Healthcare Professionals’ Attitudes toward Rapid Whole Genome Sequencing in Pediatric Acute Care
Abstract
:1. Introduction
2. Materials and Methods
2.1. Project Start-up and Participants
2.2. Survey
2.3. Statistical Analysis
3. Results
3.1. Participant Characteristics
3.2. rWGS Education
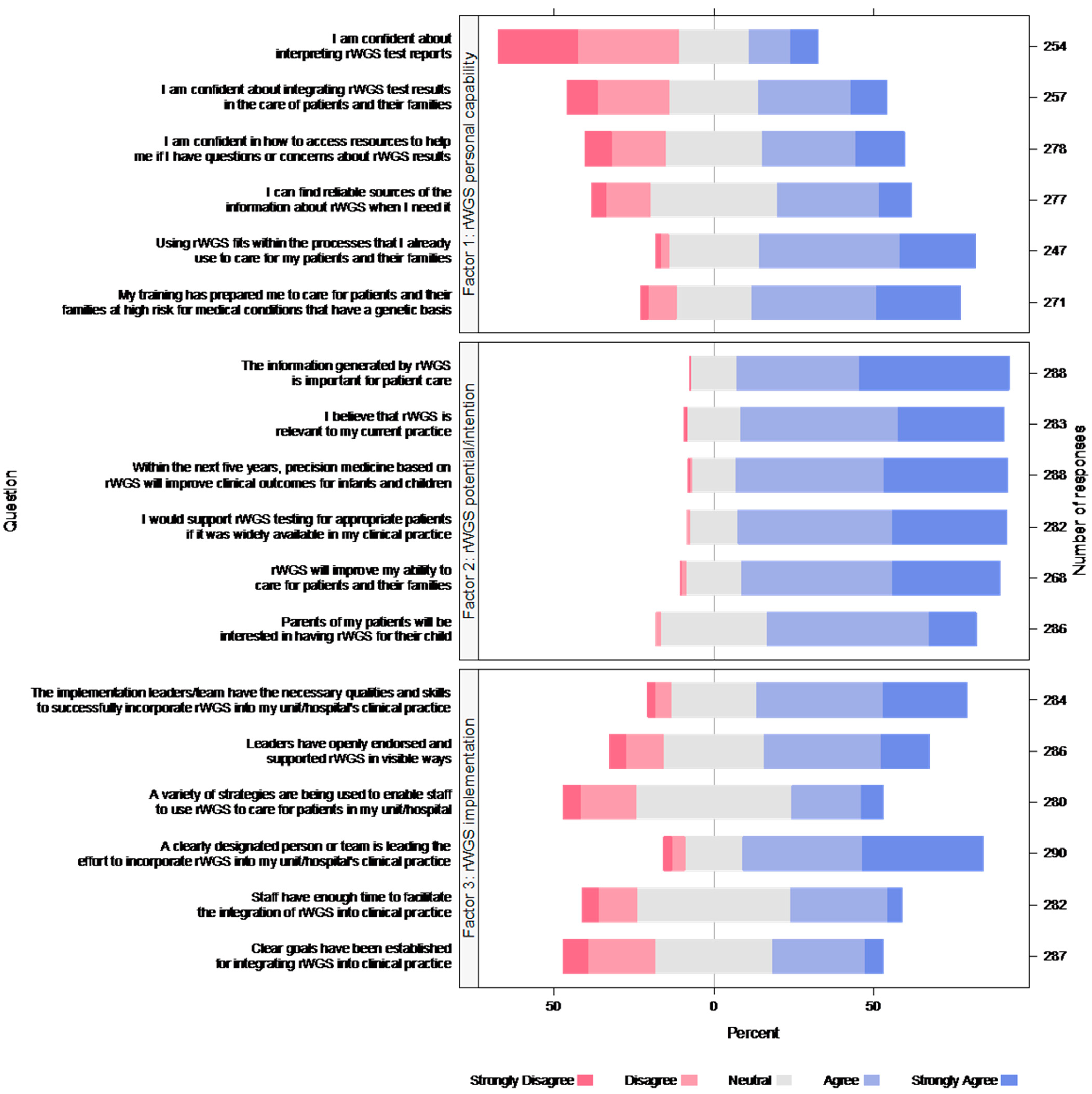
3.3. Factor Analysis—rWGS Attitudes Scale
3.4. Views about rWGS Implementation and Implications for Clinical Practice
3.5. Variables Associated with rWGS Factor and Total Attitudes Scores
3.6. Respondent Comments about rWGS Implementation and Wider Adoption
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dimmock, D.P.; Clark, M.M.; Gaughran, M.; Cakici, J.A.; Caylor, S.A.; Clarke, C.; Feddock, M.; Chowdhury, S.; Salz, L.; Cheung, C.; et al. An RCT of rapid genomic sequencing among seriously ill infants results in high clinical utility, changes in management, and low perceived harm. Am. J. Hum. Genet. 2020, 107, 942–952. [Google Scholar] [CrossRef] [PubMed]
- Kingsmore, S.F.; Henderson, A.; Owen, M.J.; Clark, M.M.; Hansen, C.; Dimmock, D.; Chambers, C.D.; Jeliffe-Pawlowski, L.L.; Hobbs, C. Measurement of genetic diseases as a cause of mortality in infants receiving whole genome sequencing. NPJ Genom. Med. 2020, 5, 49. [Google Scholar] [CrossRef] [PubMed]
- Kingsmore, S.F.; Ramchandar, N.; James, K.; Niemi, A.-K.; Feigenbaum, A.; Ding, Y.; Benson, W.; Hobbs, C.; Nahas, S.; Chowdhury, S.; et al. Mortality in a neonate with molybdenum cofactor deficiency illustrates the need for a comprehensive rapid precision medicine system. Cold Spring Harb. Mol. Case Stud. 2020, 6, a004705. [Google Scholar] [CrossRef] [PubMed]
- Meng, L.; Pammi, M.; Saronwala, A.; Magoulas, P.; Ghazi, A.R.; Vetrini, F.; Zhang, J.; He, W.; Dharmadhikari, A.V.; Qu, C.; et al. Use of exome sequencing for infants in intensive care units: Ascertainment of severe single-gene disorders and effect on medical management. JAMA Pediatr. 2017, 171, e173438. [Google Scholar] [CrossRef]
- Stark, Z.; Lunke, S.; Brett, G.R.; Tan, N.B.; Stapleton, R.; Kumble, S.; Melbourne Genomics Health Alliance. Meeting the challenges of implementing rapid genomic testing in acute pediatric care. Genet. Med. 2018, 20, 1554–1563. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mestek-Boukhibar, L.; Clement, E.; Jones, W.D.; Drury, S.; Ocaka, L.; Gagunashvili, A.; le Quesne Stabej, P.; Bacchelli, C.; Jani, N.; Rahman, S.; et al. Rapid Paediatric Sequencing (RaPS): Comprehensive real-life workflow for rapid diagnosis of critically ill children. J. Med. Genet. 2018, 55, 721–728. [Google Scholar] [CrossRef] [Green Version]
- Kingsmore, S.F.; Cakici, J.A.; Clark, M.M.; Gaughran, M.; Feddock, M.; Batalov, S.; Bainbridge, M.N.; Carroll, J.; Caylor, S.A.; Clarke, C.; et al. A Randomized, controlled trial of the analytic and diagnostic performance of singleton and trio, rapid genome and exome sequencing in ill infants. Am. J. Hum. Genet. 2019, 105, 719–733. [Google Scholar] [CrossRef]
- Ritter, A.; Bedoukian, E.; Berger, J.H.; Copenheaver, D.; Gray, C.; Krantz, I.; Izumi, K.; Juusola, J.; Leonard, J.; Lin, K.; et al. Clinical utility of exome sequencing in infantile heart failure. Genet. Med. 2020, 22, 423–426. [Google Scholar] [CrossRef]
- Lunke, S.; Eggers, S.; Wilson, M.; Patel, C.; Barnett, C.P.; Pinner, J.; Sandaradura, S.A.; Buckley, M.F.; Krzesinski, E.I.; De Silva, M.G.; et al. Feasibility of ultra-rapid exome sequencing in critically ill infants and children with suspected monogenic conditions in the Australian public health care system. JAMA 2020, 323, 2503–2511. [Google Scholar] [CrossRef]
- Sanford, E.F.; Clark, M.M.; Farnaes, L.; Williams, M.R.; Perry, J.C.; Ingulli, E.G.; Sweeney, M.N.; Doshi, A.; Gold, J.J.; Briggs, B.; et al. Rapid whole genome sequencing has clinical utility in children in the PICU. Pediatr. Crit. Care Med. 2019, 20, 1007–1020. [Google Scholar] [CrossRef]
- Sweeney, N.M.; Nahas, S.A.; Chowdhury, S.; Batalov, S.; Clark, M.; Caylor, S.; Cakici, J.; Nigro, J.J.; Ding, Y.; Veeraraghavan, M.; et al. The impact of rapid whole genome sequencing on medical management and resource utilization in the care of critically ill infants with congenital heart disease. Genom. Med. 2021, 6, 29. [Google Scholar] [CrossRef] [PubMed]
- Manickam, K.; McClain, M.R.; Demmer, L.A.; Biswas, S.; Kearney, H.M.; Malinowski, J.; Massingham, L.J.; Miller, D.; Yu, T.W.; Hisama, F.M.; et al. Exome and genome sequencing for pediatric patients with congenital anomalies or intellectual disability: An evidence-based clinical guideline of the American College of Medical Genetics and Genomics (ACMG). Genet. Med. 2021, 23, 2029–2037. [Google Scholar] [CrossRef] [PubMed]
- Beuschel, J.; Geyer, H.; Rich, M.; Leimanis, M.; Kampfschulte, A.; VanSickle, E.; Rajasekaran, S.; Bupp, C. Leveraging rapid genome sequencing to alter care plans for pediatric patients in a community hospital setting in the United States. J. Pediatr. 2021, 239, 235–239. [Google Scholar] [CrossRef]
- Swaggart, K.A.; Swarr, D.T.; Tolusso, L.K.; He, H.; Dawson, D.B.; Suhrie, K.R. Making a genetic diagnosis in a level IV neonatal intensive care unit population: Who, when, how, and at what cost? J. Pediatr. 2019, 213, 211–217.e4. [Google Scholar] [CrossRef]
- Krantz, I.D.; Medne, L.; Weatherly, J.M.; Wild, K.T.; Biswas, S.; Devkota, B.; Hartman, T.; Brunelli, L.; Fishler, K.P.; Abdul-Rahman, O.; et al. Effect of whole-genome sequencing on the clinical management of acutely ill infants with suspected genetic disease: A randomized clinical trial. JAMA Pediatr. 2021, 175, e213496. [Google Scholar]
- Greenhalgh, T.; Wherton, J.; Papoutsi, C.; Lynch, J.; Hughes, G.; A’Court, C.; Hinder, S.; Fahy, N.; Procter, R.; Shaw, S. Beyond adoption: A new framework for theorizing and evaluating nonadoption, abandonment, and challenges to the scale-up, spread, and sustainability of health and care technologies. J. Med. Internet Res. 2017, 19, e367. [Google Scholar] [CrossRef] [Green Version]
- Franck, L.S.; Kriz, R.M.; Rego, S.; Garman, K.; Hobbs, C.; Dimmock, D. Implementing rapid whole-genome sequencing in critical care: A qualitative study of facilitators and barriers to new technology adoption. J. Pediatr. 2021, 237, 237–243.e2. [Google Scholar] [CrossRef]
- Project Baby Deer: Precision Medicine in Michigan Saves Lives. Available online: https://radygenomics.org/wp-content/uploads/2022/01/Project-Baby-Deer_Pilot_Flyer_June-9-2021.pdf (accessed on 23 February 2022).
- NIH Collaboratory; IGNITE Toolbox. Provider Demographics and Practice Characteristics. Available online: https://dcricollab.dcri.duke.edu/sites/NIHKR/IGNITE%20Documents%20and%20Links%20to%20Content/Researchers/Data%20Collection/Provider%20Demographics%20and%20Practice%20Characteristics%20Survey.pdf (accessed on 23 February 2022).
- NIH Collaboratory; IGNITE Toolbox. Provider Baseline Genetics Knowledge Survey, CFIR-Based Provider Items. Available online: https://dcricollab.dcri.duke.edu/sites/NIHKR/IGNITE%20Documents%20and%20Links%20to%20Content/Researchers/Data%20Collection/Provider%20Baseline%20Knowledge%20of%20Genetic%20Testing%20Survey.pdf (accessed on 23 February 2022).
- NIH Collaboratory; IGNITE Toolbox. Provider Knowledge after Genetics Testing Implementation, APOL1 Provider Follow-Up Survey Questions. Available online: https://dcricollab.dcri.duke.edu/sites/NIHKR/IGNITE%20Documents%20and%20Links%20to%20Content/Researchers/Data%20Collection/Provider%20Knowledge%20of%20Genetic%20Testing%20after%20Implementation.pdf (accessed on 23 February 2022).
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021; Available online: https://www.R-project.org/ (accessed on 23 February 2022).
- Heiberger, R.M. HH: Statistical Analysis and Data Display: In Heiberger RM and Holland. R package Version 3.1-43. 2020. Available online: https://CRAN.R-project.org/package=HH (accessed on 23 February 2022).
- Van Buuren, S.; Groothuis-Oudshoorn, K. Mice: Multivariate imputation by chained equations in R. J. Stat. Softw. 2011, 45, 1–67. [Google Scholar] [CrossRef] [Green Version]
- Fox, J. Polycor: Polychoric and Polyserial Correlations. R Package Version 0.7-10. 2019. Available online: https://CRAN.R-project.org/package=polycor (accessed on 23 February 2022).
- Nassiri, V.; Lovik, A.; Molenberghs, G.; Verbeke, G. On using multiple imputation for exploratory factor analysis of incomplete data. Behav Res. Methods 2018, 50, 501–517. [Google Scholar] [CrossRef]
- Hsieh, H.F.; Shannon, S.E. Three approaches to qualitative content analysis. Qual. Health Res. 2005, 15, 1277–1288. [Google Scholar] [CrossRef]
- Lemke, A.A.; Bick, D.; Dimmock, D.; Simpson, P.; Veith, R. Perspectives of clinical genetics professionals toward genome sequencing and incidental findings: A survey study. Clin. Genet. 2013, 84, 230–236. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pereira, S.; Robinson, J.O.; Gutierrez, A.M.; Petersen, D.K.; Hsu, R.L.; Lee, C.H.; Schwartz, T.S.; Holm, I.A.; Beggs, A.H.; Green, R.C.; et al. Perceived benefits, risks, and utility of newborn genomic sequencing in the BabySeq project. Pediatrics 2019, 143 (Suppl. S1), S6–S13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deuitch, N.; Soo-Jin Lee, S.; Char, D. Translating genomic testing results for pediatric critical care: Opportunities for genetic counselors. J. Genet. Couns. 2020, 29, 78–87. [Google Scholar] [CrossRef] [PubMed]
- Knapp, B.; Decker, C.; Lantos, J.D. Neonatologists’ attitudes about diagnostic whole-genome sequencing in the NICU. Pediatrics 2019, 143, S54–S57. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stark, Z.; Nisselle, A.; McClaren, B.; Lynch, F.; Best, S.; Long, J.C.; Martyn, M.; Patel, C.; Schlapbach, L.J.; Barnett, C.; et al. Attitudes of Australian health professionals towards rapid genomic testing in neonatal and paediatric intensive care. Eur. J. Hum. Genet. 2019, 27, 1493–1501. [Google Scholar] [CrossRef] [PubMed]
- Jayasinghe, K.; Quinlan, C.; Mallett, A.J.; Kerr, P.G.; McClaren, B.; Nisselle, A.; Mallawaarachchi, A.; Polkinghorne, K.R.; Patel, C.; Best, S.; et al. Attitudes and practices of Australian nephrologists toward implementation of clinical genomics. Kidney Int. Rep. 2020, 6, 272–283. [Google Scholar] [CrossRef]
- Zebrowski, A.M.; Ellis, D.E.; Barg, F.K.; Sperber, N.R.; Bernhardt, B.A.; Denny, J.C.; Dexter, P.R.; Ginsburg, G.S.; Horowitz, C.R.; Johnson, J.A.; et al. Qualitative study of system-level factors related to genomic implementation. Genet. Med. 2019, 21, 1534–1540. [Google Scholar] [CrossRef]
- Michigan Department of Health and Human Services. Coverage of Rapid Whole Genome Sequencing (rWGS) Testing. 2021. Available online: https://www.michigan.gov/documents/mdhhs/MSA_21-33_732848_7.pdf (accessed on 23 February 2022).
| Characteristic (n) | Mean (SD) or n (%) |
|---|---|
| Age in years (n = 302) | 42.12 (12.77) |
| Years in practice (n = 304) | 15.25 (12.77) |
| Gender (n = 305) | |
| Female | 251 (82%) |
| Race (n = 298) | |
| White | 262 (88%) |
| South Asian | 18 (6%) |
| African American | 8 (3%) |
| Hispanic | 5 (2%) |
| Multiracial/Other | 4 (1%) |
| Primary position | |
| Physician—attending | 80 (26%) |
| Physician—resident | 26 (9%) |
| Nurse practitioner | 19 (6%) |
| Genetic counselor | 13 (4%) |
| Nurse (direct patient care) | 130 (42%) |
| Pharmacist, Therapist, Social worker, Parent liaison | 12 (4%) |
| Laboratory director | 3 (1%) |
| Laboratory staff | 6 (2%) |
| Hospital administrator | 8 (3%) |
| Nursing leader 1 | 10 (3%) |
| Attending physician (n = 76) | |
| Neonatologist/ Intensivist | 31 (41%) |
| Pediatrician/ Hospitalist | 19 (25%) |
| Pediatric subspecialty 2 | 14 (18%) |
| Geneticist | 12 (16%) |
| Primary Unit (n = 304) | |
| NICU | 141 (46%) |
| Multiple units/hospital wide | 58 (19%) |
| PICU | 37 (12%) |
| Medical surgical | 21 (9%) |
| Outpatient clinic | 27 (7%) |
| Non-clinical | 14 (5%) |
| Laboratory | 3 (1%) |
| Emergency room | 3 (1%) |
| Type of Education | n (%) |
|---|---|
| On-the-job training | 164 (53) |
| Genetics course in initial professional training | 130 (42) |
| Hospital supported training | 112 (37) |
| Self-directed education (journal articles, etc.) | 97 (32) |
| Continuing education courses in genetics | 79 (26) |
| Genetics course in graduate school | 62 (20) |
| Seminar/workshops in genetics | 44 (14) |
| Genetics conferences | 37 (12) |
| Advanced training in genetics | 20 (7) |
| No specific training | 76 (25) |
| Item | Factor 1: Personal Capability | Factor 2: Potential/ Intention | Factor 3: Implementation |
|---|---|---|---|
| I am confident about interpreting rWGS test reports | 0.935 | −0.154 | −0.0602 |
| I am confident about integrating rWGS test results in the care of patients and their families | 0.768 | 0.125 | −0.075 |
| I am confident in how to access resources to help me if I have questions or concerns about rWGS results | 0.72 | 0.129 | −0.00583 |
| I can find reliable sources of the information about rWGS when I need it | 0.611 | −0.092 | 0.27 |
| Using rWGS fits within the processes that I already use to care for my patients and their families | 0.452 | 0.147 | 0.211 |
| My training has prepared me to care for patients and their families at high risk for medical conditions that have a genetic basis | 0.428 | 0.212 | 0.0334 |
| The information generated by rWGS is important for patient care | −0.243 | 0.806 | 0.178 |
| I believe that rWGS is relevant to my current practice | 0.141 | 0.748 | −0.0343 |
| Within the next five years, precision medicine based on rWGS will improve clinical outcomes for infants and children | −0.0144 | 0.739 | 0.032 |
| I would support rWGS testing for appropriate patients if it was widely available in my clinical practice | −0.0109 | 0.684 | −0.00894 |
| rWGS will improve my ability to care for patients and their families | 0.189 | 0.676 | −0.0253 |
| Parents of my patients will be interested in having rWGS for their child | 0.291 | 0.506 | −0.136 |
| The implementation leaders/team have the necessary qualities and skills to successfully incorporate rWGS into my unit/ hospital’s clinical practice | −0.188 | 0.231 | 0.763 |
| Leaders have openly endorsed and supported rWGS in visible ways | 0.0379 | 0.00119 | 0.75 |
| A variety of strategies are being used to enable staff to use rWGS to care for patients in my unit/hospital | 0.277 | −0.132 | 0.611 |
| A clearly designated person or team is leading the effort to incorporate rWGS into my unit/hospital’s clinical practice | −0.213 | 0.336 | 0.585 |
| Staff have enough time to facilitate the integration of rWGS into clinical practice | 0.204 | −0.118 | 0.584 |
| Clear goals have been established for integrating rWGS into clinical practice | 0.399 | −0.117 | 0.527 |
| Term | Factor 1: Personal Capability | Factor 2: Potential/Intention | Factor 3: Implementation | Total Attitudes Score |
|---|---|---|---|---|
| Self-rated level of knowledge about rWGS | 0.505 (0.415, 0.596) (>0.001) | 0.44 (0.357, 0.523) (>0.001) | 0.378 (0.273, 0.483) (>0.001) | 5.62 (4.58, 6.66) (>0.001) |
| Clinical role (vs. non-clinical) | 0.326 (0.114, 0.538) (0.0028) | 3.7 (1.62, 5.78) (>0.001) | ||
| Confidence about future insurance coverage for rWGS | 0.219 (0.112, 0.326) (>0.001) | 0.223 (0.119, 0.326) (>0.001) | 0.208 (0.0928, 0.324) (>0.001) | 3.01 (1.87, 4.15) (>0.001) |
| Concerns about potential long-term effects of genomic testing on patients/ families | −0.266 (0.174, 0.358) (>0.001) | −1.95 (0.934, 2.97) (>0.001) | ||
| NICU/PICU (vs. other unit) | −0.285 (−0.464, −0.107) (0.0019) | |||
| Concern about racial disparities in use of genomic testing | −0.265 (0.167, 0.363) (>0.001) | |||
| Site2 (ref = Site 1) | 0.419 (0.216, 0.622) (>0.001) | |||
| Adjusted R2 | 0.43 | 0.44 | 0.32 | 0.51 |
| Themes/Subthemes | Quotes |
|---|---|
| Enthusiasm for rWGS in current or future patient care | |
| Potential to improve diagnostic accuracy and speed, leading to more effective treatment | “This tool will change how we care for patients; it can offer treatments and early diagnosis to conditions that would have otherwise taken a very long time and additional costs.” (Physician) “Very important to help families get answers sooner and help determine if there are any treatments available that may help.” (Genetic Counselor) “There are definitely some patients who could benefit, especially those whose clinical conditions are hard to explain for other reasons.” (Direct care nurse) |
| Qualified enthusiasm | “Hoping this test becomes a covered benefit from insurers so no child in need has to go without access.” (Physician) “Cost, resources and utility of results in the near term are not yet certain although expect this to be more obvious in the future.” (Hospital administrator) |
| Implementation of rWGS | |
| Satisfaction with implementation | “It is well run with our clinical champions coordinating and providing the counseling and education.” (Physician) |
| Limited experience; interest in learning more | “It has been so great to be able to do rWGS but more education needed for staff and provider understanding/comfort.” (Nurse Practitioner) “I have heard it mentioned while on multidisciplinary rounds, but as a resident, I feel disconnected and not sure where to turn to (aside from our Geneticist) on a day-to-day basis for rWGS testing and information. I do not feel confident in talking about rWGS alone with my patients and would appreciate more education to help this project take flight at my institution.” (Resident) |
| Role of genetics service | “Our genetic team has been a key for this project and we rely on them.” (Physician) “Until now this has been monopolized by the genetics department. I feel that this should be a more widely accessible test and based on results or concerns then genetics can be contacted. This reminds me of not ordering an echo till a patient is seen by cardiology. The reality is we order echo and work-ups including troponins and BNP etc., then contact cardiology.” (Physician) |
| Concerns about wider use of rWGS/genomic testing | |
| “I think case specific rWGS has merit, but the extraneous data that may impact insurability must be non-discoverable by insurance companies, and extreme caution should be used by providers to be sure patients are not overwhelmed by unexpected information.” (Physician) “I feel this is an option for parents but also feel it is a decision they need to make after all the appropriate education is given to them.” (Nurse Practitioner) | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Franck, L.S.; Scheurer-Monaghan, A.; Bupp, C.P.; Fakhoury, J.D.; Hoffmann, T.J.; Deshpandey, M.; Arenchild, M.; Dimmock, D.P. Healthcare Professionals’ Attitudes toward Rapid Whole Genome Sequencing in Pediatric Acute Care. Children 2022, 9, 357. https://doi.org/10.3390/children9030357
Franck LS, Scheurer-Monaghan A, Bupp CP, Fakhoury JD, Hoffmann TJ, Deshpandey M, Arenchild M, Dimmock DP. Healthcare Professionals’ Attitudes toward Rapid Whole Genome Sequencing in Pediatric Acute Care. Children. 2022; 9(3):357. https://doi.org/10.3390/children9030357
Chicago/Turabian StyleFranck, Linda S., Andrea Scheurer-Monaghan, Caleb P. Bupp, Joseph D. Fakhoury, Thomas J. Hoffmann, Manasi Deshpandey, Madison Arenchild, and David P. Dimmock. 2022. "Healthcare Professionals’ Attitudes toward Rapid Whole Genome Sequencing in Pediatric Acute Care" Children 9, no. 3: 357. https://doi.org/10.3390/children9030357
APA StyleFranck, L. S., Scheurer-Monaghan, A., Bupp, C. P., Fakhoury, J. D., Hoffmann, T. J., Deshpandey, M., Arenchild, M., & Dimmock, D. P. (2022). Healthcare Professionals’ Attitudes toward Rapid Whole Genome Sequencing in Pediatric Acute Care. Children, 9(3), 357. https://doi.org/10.3390/children9030357









