Enhancing Small Medical Dataset Classification Performance Using GAN
Abstract
:1. Introduction
- We proposed a general methodology for small medical data classification that deploys an augmentation technique and a feature selection strategy, followed by appropriate training of a suitable classifier algorithm.
- Brouta feature selection is utilized as a feature selection strategy with multiple machine learning algorithms.
- We presented highly accurate classifiers for 13 different diseases and suggested a generalized augmentation approach that should perform well for other similar datasets.
2. Related Works
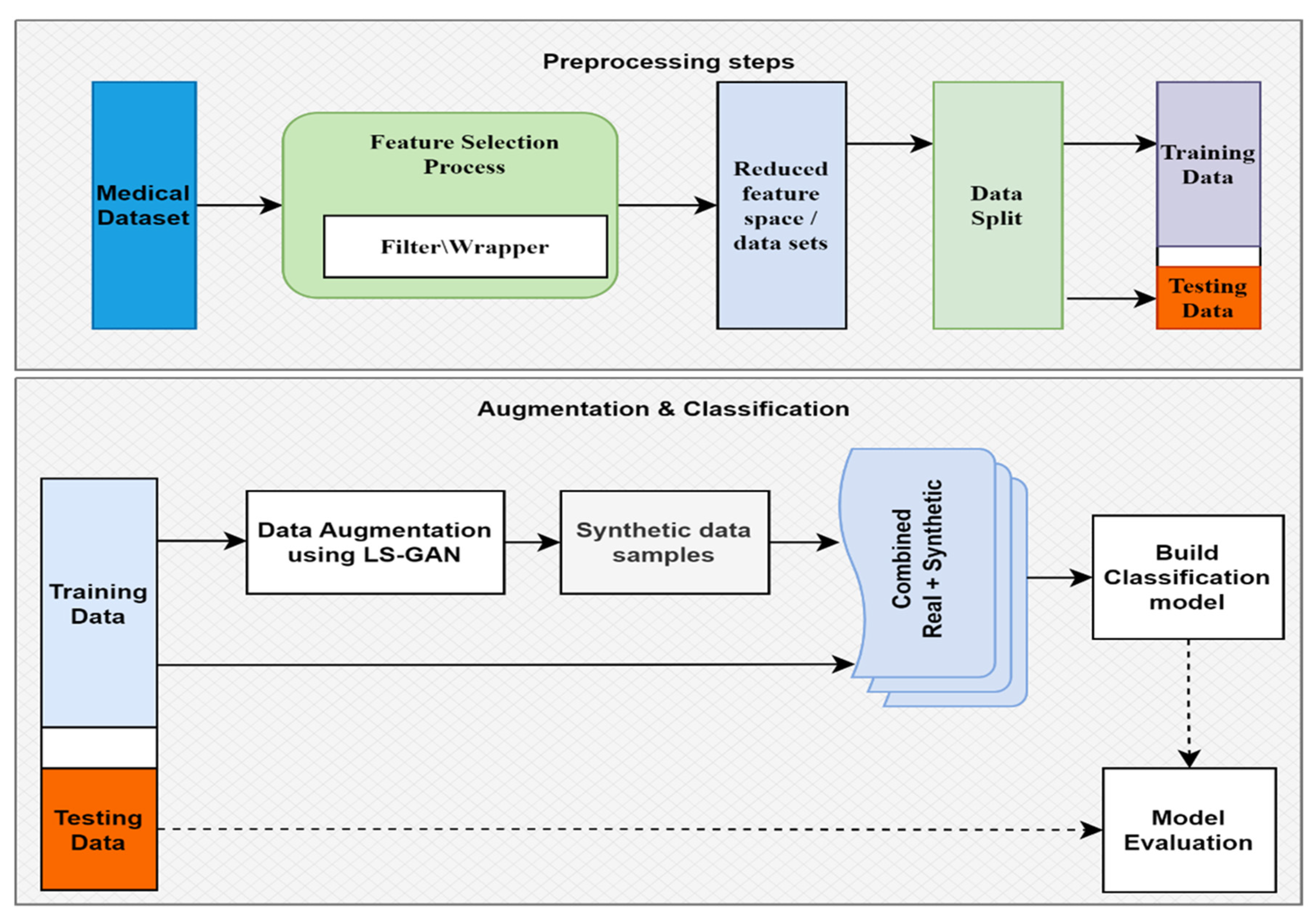
3. Materials and Methods
3.1. Datasets
3.2. Model Construction Overview
3.3. Feature Selection
3.4. Data Augmentation
- Define the networks: the generator G and the discriminator D.
- Initialize the generator and discriminator networks with random weights.
- Sample a random noise vector z from a noise distribution p(z).
- Generate a fake sample x′ = G(z) using the generator network G.
- Sample a real sample x from the real data distribution.
- Pass x and x′ through the discriminator network D to get the output values D(x) and D(x′).
- Calculate the least-squares loss function for the generator and the discriminator:
- 8.
- Update the generator and discriminator networks using the gradients of the loss functions with respect to the network parameters.
- 9.
- Repeat steps 2–7 for a fixed number of iterations or until the desired level of performance is reached.
4. Machine Learning Techniques
5. Results and Discussion
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Alam, M.Z.; Rahman, M.S.; Rahman, M.S. A Random Forest based predictor for medical data classification using feature ranking. Inform. Med. Unlocked 2019, 15, 100180. [Google Scholar] [CrossRef]
- Azadi, A.; García-Peñalvo, F.J. Synergistic Effect of Medical Information Systems Integration: To What Extent Will It Affect the Accuracy Level in the Reports and Decision-Making Systems? Informatics 2023, 10, 12. [Google Scholar] [CrossRef]
- Rashid, K.M.; Louis, J. Times-series data augmentation and deep learning for construction equipment activity recognition. Adv. Eng. Inform. 2019, 42, 100944. [Google Scholar] [CrossRef]
- Al-Antari, M.A. Artificial Intelligence for Medical Diagnostics—Existing and Future AI Technology! Diagnostics 2023, 13, 688. [Google Scholar] [PubMed]
- Oza, P.; Sharma, P.; Patel, S. Deep ensemble transfer learning-based framework for mammographic image classification. J. Supercomput. 2022, 1–22. [Google Scholar] [CrossRef]
- Oza, P.; Sharma, P.; Patel, S.; Kumar, P. Deep convolutional neural networks for computer-aided breast cancer diagnostic: A survey. Neural Comput. Appl. 2022, 34, 1815–1836. [Google Scholar] [CrossRef]
- Shorten, C.; Khoshgoftaar, T.M. A survey on image data augmentation for deep learning. J. Big Data 2019, 6, 60. [Google Scholar] [CrossRef]
- Coutinho-Almeida, J.; Rodrigues, P.P.; Cruz-Correia, R.J. GANs for Tabular Healthcare Data Generation: A Review on Utility and Privacy. In Discovery Science, Proceedings of the 24th International Conference, DS 2021, Halifax, NS, Canada, 11–13 October 2021; Springer International Publishing: Cham, Switzerland, 2021; pp. 282–291. [Google Scholar]
- Chatterjee, S.; Hazra, D.; Byun, Y.-C.; Kim, Y.-W. Enhancement of Image Classification Using Transfer Learning and GAN-Based Synthetic Data Augmentation. Mathematics 2022, 10, 1541. [Google Scholar] [CrossRef]
- Kim, J.-H.; Hwang, Y. Gan-based synthetic data augmentation for infrared small target detection. IEEE Trans. Geosci. Remote Sens. 2022, 60, 1541. [Google Scholar] [CrossRef]
- Imran, A.S.; Yang, R.; Kastrati, Z.; Daudpota, S.M.; Shaikh, S. The impact of synthetic text generation for sentiment analysis using GAN based models. Egypt. Inform. J. 2022, 23, 547–557. [Google Scholar] [CrossRef]
- Oza, P.; Sharma, P.; Patel, S.; Kumar, P. Computer-Aided Breast Cancer Diagnosis: Comparative Analysis of Breast Imaging Modalities and Mammogram Repositories. Curr. Med. Imaging 2023, 19, 456–468. [Google Scholar] [CrossRef]
- Oza, P.; Sharma, P.; Patel, S.; Adedoyin, F.; Bruno, A. Image Augmentation Techniques for Mammogram Analysis. J. Imaging 2022, 8, 141. [Google Scholar] [CrossRef] [PubMed]
- Abedi, M.; Hempel, L.; Sadeghi, S.; Kirsten, T. GAN-Based Approaches for Generating Structured Data in the Medical Domain. Appl. Sci. 2022, 12, 7075. [Google Scholar] [CrossRef]
- Oza, P.; Sharma, P.; Patel, S.; Bruno, A. A bottom-up review of image analysis methods for suspicious region detection in mammograms. J. Imaging 2021, 7, 190. [Google Scholar] [CrossRef] [PubMed]
- Abbass, H.A. An evolutionary artificial neural networks approach for breast cancer diagnosis. Artif. Intell. Med. 2002, 25, 265–281. [Google Scholar] [CrossRef] [PubMed]
- Kiyan, T.; Yildirim, T. Breast cancer diagnosis using statistical neural networks. IU-J. Electr. Electron. Eng. 2004, 4, 1149–1153. [Google Scholar]
- Karabatak, M.; Ince, M.C. An expert system for detection of breast cancer based on association rules and neural network. Expert Syst. Appl. 2009, 36, 3465–3469. [Google Scholar] [CrossRef]
- Peng, Y.; Wu, Z.; Jiang, J. A novel feature selection approach for biomedical data classification. J. Biomed. Inform. 2010, 43, 15–23. [Google Scholar] [CrossRef] [Green Version]
- Fan, C.-Y.; Chang, P.-C.; Lin, J.-J.; Hsieh, J. A hybrid model combining case-based reasoning and fuzzy decision tree for medical data classification. Appl. Soft Comput. 2011, 11, 632–644. [Google Scholar] [CrossRef]
- Azar, A.T.; El-Said, S.A. Performance analysis of support vector machines classifiers in breast cancer mammography recognition. Neural Comput. Appl. 2014, 24, 1163–1177. [Google Scholar] [CrossRef]
- Cubuk, E.D.; Zoph, B.; Mane, D.; Vasudevan, V.; Le, Q.V. Autoaugment: Learning augmentation strategies from data. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Long Beach, CA, USA, 16–17 June 2019; pp. 113–123. [Google Scholar]
- Iizuka, S.; Simo-Serra, E.; Ishikawa, H. Globally and locally consistent image completion. ACM Trans. Graph. (ToG) 2017, 36, 1–14. [Google Scholar] [CrossRef]
- Xie, Q.; Dai, Z.; Hovy, E.; Luong, T.; Le, Q. Unsupervised data augmentation for consistency training. Adv. Neural Inf. Process. Syst. 2020, 33, 6256–6268. [Google Scholar]
- Tran, T.; Pham, T.; Carneiro, G.; Palmer, L.; Reid, I. A bayesian data augmentation approach for learning deep models. Adv. Neural Inf. Process. Syst. 2017, 30, 2797–2806. [Google Scholar]
- Lim, S.; Kim, I.; Kim, T.; Kim, C.; Kim, S. Fast autoaugment. Adv. Neural Inf. Process. Syst. 2019, 32, 6665–6675. [Google Scholar]
- Ho, D.; Liang, E.; Chen, X.; Stoica, I.; Abbeel, P. Population based augmentation: Efficient learning of augmentation policy schedules. In Proceedings of the International Conference on Machine Learning, Long Beach, CA, USA, 9–15 June 2019; pp. 2731–2741. [Google Scholar]
- Raja, K.C.; Kannimuthu, S. Conditional Generative Adversarial Network Approach for Autism Prediction. Comput. Syst. Sci. Eng. 2023, 44, 741–755. [Google Scholar] [CrossRef]
- Zhong, Z.; Zheng, L.; Kang, G.; Li, S.; Yang, Y. Random erasing data augmentation. In Proceedings of the AAAI Conference on Artificial Intelligence, New York, NY, USA, 7–12 February 2020; pp. 13001–13008. [Google Scholar]
- Frank, A. UCI Machine Learning Repository. 2010. Available online: http://archive.ics.uci.edu/ml (accessed on 15 September 2022).
- Derrac, J.; Garcia, S.; Sanchez, L.; Herrera, F. Keel data-mining software tool: Data set repository, integration of algorithms and experimental analysis framework. J. Mult. Valued Log. Soft Comput 2011, 17, 255–287. [Google Scholar]
- Kursa, M.B.; Jankowski, A.; Rudnicki, W.R. Boruta–a system for feature selection. Fundam. Inform. 2010, 101, 271–285. [Google Scholar] [CrossRef]
- Mao, X.; Li, Q.; Xie, H.; Lau, R.Y.; Wang, Z.; Paul Smolley, S. Least squares generative adversarial networks. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 2794–2802. [Google Scholar]
- Randhawa, R.H.; Aslam, N.; Alauthman, M.; Rafiq, H.; Comeau, F. Security hardening of botnet detectors using generative adversarial networks. IEEE Access 2021, 9, 78276–78292. [Google Scholar] [CrossRef]
- Mitchell, T.M. Machine Learning; McGraw-Hill: New York, NY, USA, 2007; Volume 1. [Google Scholar]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef] [Green Version]
- McCallum, A.; Nigam, K. A comparison of event models for naive bayes text classification. In Proceedings of the AAAI-98 Workshop on Learning for Text Categorization, Madison, WI, USA, 26 July 1998; pp. 41–48. [Google Scholar]
- Kleinbaum, D.; Klein, M.; Regression, L. A Self-Learning Text; Springer: New York, NY, USA, 2005; Volume 21, p. 22. [Google Scholar]
- Cortes, C.; Vapnik, V. Support-vector networks. Mach. Learn. 1995, 20, 273–297. [Google Scholar] [CrossRef]
- Haykin, S.; Network, N. A comprehensive foundation. Neural Netw. 2004, 2, 41. [Google Scholar]
- García, S.; Fernández, A.; Luengo, J.; Herrera, F. A study of statistical techniques and performance measures for genetics-based machine learning: Accuracy and interpretability. Soft Comput. 2009, 13, 959–977. [Google Scholar] [CrossRef]





| No. | Research | Datasets | Method and Results |
|---|---|---|---|
| 1 | Daniel et al. [27]: Population Augmentation policy | SVHN | Regression and the error: 0.1 |
| 2 | Zhong et al. [29]: Augmentation by Data Random Erasing | CIFAR-10 | Regression and the error: 0.31 |
| 3 | Lim et al. [26]: Fast AutoAugment. | CIFAR-10 | Regression and the error: 0.20 |
| 4 | Cubuk et al. [22]: AutoAugment data strategies. | SVHN and ImageNet | Classification accuracy rates: 0.835 Regression and the error: 0.10% |
| 5 | Xie et al. [24]: Unsupervised Data Augmentation. | CIFAR-10 | Classification accuracy rates: 0.79 Regression and the error rate: 0.3 |
| 6 | Tran et al. [25]:A Bayesian Data Augmentation. | CIFAR-10 | Classification and Accuracy: 0.93 |
| Dataset | Samples | Attribute | Train Set | Test Set |
|---|---|---|---|---|
| Liver | 345 | 7 | 225 | 120 |
| Blood | 748 | 5 | 498 | 250 |
| Haberman | 306 | 4 | 206 | 100 |
| Diabetes | 768 | 8 | 508 | 260 |
| Hepatitis | 155 | 11 | 105 | 50 |
| Breast | 699 | 9 | 459 | 240 |
| Heart | 270 | 14 | 180 | 90 |
| Parkinsons | 195 | 23 | 125 | 70 |
| Phoneme | 5404 | 6 | 3604 | 1800 |
| planningRelax | 182 | 13 | 122 | 60 |
| Saheart | 462 | 9 | 302 | 160 |
| Spectf | 267 | 45 | 177 | 90 |
| WDBC | 569 | 30 | 379 | 190 |
| Dataset | Augmented Ratio | Size of Data after Augmentation |
|---|---|---|
| Blood | 0.7 | 523 |
| Breast | 0.8 | 559 |
| Diabetes | 0.9 | 692 |
| Haberman | 0.1 | 136 |
| Habitat | 0.1 | 155 |
| Heart | 0.5 | 405 |
| Liver | 0.1 | 345 |
| Parkinsons | 0.5 | 292 |
| Phoneme | 1 | 5404 |
| PlanningRelax | 0.1 | 182 |
| Saheart | 0.1 | 462 |
| Spectf | 0.3 | 400 |
| Wdbc | 0.5 | 854 |
| Dataset | Measure | SVM | ANN | RF | NB | LR |
|---|---|---|---|---|---|---|
| Blood | Avg | 0.7978 | 0.7844 | 0.8035 | 0.8005 | 0.7982 |
| Stdv | 0.0075 | 0.0029 | 0.0057 | 0.0087 | 0.008 | |
| Best | 0.8089 | 0.7854 | 0.8129 | 0.8168 | 0.8089 | |
| Worse | 0.7815 | 0.7815 | 0.7893 | 0.7815 | 0.7854 | |
| Augmented Ratio | 0.7 | 1 | 0.2 | 0.2 | 0.6 | |
| Breast | Avg | 0.9738 | 0.9627 | 0.9741 | 0.9629 | 0.9757 |
| Stdv | 0.0056 | 0.0078 | 0.0075 | 0.0128 | 0.006 | |
| Best | 0.9801 | 0.9675 | 0.9843 | 0.9885 | 0.9843 | |
| Worse | 0.9591 | 0.9465 | 0.9633 | 0.9423 | 0.9633 | |
| Augmented Ratio | 0.8 | 0.5 | 0.5 | 0.8 | 0.8 | |
| Diabetes | Avg | 0.7726 | 0.7258 | 0.7659 | 0.7554 | 0.7636 |
| Stdv | 0.0128 | 0.0088 | 0.013 | 0.0203 | 0.0148 | |
| Best | 0.7824 | 0.7377 | 0.7928 | 0.7876 | 0.798 | |
| Worse | 0.7407 | 0.6996 | 0.7407 | 0.7094 | 0.7355 | |
| Augmented Ratio | 0.9 | 1 | 0.8 | 0.6 | 0.7 | |
| Haberman | Avg | 0.7313 | 0.7306 | 0.7319 | 0.7281 | 0.73 |
| Stdv | 0.0057 | 0.007 | 0.0101 | 0.0208 | 0.0093 | |
| Best | 0.7344 | 0.7344 | 0.7535 | 0.763 | 0.744 | |
| Worse | 0.7249 | 0.7154 | 0.7154 | 0.6963 | 0.7154 | |
| Augmented Ratio | 0.1 | 0.9 | 0.1 | 0.3 | 0.2 | |
| Hepatitis | Avg | 0.9124 | 0.8759 | 0.8747 | 0.8489 | 0.847 |
| Stdv | 0.0204 | 0.0193 | 0.0279 | 0.035 | 0.0329 | |
| Best | 0.9256 | 0.9256 | 0.9256 | 0.9256 | 0.9445 | |
| Worse | 0.869 | 0.8502 | 0.8313 | 0.7936 | 0.8124 | |
| Augmented Ratio | 0.1 | 0.4 | 0.1 | 0.1 | 0.2 | |
| Heart | Avg | 0.8623 | 0.8413 | 0.7978 | 0.7754 | 0.8478 |
| Stdv | 0.033 | 0.0348 | 0.0335 | 0.0414 | 0.0183 | |
| Best | 0.8924 | 0.8707 | 0.8598 | 0.8598 | 0.8707 | |
| Worse | 0.8163 | 0.7837 | 0.7185 | 0.6859 | 0.8055 | |
| Augmented Ratio | 0.5 | 0.5 | 0.2 | 0.4 | 0.3 | |
| Liver | Avg | 0.7333 | 0.6921 | 0.7014 | 0.6972 | 0.7203 |
| Stdv | 0.0369 | 0.0356 | 0.038 | 0.0513 | 0.0273 | |
| Best | 0.7892 | 0.713 | 0.7808 | 0.7638 | 0.7723 | |
| Worse | 0.6197 | 0.5859 | 0.6113 | 0.5689 | 0.6621 | |
| Augmented Ratio | 0.1 | 0.2 | 0.5 | 0.1 | 0.3 | |
| Parkinsons | Avg | 0.9071 | 0.8902 | 0.8882 | 0.8543 | 0.8717 |
| Stdv | 0.013 | 0.0264 | 0.0242 | 0.0375 | 0.0263 | |
| Best | 0.9265 | 0.9116 | 0.9265 | 0.8966 | 0.9116 | |
| Worse | 0.8668 | 0.8519 | 0.8071 | 0.7474 | 0.822 | |
| Augmented Ratio | 0.5 | 0.7 | 0.6 | 0.7 | 0.5 | |
| Phoneme | Avg | 0.8357 | 0.8325 | 0.836 | 0.8387 | 0.8243 |
| Stdv | 0.007 | 0.0091 | 0.0074 | 0.0078 | 0.0088 | |
| Best | 0.846 | 0.839 | 0.8504 | 0.8482 | 0.8373 | |
| Worse | 0.8205 | 0.8118 | 0.8232 | 0.8259 | 0.7993 | |
| Augmented Ratio | 1 | 1 | 1 | 1 | 1 | |
| PlanningRelax | Avg | 0.6565 | 0.6549 | 0.6484 | 0.6221 | 0.6554 |
| Stdv | 0.009 | 0.0093 | 0.0179 | 0.0354 | 0.0121 | |
| Best | 0.6624 | 0.6624 | 0.6785 | 0.6785 | 0.6785 | |
| Worse | 0.6463 | 0.6463 | 0.614 | 0.5334 | 0.6301 | |
| Augmented Ratio | 0.1 | 0.7 | 0.1 | 0.2 | 0.2 | |
| Saheart | Avg | 0.7541 | 0.7553 | 0.7342 | 0.7332 | 0.7484 |
| Stdv | 0.0115 | 0.0099 | 0.0228 | 0.0235 | 0.0161 | |
| Best | 0.7669 | 0.7606 | 0.7733 | 0.7733 | 0.7796 | |
| Worse | 0.729 | 0.7353 | 0.6973 | 0.6846 | 0.7226 | |
| Augmented Ratio | 0.1 | 0.8 | 0.1 | 0.1 | 0.8 | |
| Spectf | Avg | 0.7835 | 0.7872 | 0.7777 | 0.767 | 0.7828 |
| Stdv | 0.0173 | 0.0067 | 0.0201 | 0.0244 | 0.0067 | |
| Best | 0.7923 | 0.7923 | 0.8033 | 0.8143 | 0.7923 | |
| Worse | 0.7374 | 0.7813 | 0.7264 | 0.7154 | 0.7703 | |
| Augmented Ratio | 0.3 | 0.4 | 0.1 | 1 | 0.3 | |
| Wdbc | Avg | 0.9624 | 0.9513 | 0.9549 | 0.9528 | 0.9588 |
| Stdv | 0.0059 | 0.0123 | 0.0117 | 0.0125 | 0.0113 | |
| Best | 0.9753 | 0.9599 | 0.9856 | 0.9753 | 0.9753 | |
| Worse | 0.9547 | 0.9289 | 0.9341 | 0.9186 | 0.9392 | |
| Augmented Ratio | 0.5 | 0.7 | 0.4 | 0.5 | 0.5 |
| Dataset | SVM-ANN | SVM-NB | SVM-RF | SVM-LS |
|---|---|---|---|---|
| Blood | 0.0016 | 0.1023 | 0.8088 | |
| Breast | 0.9506 | 0.0095 | 0.0949 | |
| Diabetes | 0.0030 | 0.0015 | 0.0024 | |
| Hepatitis | 0.001 | 0.0 | 0 | |
| Heart | 0.0 | 0.0 | 0.010933 | 0.1205 |
| Liver | 0.0132 | 0.0101 | 0.1316 | |
| Parkinsons | 0.0015 | 0.0 | 0.0043 | 0.0 |
| Phoneme | 0.930 | 0.0933 | 0.1184 | 0.0 |
| planningRelax | 0.0 | 0 | 0.4411 | 0.6013 |
| Saheart | 0.0027 | 0.0054 | 0.79501 | 0.0506 |
| Wdbc | 0.0042 | 0.0037 | 0.1495 | |
| Spectf | 0.1060 | 0.0060 | 0.6535 | 0.018 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alauthman, M.; Al-qerem, A.; Sowan, B.; Alsarhan, A.; Eshtay, M.; Aldweesh, A.; Aslam, N. Enhancing Small Medical Dataset Classification Performance Using GAN. Informatics 2023, 10, 28. https://doi.org/10.3390/informatics10010028
Alauthman M, Al-qerem A, Sowan B, Alsarhan A, Eshtay M, Aldweesh A, Aslam N. Enhancing Small Medical Dataset Classification Performance Using GAN. Informatics. 2023; 10(1):28. https://doi.org/10.3390/informatics10010028
Chicago/Turabian StyleAlauthman, Mohammad, Ahmad Al-qerem, Bilal Sowan, Ayoub Alsarhan, Mohammed Eshtay, Amjad Aldweesh, and Nauman Aslam. 2023. "Enhancing Small Medical Dataset Classification Performance Using GAN" Informatics 10, no. 1: 28. https://doi.org/10.3390/informatics10010028
APA StyleAlauthman, M., Al-qerem, A., Sowan, B., Alsarhan, A., Eshtay, M., Aldweesh, A., & Aslam, N. (2023). Enhancing Small Medical Dataset Classification Performance Using GAN. Informatics, 10(1), 28. https://doi.org/10.3390/informatics10010028


_Bryant.png)




