Wolf-Hirschhorn Syndrome: Clinical and Genetic Study of 7 New Cases, and Mini Review
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethical Compliance
2.2. Patient Recruitment
2.3. Research Methodology
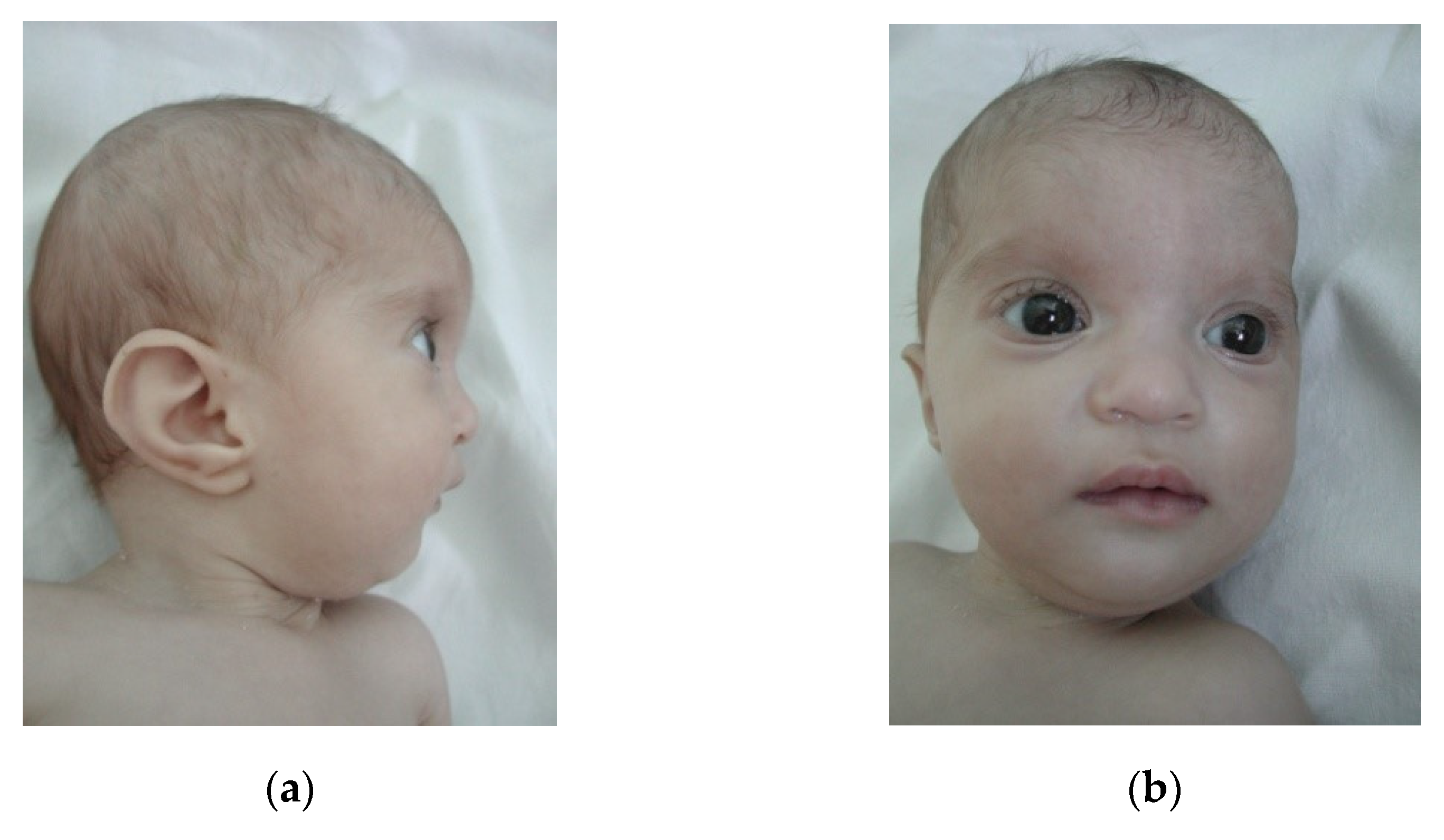
3. Results
3.1. Small Deletions (<3 Mb)
3.2. Large Deletions (>3 Mb)
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Paradowska-Stolarz, A.M. Wolf-Hirschhorn Syndrome (WHS)—Literature review on the features of the syndrome. Adv. Clin. Exp. Med. 2014, 23, 485–489. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bailey, R. Wolf-hirschhorn syndrome: A case study and disease overview. Adv. Neonatal Care 2014, 14, 318–321. [Google Scholar] [CrossRef] [PubMed]
- Battaglia, A.; Carey, J.C.; South, S.T. Wolf-Hirschhorn syndrome: A review and update. Am. J. Med. Genet. Part. C Semin. Med. Genet. 2015, 169, 216–223. [Google Scholar] [CrossRef] [PubMed]
- Nevado, J.; Ho, K.S.; Zollino, M.; Blanco, R.; Cobaleda, C.; Golzio, C.; Beaudry-Bellefeuille, I.; Berrocoso, S.; Limeres, J.; Barrúz, P.; et al. International meeting on Wolf-Hirschhorn syndrome: Update on the nosology and new insights on the pathogenic mechanisms for seizures and growth delay. Proc. Am. J. Med. Genet. Part A 2020, 182, 257–267. [Google Scholar] [CrossRef]
- Andersen, E.F.; Carey, J.C.; Earl, D.L.; Corzo, D.; Suttie, M.; Hammond, P.; South, S.T. Deletions involving genes WHSC1 and LETM1 may be necessary, but are not sufficient to cause Wolf-Hirschhorn Syndrome. Eur. J. Hum. Genet. 2014, 22, 464–470. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Sun, H.; Lin, Q.; Wang, Z.; Wang, G.; Wang, J.; Jiang, F.; Yao, R. De novo truncating variant in NSD2gene leading to atypical Wolf-Hirschhorn syndrome phenotype. BMC Med. Genet. 2019, 20, 134. [Google Scholar] [CrossRef] [Green Version]
- Sukarova-Angelovska, E.; Kocova, M.; Sabolich, V.; Palcevska, S.; Angelkova, N. Phenotypic variations in Wolf-Hirschhorn syndrome. Balk. J. Med. Genet. 2014, 17, 23–30. [Google Scholar] [CrossRef] [Green Version]
- Schouten, J.P.; McElgunn, C.J.; Waaijer, R.; Zwijnenburg, D.; Diepvens, F.; Pals, G. Relative quantification of 40 nucleic acid sequences by multiplex ligation-dependent probe amplification. Nucleic Acids Res. 2002, 30, e57. [Google Scholar] [CrossRef] [Green Version]
- Stuppia, L.; Antonucci, I.; Palka, G.; Gatta, V. Use of the MLPA assay in the molecular diagnosis of gene copy number alterations in human genetic diseases. Int. J. Mol. Sci. 2012, 13, 3245–3276. [Google Scholar] [CrossRef]
- Zollino, M.; Murdolo, M.; Marangi, G.; Pecile, V.; Galasso, C.; Mazzanti, L.; Neri, G. On the nosology and pathogenesis of Wolf-Hirschhorn syndrome: Genotype-phenotype correlation analysis of 80 patients and literature review. Am. J. Med. Genet. Part. C Semin. Med. Genet. 2008, 148C, 257–269. [Google Scholar] [CrossRef]
- Sheth, F.; Akinde, O.R.; Datar, C.; Adeteye, O.V.; Sheth, J. Genotype-Phenotype Characterization of Wolf-Hirschhorn Syndrome Confirmed by FISH: Case Reports. Case Rep. Genet. 2012, 2012, 1–5. [Google Scholar] [CrossRef]
- Bi, W.; Cheung, S.-W.; Breman, A.M.; Bacino, C.A. 4p16.3 microdeletions and microduplications detected by chromosomal microarray analysis: New insights into mechanisms and critical regions. Am. J. Med. Genet. Part A 2016, 170, 2540–2550. [Google Scholar] [CrossRef]
- Catela, C.; Bilbao-Cortes, D.; Slonimsky, E.; Kratsios, P.; Rosenthal, N.; Te Welscher, P. Multiple congenital malformations of Wolf-Hirschhorn syndrome are recapitulated in Fgfrl1 null mice. DMM Dis. Model. Mech. 2009, 2, 283–294. [Google Scholar] [CrossRef] [Green Version]
- South, S.T.; Whitby, H.; Battaglia, A.; Carey, J.C.; Brothman, A.R. Comprehensive analysis of Wolf-Hirschhorn syndrome using array CGH indicates a high prevalence of translocations. Eur. J. Hum. Genet. 2008, 16, 45–52. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Venegas-Vega, C.A.; Fernández-Ramírez, F.; Zepeda, L.M.; Nieto-Martínez, K.; Gómez-Laguna, L.; Garduño-Zarazúa, L.M.; Berumen, J.; Kofman, S.; Cervantes, A. Diagnosis of familial wolf-hirschhorn syndrome due to a paternal cryptic chromosomal rearrangement by conventional and molecular cytogenetic techniques. BioMed Res. Int. 2013, 2013, 209204. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Oh, P.S.; Na, H.Y.; Kim, S.H.; Cho, H.C. A case of mosaic ring chromosome 4 with subtelomeric 4p deletion. Korean J. Lab. Med. 2009, 29, 77–81. [Google Scholar] [CrossRef] [Green Version]
- Battaglia, A.; Filippi, T.; Carey, J.C. Update on the clinical features and natural history of Wolf-Hirschhorn (4p-) syndrome: Experience with 87 patients and recommendations for routine health supervision. Am. J. Med. Genet. Part C Semin. Med. Genet. 2008, 148C, 246–251. [Google Scholar] [CrossRef] [PubMed]
- Sifakis, S.; Manolakos, E.; Vetro, A.; Kappou, D.; Peitsidis, P.; Kontodiou, M.; Garas, A.; Vrachnis, N.; Konstandinidou, A.; Zuffardi, O.; et al. Prenatal diagnosis of Wolf-Hirschhorn syndrome confirmed by comparative genomic hybridization array: Report of two cases and review of the literature. Mol. Cytogenet. 2012, 5, 12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wright, T.J.; Ricke, D.O.; Denison, K.; Abmayr, S.; Cotter, P.D.; Hirschhorn, K.; Keinänen, M.; McDonald-McGinn, D.; Somer, M.; Spinner, N.; et al. A transcript map of the newly defined 165 kb Wolf-Hirschhorn syndrome critical region. Hum. Mol. Genet. 1997, 6, 317–324. [Google Scholar] [CrossRef] [PubMed]
- Boczek, N.J.; Lahner, C.A.; Nguyen, T.; Ferber, M.J.; Hasadsri, L.; Thorland, E.C.; Niu, Z.; Gavrilova, R.H. Developmental delay and failure to thrive associated with a loss-of-function variant in WHSC1 (NSD2). Am. J. Med. Genet. Part A 2018, 176, 2798–2802. [Google Scholar] [CrossRef]
- Zollino, M.; Stefano, C.D.; Zampino, G.; Mastroiacovo, P.; Wright, T.J.; Sorge, G.; Selicorni, A.; Tenconi, R.; Zappal, A.; Battaglia, A.; et al. Genotype-phenotype correlations and clinical diagnostic criteria in Wolf-Hirschhorn syndrome. Am. J. Med. Genet. 2000, 94, 254–261. [Google Scholar] [CrossRef]
- Zollino, M.; Lecce, R.; Fischetto, R.; Murdolo, M.; Faravelli, F.; Selicorni, A.; Buttè, C.; Memo, L.; Capovilla, G.; Neri, G. Mapping the Wolf-Hirschhorn syndrome phenotype outside the currently accepted WHS critical region and defining a new critical region, WHSCR-2. Am. J. Hum. Genet. 2003, 72, 590–597. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Limeres, J.; Serrano, C.; De Nova, J.M.; Silvestre-Rangil, J.; Machuca, G.; Maura, I.; Villandiego, J.C.R.; Diz, P.; Lago, R.B.; Nevado, J.; et al. Oral Manifestations of Wolf-Hirschhorn Syndrome: Genotype-Phenotype Correlation Analysis. J. Clin. Med. 2020, 9, 3556. [Google Scholar] [CrossRef] [PubMed]




| Name | Case 1 | Case 2 | Case 3 | Case 4 | Case 5 | Case 6 | Case 7 |
|---|---|---|---|---|---|---|---|
| Age (years) | 13 | 4 | 2 | 3 | 1 | 5 | 1 |
| Sex | M | F | M | M | M | F | F |
| Uneventful pregnancy | + | − | − | − | + | − | − |
| Weight at birth (g) | 3600 | 2500 | 1400 | 2600 | 2700 | 930 | 1600 |
| Weight last exam (SD) | −0.89 | −3.2 | −7.09 | −5.61 | −5.75 | −2.6 | −5.19 |
| Height at birth (cm) | 52 | 48 | 30 | 49 | - | 38 | 45 |
| Height last exam (SD) | −2.39 | −4.15 | −5.75 | −3.78 | - | −4.08 | −3.19 |
| OFC at birth (cm) | 36.5 | 31.5 | 28 | 33 | - | 36 | 29 |
| OFC last exam (SD | −1.23 | −4.66 | −4.81 | −2.75 | −3.91 | −7.41 | −2.81 |
| Prenatal growth delay | − | − | + | + | − | + | − |
| Postnatal development delay | + | + | + | + | + | + | + |
| Born at term | + | + | + | + | + | − | + |
| IUGR | − | +/− | + | − | n.a | + | + |
| Karyotype | 46,XY | 46,XX | 46,XY,r(4)(p15.1q35)/46,XY,−4,+mar/47,XY,r(4)(p15.1q35),+mar/46,XY,der(4),+mar/46,XY | 46,XY | 46,XY,del(4)(p16.3),del(22)(q11.23) | 46,XX,del(4)(p16.1-pter) | 46,XX,del(4)(p15.2-pter) |
| Size of deletion | ~2 MB | ~2 MB | ~2.85 MB | ~2 MB | ~2 MB | ~8 MB | ~22 MB |
| Dysmorphic face | + | + | + | atypical | +/− | + | + |
| Microcephaly | − | + | + | +/− | +/− | +++ | +/− |
| Dental anomalies | + | + | − | − | − | + | − |
| Delayed tooth eruption | + | + | |||||
| Anodontia | + | ||||||
| Ear anomalies | + | + | + | + | − | + | + |
| Left preauricular pit | + | ||||||
| Hearing loss | − | − | − | − | − | + | + |
| Ocular defects | + | + | − | + | − | + | − |
| Defect of lacrimal system | + | ||||||
| Congenital cataract | + | + | |||||
| Tuberous hemangiomas | + | ||||||
| Bilateral optic atrophy | + | ||||||
| Iris coloboma | + | + | |||||
| Intellectual disability | Severe | Mild | Severe | Severe | n.a. | Severe | n.a. |
| Brain anomalies | + | − | + | + | − | − | − |
| Spastic quadriplegia | + | ||||||
| Hypoplastic corpus callosum | + | + | |||||
| Corpus callosum cyst | + | ||||||
| Seizures | − | + | − | + | + | − | − |
| Jacksonian seizures | + | ||||||
| Immunodeficiency | + | − | − | + | + | − | + |
| Cardiac defects | + | + | + | + | + | + | + |
| ASD | + | + | + | + | + | + | + |
| VSD | + | + | |||||
| PDA | + | ||||||
| Tricuspid insufficiency | + | ||||||
| Mitral insufficiency | + | ||||||
| Renal anomalies | − | − | − | + | + | − | − |
| Pulmonary defects | + | + | + | + | + | + | |
| Pulmonary insufficiency | + | ||||||
| Pulmonary stenosis | + | + | |||||
| Infections | + | + | + | + | |||
| Hypotonia | + | + | + | + | + | ||
| Diastasis recti, hypospadias, undescended testes | + | + | + |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gavril, E.-C.; Luca, A.C.; Curpan, A.-S.; Popescu, R.; Resmerita, I.; Panzaru, M.C.; Butnariu, L.I.; Gorduza, E.V.; Gramescu, M.; Rusu, C. Wolf-Hirschhorn Syndrome: Clinical and Genetic Study of 7 New Cases, and Mini Review. Children 2021, 8, 751. https://doi.org/10.3390/children8090751
Gavril E-C, Luca AC, Curpan A-S, Popescu R, Resmerita I, Panzaru MC, Butnariu LI, Gorduza EV, Gramescu M, Rusu C. Wolf-Hirschhorn Syndrome: Clinical and Genetic Study of 7 New Cases, and Mini Review. Children. 2021; 8(9):751. https://doi.org/10.3390/children8090751
Chicago/Turabian StyleGavril, Eva-Cristiana, Alina Costina Luca, Alexandrina-Stefania Curpan, Roxana Popescu, Irina Resmerita, Monica Cristina Panzaru, Lacramioara Ionela Butnariu, Eusebiu Vlad Gorduza, Mihaela Gramescu, and Cristina Rusu. 2021. "Wolf-Hirschhorn Syndrome: Clinical and Genetic Study of 7 New Cases, and Mini Review" Children 8, no. 9: 751. https://doi.org/10.3390/children8090751
APA StyleGavril, E.-C., Luca, A. C., Curpan, A.-S., Popescu, R., Resmerita, I., Panzaru, M. C., Butnariu, L. I., Gorduza, E. V., Gramescu, M., & Rusu, C. (2021). Wolf-Hirschhorn Syndrome: Clinical and Genetic Study of 7 New Cases, and Mini Review. Children, 8(9), 751. https://doi.org/10.3390/children8090751






