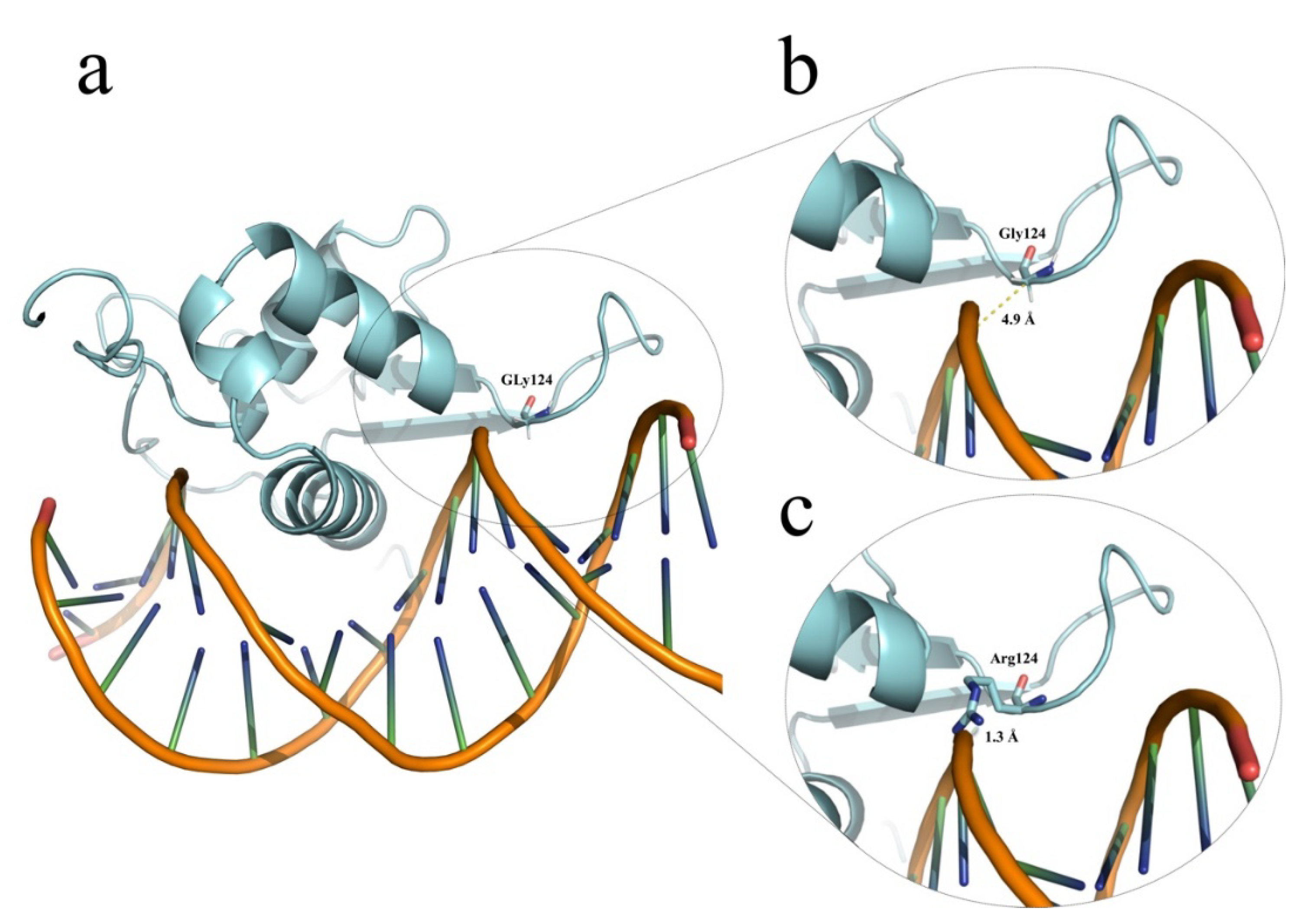
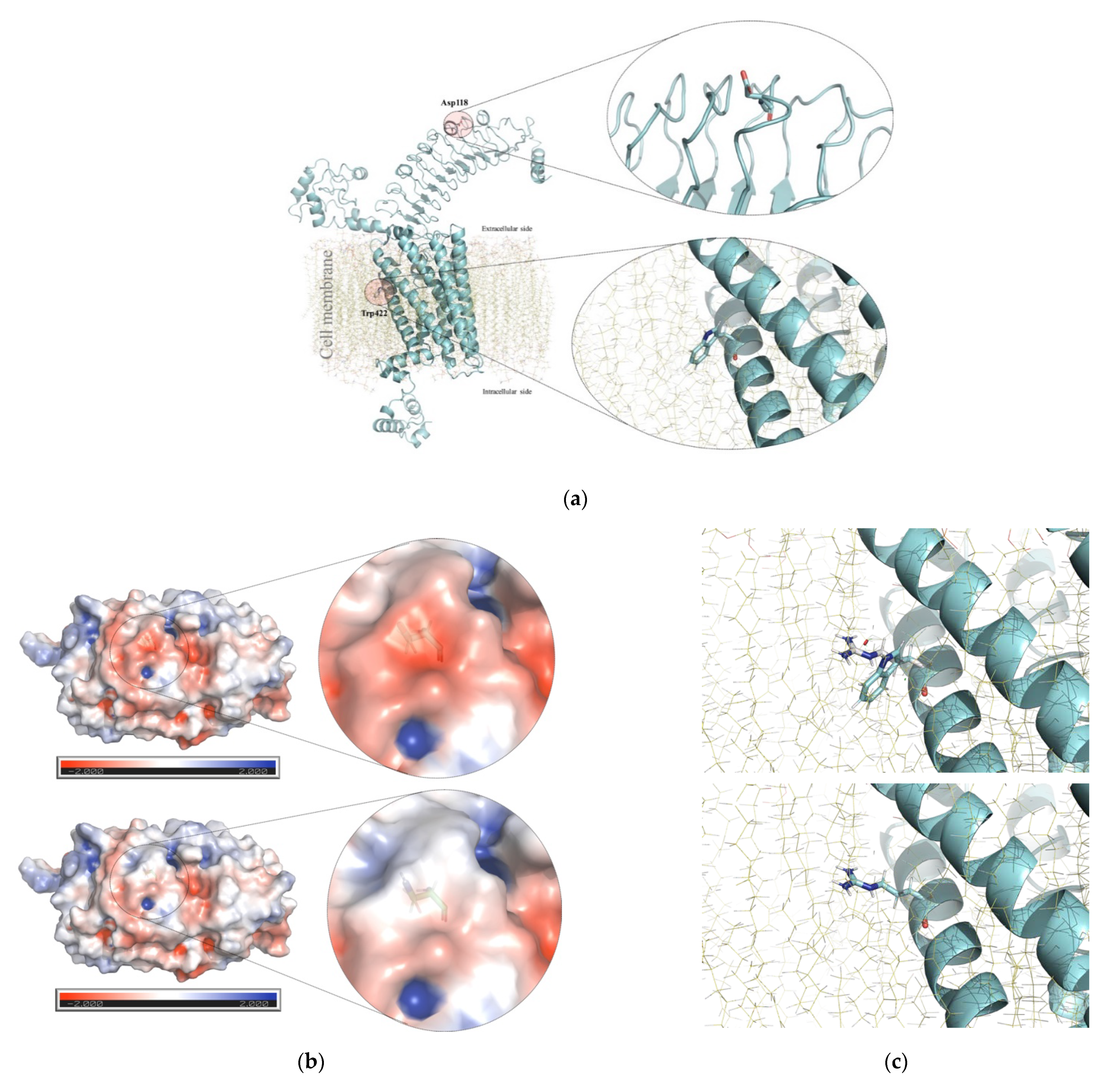
| Patient ID (Gender) | Genotype | ACMG/AMP 1 Criteria and Variant Classification GnomAD Allele Frequencies 2 | Thyroid and Clinical Phenotype | Relevant Familial History |
|---|---|---|---|---|
| HC-266 (female) | Homozygous NM_004473.3 (FOXE1):c.370G>C or p.(Gly124Arg) [rs774035532] at forkhead domain. Homozygous for 16 alanine polyAla tract. | PM1, PM2, PP2, PP3. Likely pathogenic (V). 0.000016 | Thyroid hypoplasia (scintigraphy) without other birth defects or clinical data suggestive of Bamforth-Lazarus syndrome (MIM#241850). | No familial history of CH. Healthy apparently non-consanguineous parents with normal thyroid profiles. Heterozygous for p.(Gly124Arg) and 14/16 alanine polyAla FOXE1 genotypes. |
| HC-215 (female) | Heterozygous NM_004473.3 (FOXE1):c.1004C>G or p.(Ala335Gly) [rs543372757]. Heterozygous for 14/16 alanine tract. | PM2, PP2. Variant of unknown significance. 0.0000076 | Thyroid ectopy (scintigraphy) without other birth defects. | No familial history of CH. Healthy non-consanguineous parents with normal thyroid profile. Mother heterozygous for c.1004C>G or p.(Ala335Gly). |
| HC-321 (male) | Heterozygous NM_004387.3 (NKX2-5):c.355G>T or p.(Ala119Ser) [rs137852684]. | PM2, PP2, BP6, BS2, BS4. Benign (II). 0.00097 | Thyroid ectopy (scintigraphy) without other birth defect. | No familial history of CH or congenital heart disease. Healthy non-consanguineous parents with normal thyroid profile. Father heterozygous for c.355G>T or p.(Ala119Ser) without any clinical data suggestive of heart disease. |
| HC-324 (female) | Compound heterozygous NM_000369.2(TSHR) c.[352G>A];[1264T>C] or p.[Asp118Asn];[Trp422Arg] (rs1414102266 and rs746029360). | p.(Asp118Asn): PM2, PP2, BP4. Variant of unknown significance. 0.0000039 p.(Trp422Arg): PM1, PM2, PP2, PP3. Likely pathogenic (V). 0.000015 | Thyroid agenesis (ultrasonography); serum thyroglobulin levels not available. | No familial history of CH. Healthy non-consanguineous parents not available for thyroid profile evaluation or TSHR genotyping. |
| PolyAla Allele | CH-TD Alleles (N = 256) | Allelic Frequencies | Healthy Control Alleles (N = 232) | Allelic Frequencies | OR (95% CI) | p-Value |
|---|---|---|---|---|---|---|
| <14 alanines | 8 | 0.03125 | 2 | 0.0172 | 3.67 (0.77–17.5) | 0.110 |
| 14 alanines (reference) | 222 | 0.8671 | 204 | 0.8793 | ||
| >14 alanines | 26 | 0.1015 | 26 | 0.1120 | 0.91 (0.51–1.63) | 0.883 |
| PolyAla genotype | CH-TD patients (N = 128) | Frequency (%) | Healthy controls (N = 116) | Frequency | OR (95% CI) | p-Value |
| ≤14/≤14 alanines | 7 | 5.5 | 2 | 1.7 | 3.31 (0.67–16.39) | 0.174 |
| 14/14 alanines (reference) | 96 | 75 | 91 | 78.5 | ||
| ≥14/≥14 alanines | 25 | 19.5 | 23 | 19.8 | 0.91 (0.54–1.94) | 1.0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alcántara-Ortigoza, M.A.; Sánchez-Verdiguel, I.; Fernández-Hernández, L.; Enríquez-Flores, S.; González-Núñez, A.; Hernández-Martínez, N.L.; Sánchez, C.; González-del Angel, A. Further Evidence That Defects in Main Thyroid Dysgenesis-Related Genes Are an Uncommon Etiology for Primary Congenital Hypothyroidism in Mexican Patients: Report of Rare Variants in FOXE1, NKX2-5 and TSHR. Children 2021, 8, 457. https://doi.org/10.3390/children8060457
Alcántara-Ortigoza MA, Sánchez-Verdiguel I, Fernández-Hernández L, Enríquez-Flores S, González-Núñez A, Hernández-Martínez NL, Sánchez C, González-del Angel A. Further Evidence That Defects in Main Thyroid Dysgenesis-Related Genes Are an Uncommon Etiology for Primary Congenital Hypothyroidism in Mexican Patients: Report of Rare Variants in FOXE1, NKX2-5 and TSHR. Children. 2021; 8(6):457. https://doi.org/10.3390/children8060457
Chicago/Turabian StyleAlcántara-Ortigoza, Miguel Angel, Iraís Sánchez-Verdiguel, Liliana Fernández-Hernández, Sergio Enríquez-Flores, Aidy González-Núñez, Nancy Leticia Hernández-Martínez, Carmen Sánchez, and Ariadna González-del Angel. 2021. "Further Evidence That Defects in Main Thyroid Dysgenesis-Related Genes Are an Uncommon Etiology for Primary Congenital Hypothyroidism in Mexican Patients: Report of Rare Variants in FOXE1, NKX2-5 and TSHR" Children 8, no. 6: 457. https://doi.org/10.3390/children8060457
APA StyleAlcántara-Ortigoza, M. A., Sánchez-Verdiguel, I., Fernández-Hernández, L., Enríquez-Flores, S., González-Núñez, A., Hernández-Martínez, N. L., Sánchez, C., & González-del Angel, A. (2021). Further Evidence That Defects in Main Thyroid Dysgenesis-Related Genes Are an Uncommon Etiology for Primary Congenital Hypothyroidism in Mexican Patients: Report of Rare Variants in FOXE1, NKX2-5 and TSHR. Children, 8(6), 457. https://doi.org/10.3390/children8060457






