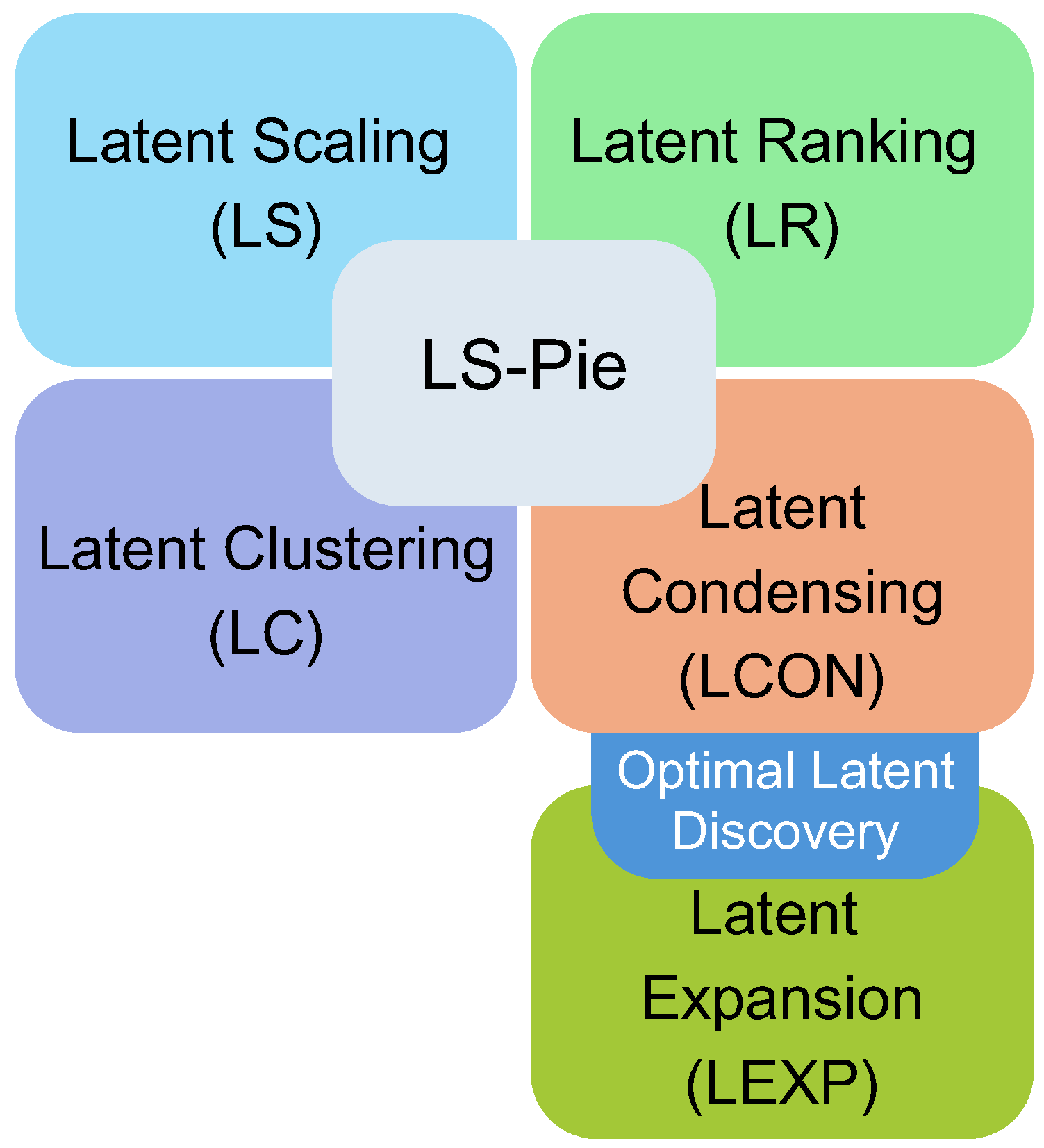
Enhancing LS-PIE’s Optimal Latent Dimensional Identification: Latent Expansion and Latent Condensation
Abstract
1. Introduction
2. Background
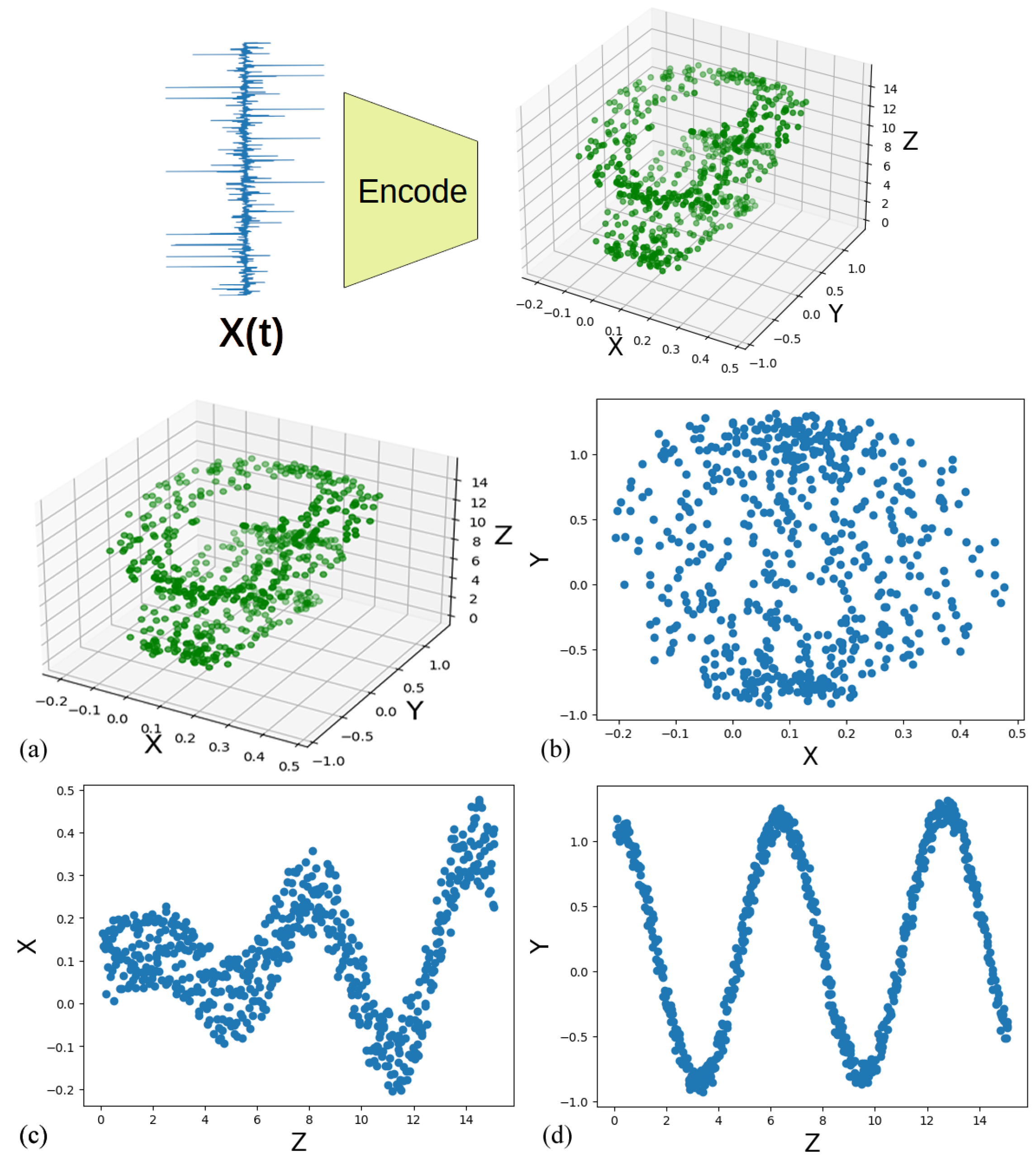
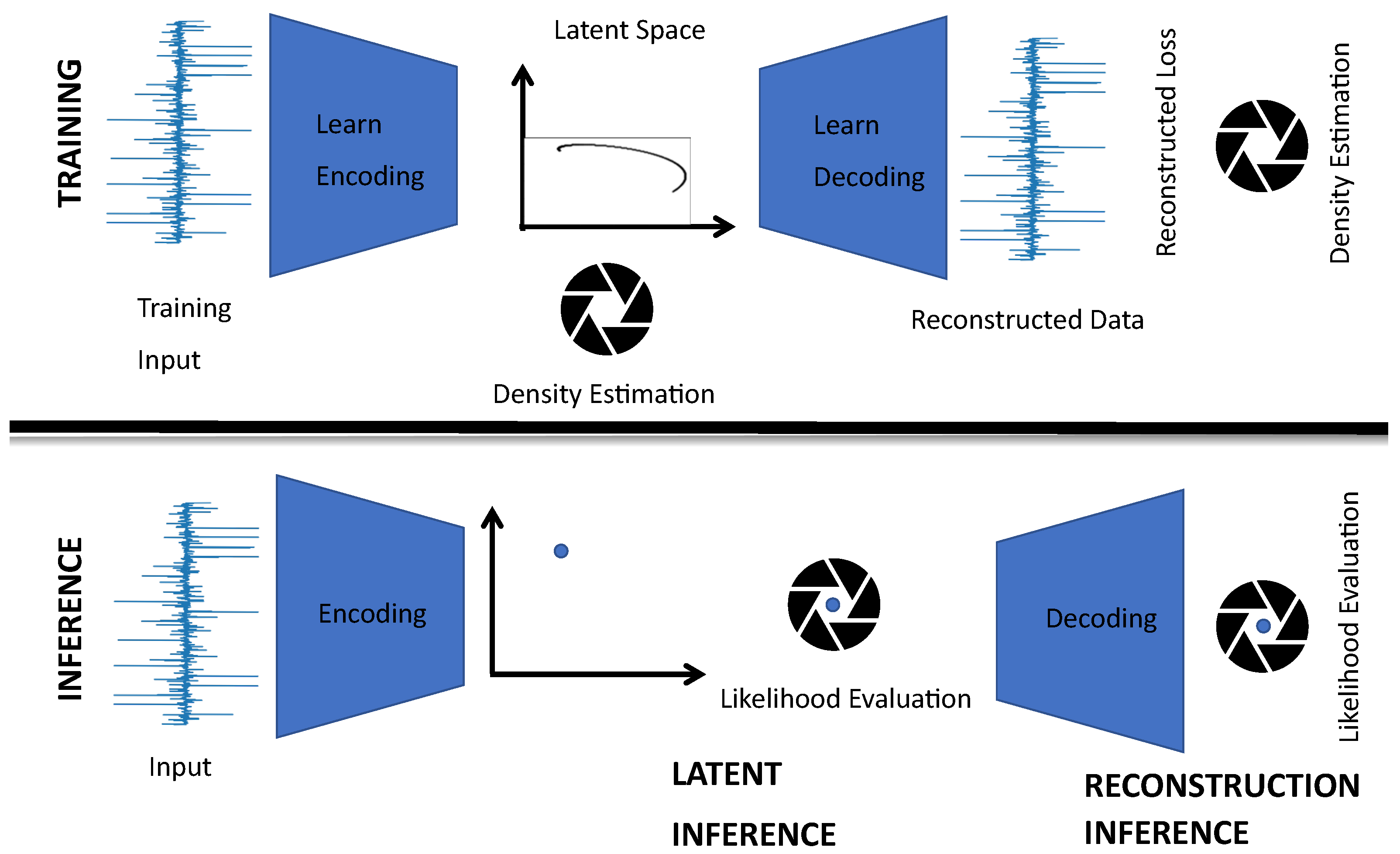
2.1. Latent Spaces
2.2. Latent Vector Models
2.2.1. PCA
2.2.2. ICA
2.3. Latent Clustering
2.4. Pre-Processing—Hankelisation
3. Materials and Methods
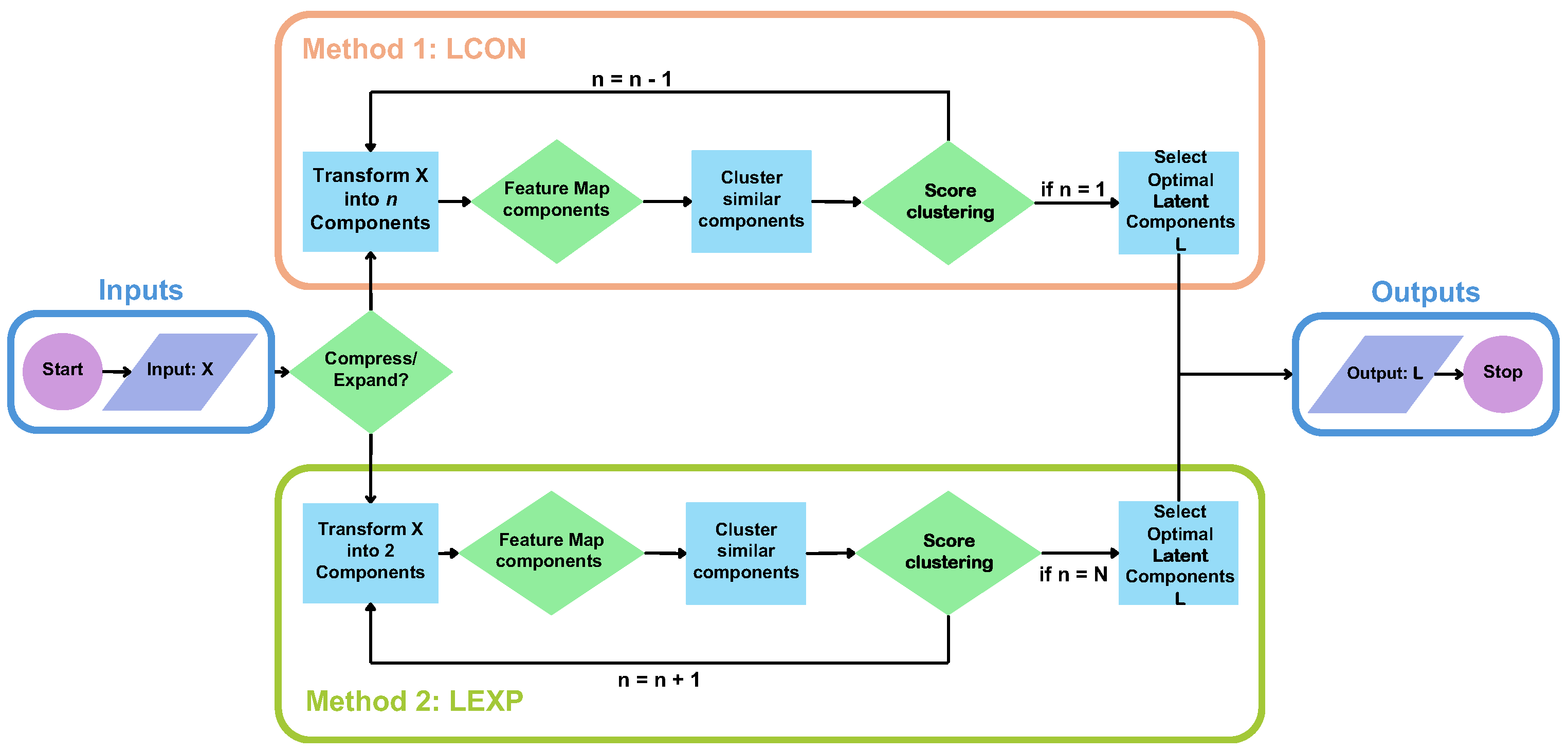
3.1. Optimal Latent Discovery Framework
3.2. Optimal Latent Discovery
3.2.1. Latent Condensing (LCON)
- selecting clustering algorithms that find the optimal number of latent clusters such as balanced iterative reducing and clustering using hierarchies (BIRCH) [24,24], density-based spatial clustering of applications with noise (DBSCAN) [25,26], Ordering Points To Identify the Clustering Structure (OPTICS) [27,28], Mean Shift [29] and Affinity Propagation [30,31];
- systematically reducing the latent dimensions and minimising or maximising a selected clustering index to find the optimal number of clusters.
| Algorithm 1 Latent Condensing (LCON) for Hankelised Time Series Data | |
| Require: Hankelised or multi-channel time series data matrix , latent variable model LVM, clus-tering algorithm C, feature mapping function f, distance metric d, cluster scoring function s, specified or maximum number of latent vectors m, clustering approach or not Ensure: Best feature clustering , cluster score and latent components
| |
| ▹ Decompose into k latent vectors ▹ Map latent vectors to feature space ▹ User-specified feature space scaling ▹ Automatic feature clustering using user-specified |
| distance metric d to find protype latent vectors | |
| ▹ Cluster scoring |
| |
|
▹ Cluster into j clusters using distance metric d ▹ Cluster scoring |
| |
|
Feature mapping function f: , where are selected individual feature functions that could include: | |
| 0. Identity: | ▹ Keeps original vector unchanged |
|
1. Variance: 2. Kurtosis: 3. Spectral centroid: 4. Entropy: , where is the probability of the k-th element in Distance metrics d for clustering: - Euclidean distance: [37] - Manhattan distance: [38] - Cosine distance: [39] - Mahalanobis distance: , where is the covariance matrix [40] Cluster scoring functions s: 1. Silhouette score: is the mean intra-cluster distance, and is the mean nearest-cluster distance 2. Variance-based: 3. Kurtosis-based: 4. Frequency-based: is the mean frequency for the j-th latent component | |
3.2.2. Latent Expansion (LEXP)
| Algorithm 2 Latent Expansion (LEXP) for Hankelised Time Series Data | |
| Require: Hankelised or multi-channel time series data matrix , latent variable model , clustering algorithm C, feature mapping function f, distance metric d, cluster scoring function s, specified or maximum number of latent vectors m Ensure: Best feature clustering , cluster score and latent components
| |
|
▹ Decompose into k latent vectors ▹ Map latent vectors to feature space ▹ User-specified feature space scaling ▹ Cluster into j clusters using distance metric d ▹ Cluster scoring |
Distance metrics d for clustering: See Algorithm 1 Cluster scoring functions s: See Algorithm 1 | |
4. Numerical Analysis
4.1. Datasets Overview
4.1.1. Foundational Problem: Single-Channel
4.1.2. Foundational Problem: Complex Mixed Signal
4.1.3. Experimental Data: ECG Heartbeat Categorisation
4.1.4. Experimental Data: Vibration Guided Wave-Based Monitoring
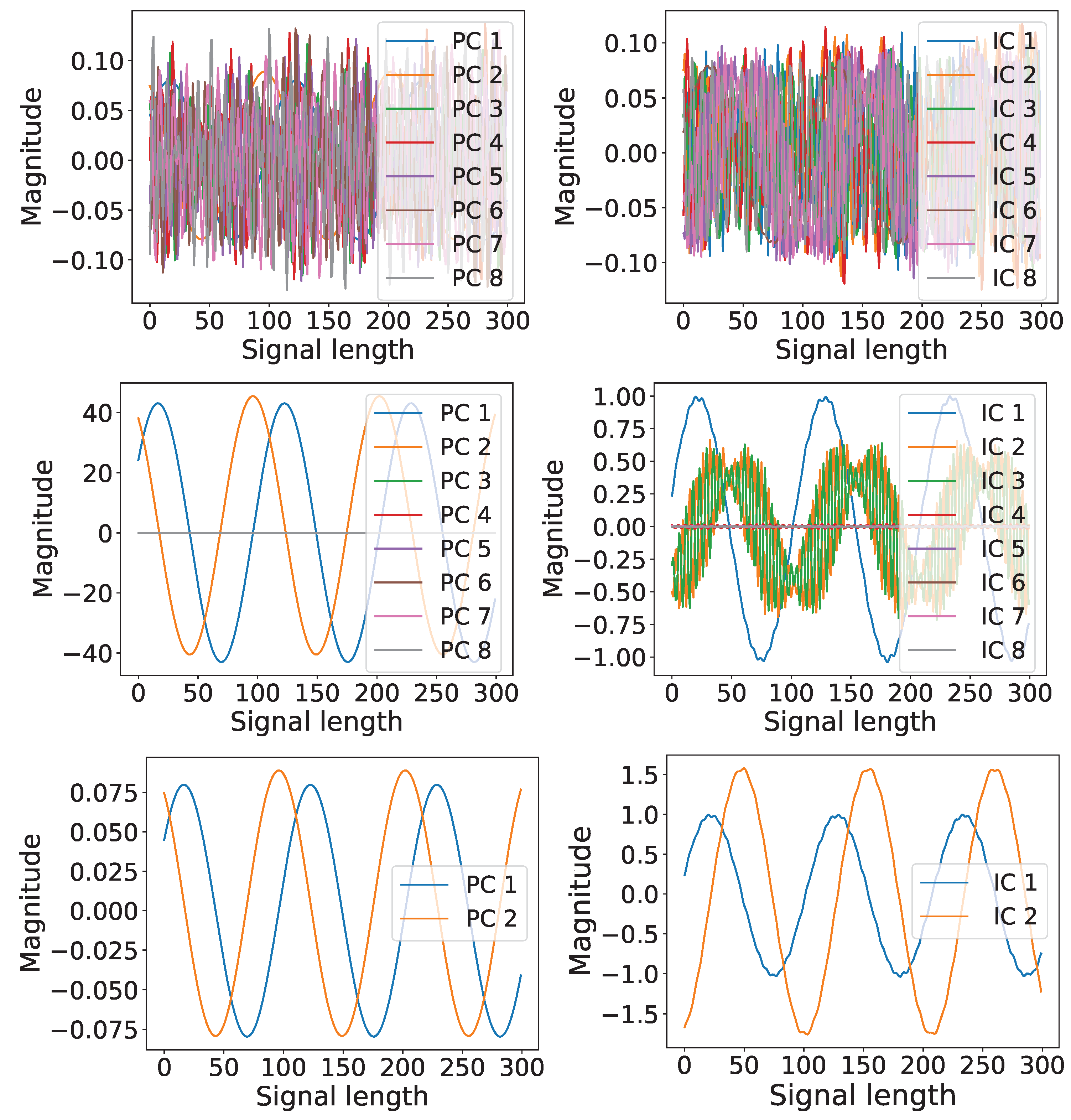
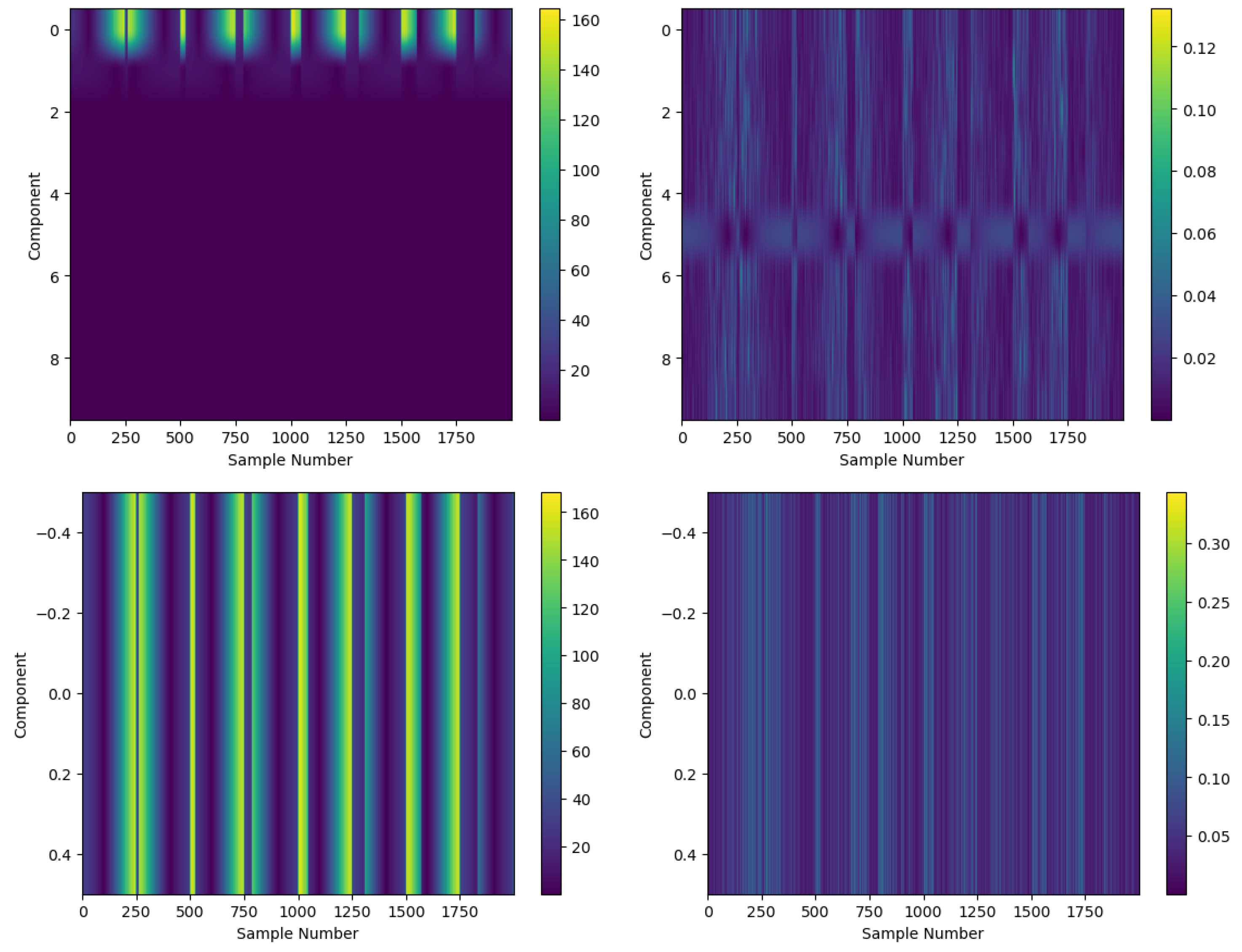
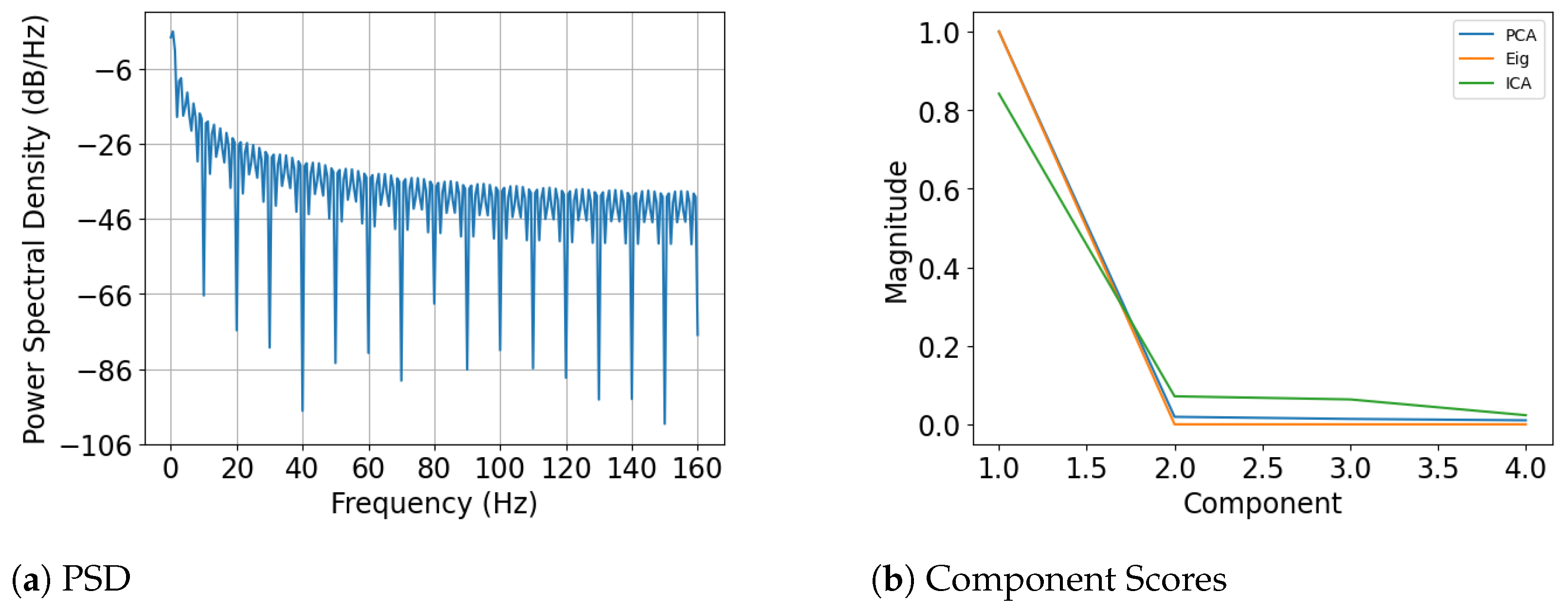
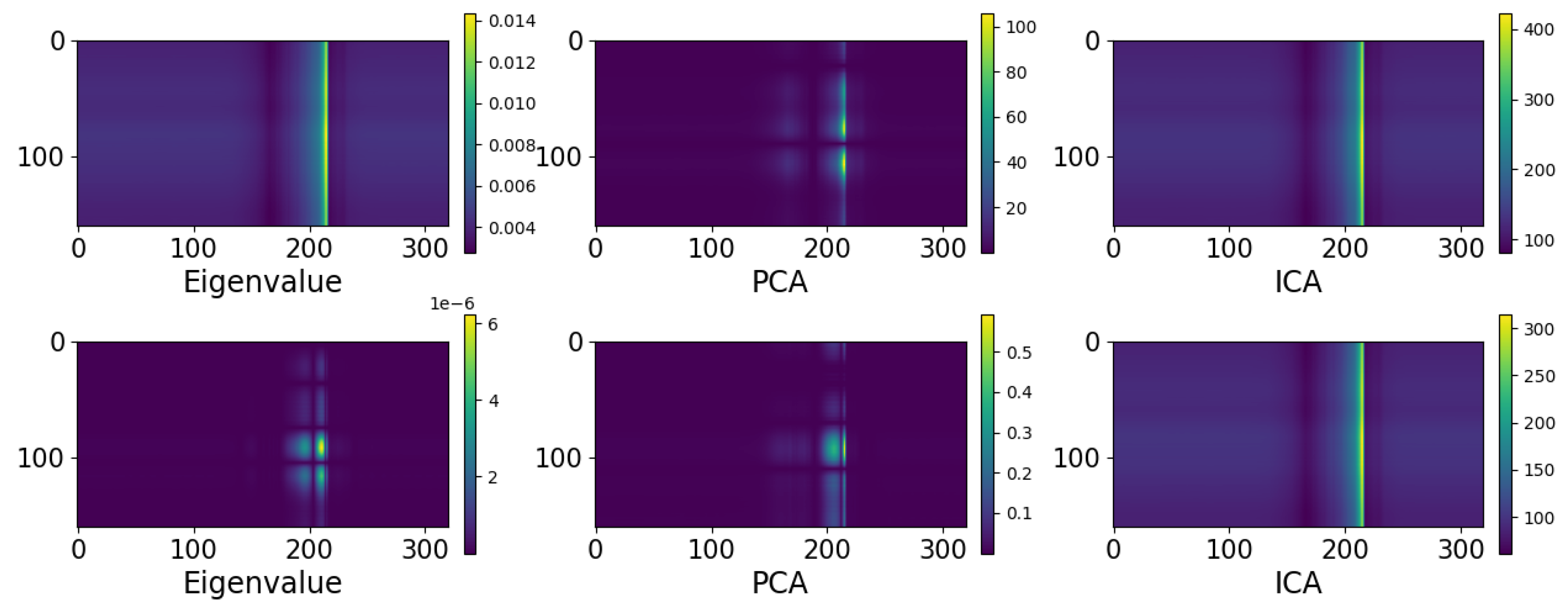
4.2. Optimal Latent Discovery: Single Channel
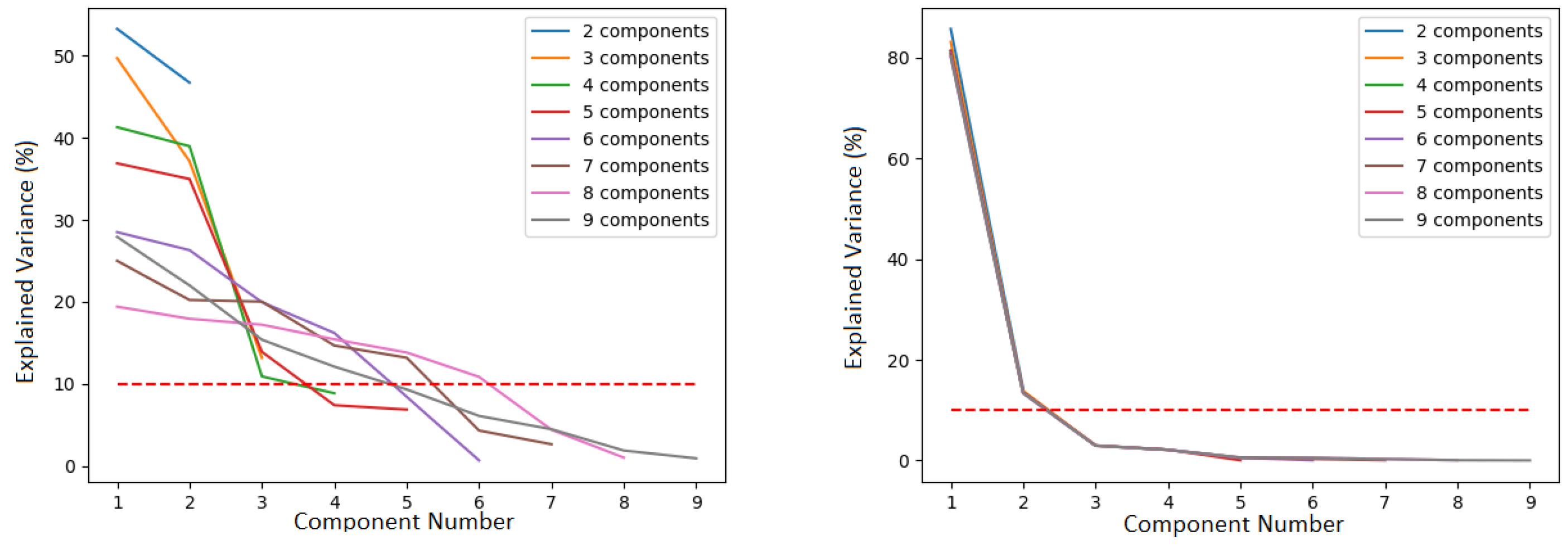
4.3. Optimal Latent Discovery: Complex Mixed Signals
Comparison: Impact of Increasing Number of Components
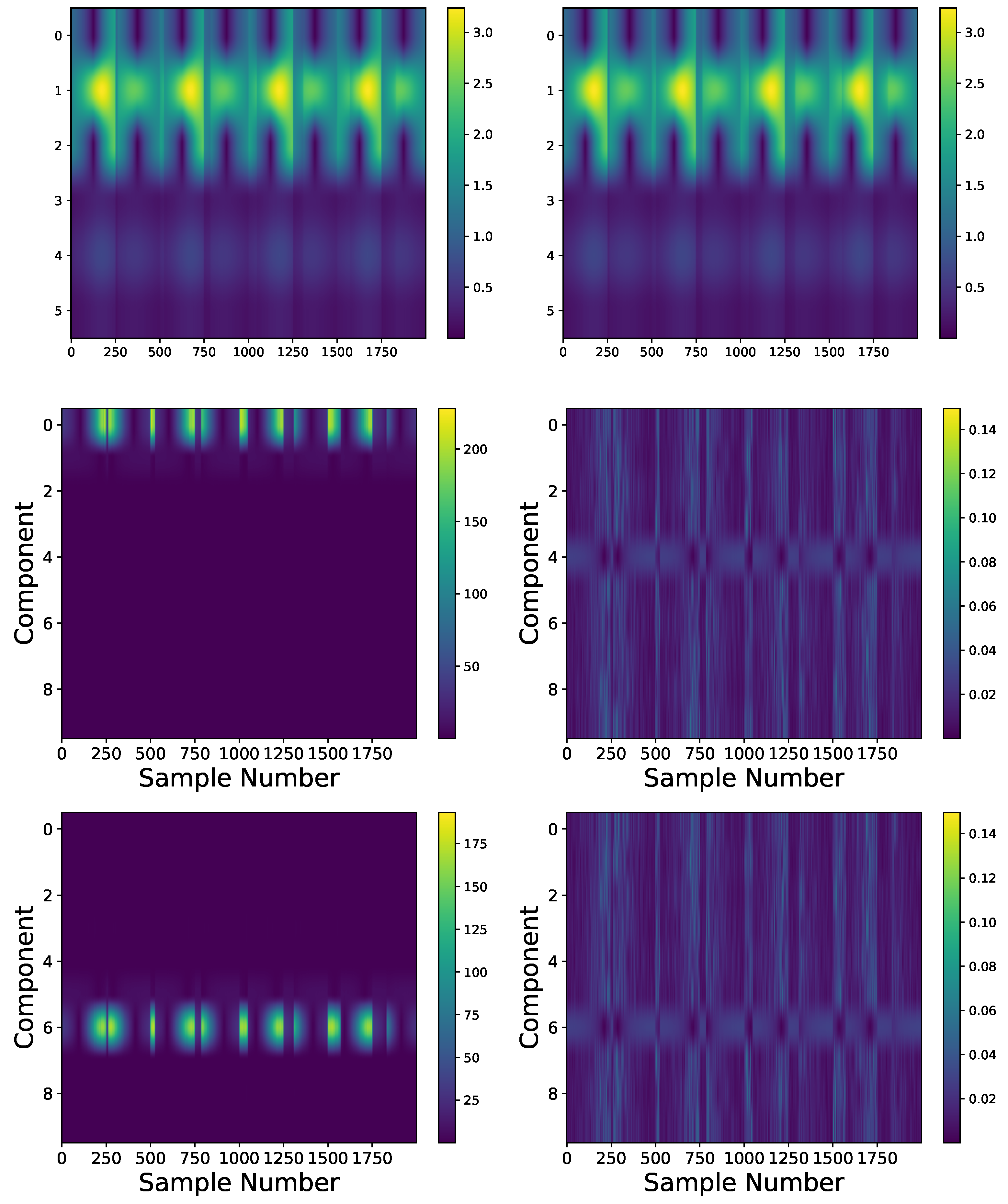
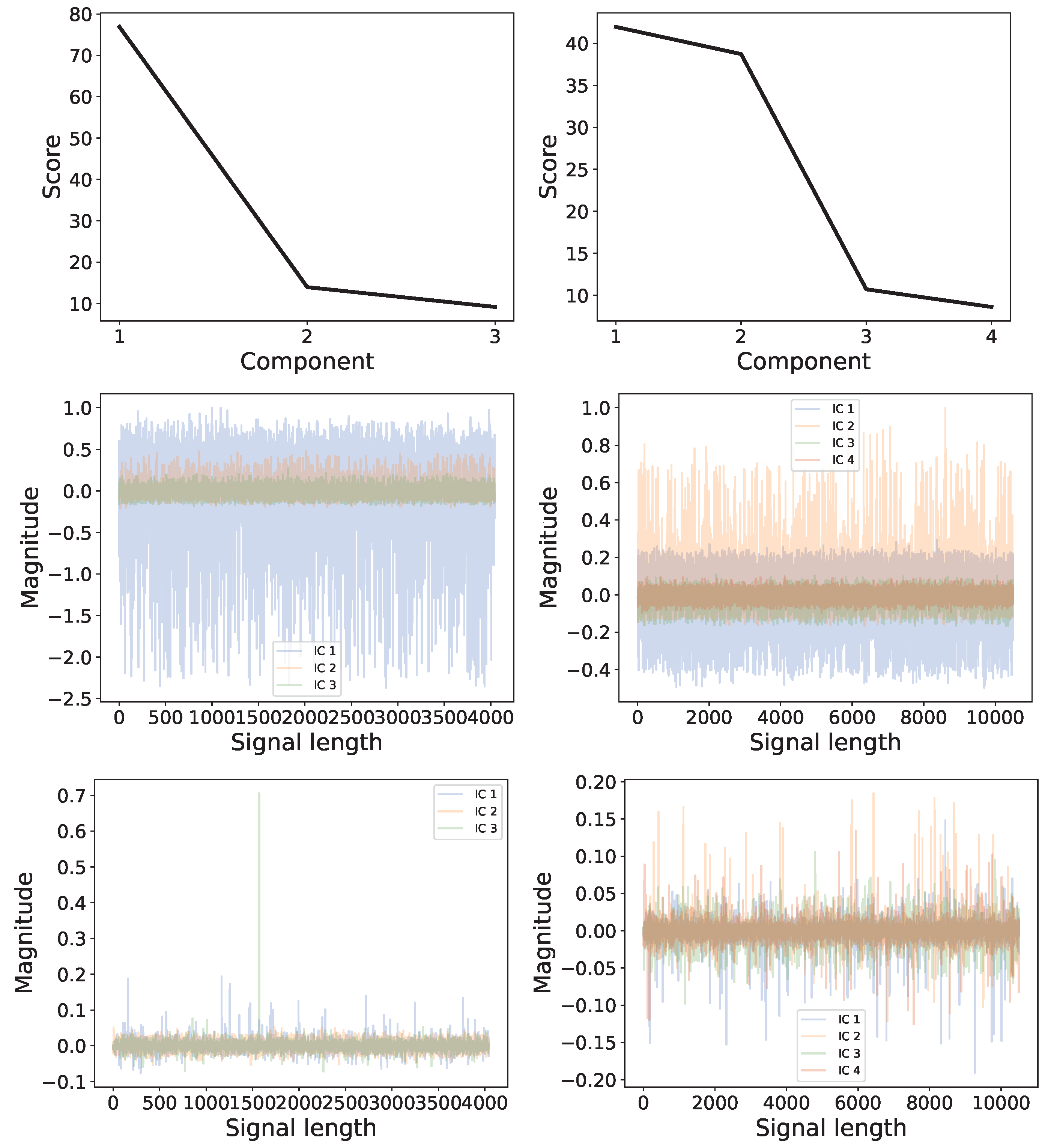
4.4. Experimental Datasets: Heartbeat Data
Comparison: LCON and LEXP
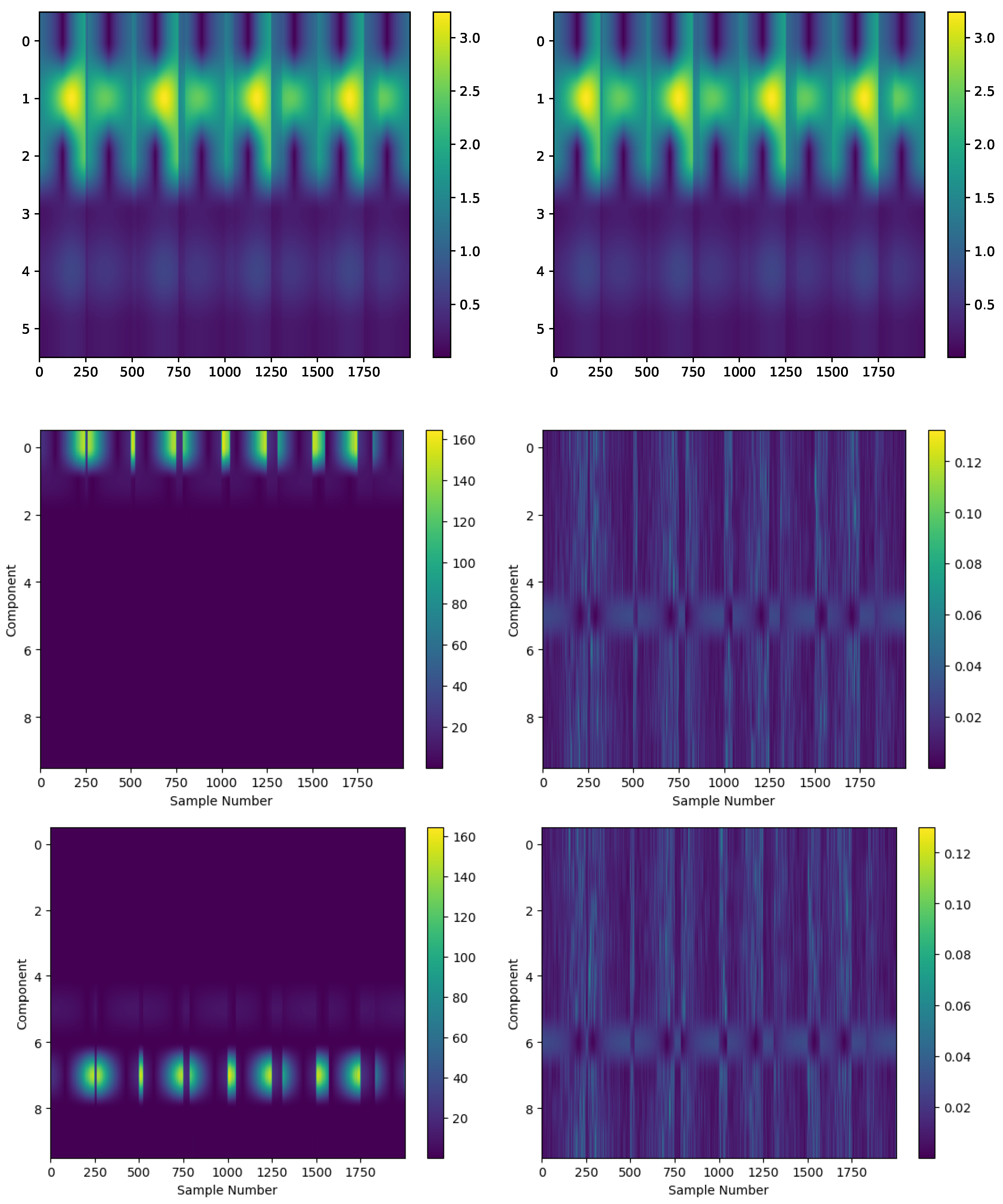
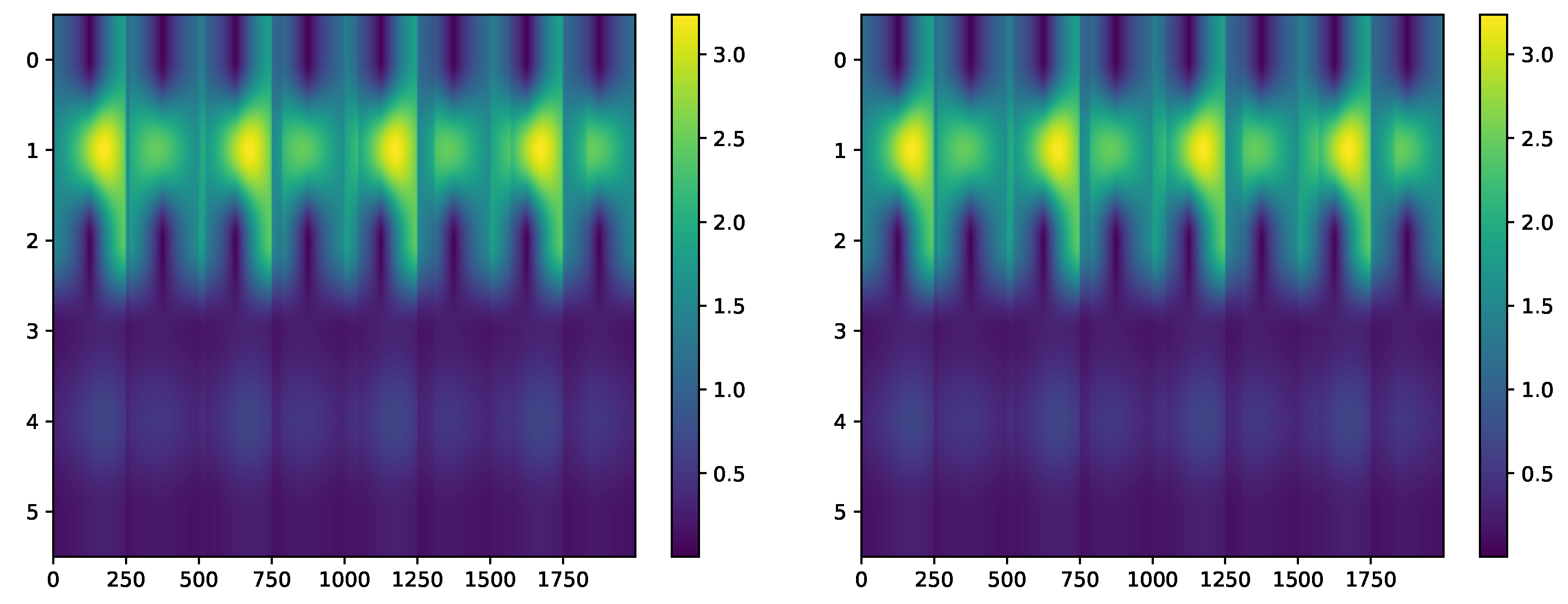
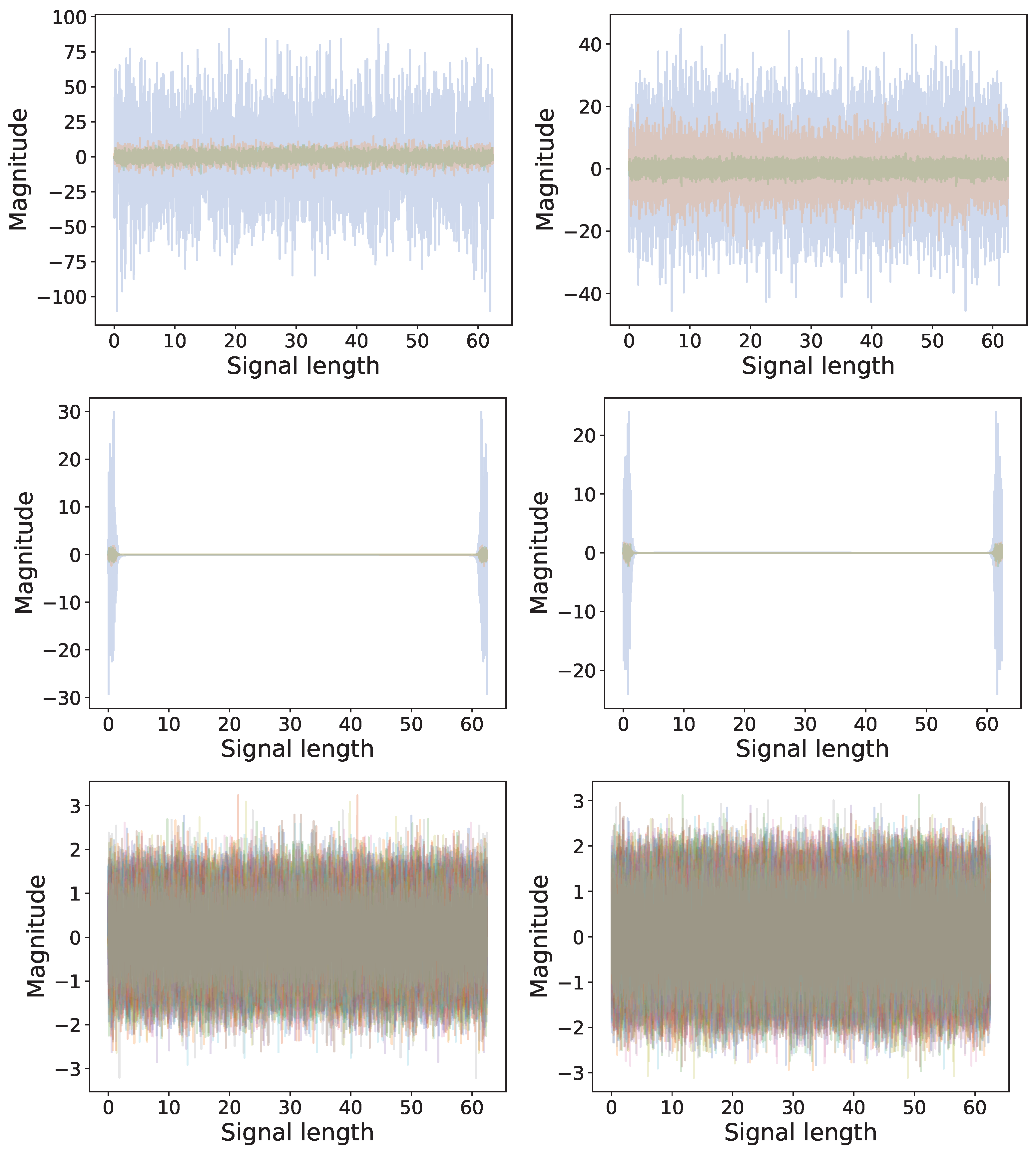
4.5. Experimental Data: Vibration Guided Wave-Based Monitoring
Comparison: LCON and LEXP
5. Results
6. Discussion
7. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| ICA | Independent Component Analysis |
| IC | Independent Component |
| PCA | Principal Component Analysis |
| LVM | Latent Variable Model |
| LS-PIE | Latent Space Perspicacity and Interpretation Enhancement |
References
- Wilke, D.N.; Heyns, P.S.; Schmidt, S. The Role of Untangled Latent Spaces in Unsupervised Learning Applied to Condition-Based Maintenance. In Modelling and Simulation of Complex Systems for Sustainable Energy Efficiency; Hammami, A., Heyns, P.S., Schmidt, S., Chaari, F., Abbes, M.S., Haddar, M., Eds.; Springer: Cham, Switzerland, 2022; pp. 38–49. [Google Scholar]
- Stevens, J.; Wilke, D.N.; Setshedi, I. Latent Space Perspicacity and Interpretation Enhancement (LS-PIE) Framework. arXiv 2023, arXiv:2307.05620. [Google Scholar]
- Liu, Y.; Li, Q. Latent Space Cartography: Visual Analysis of Vector Space Embeddings. Comput. Graph. Forum 2019, 38, 67–78. [Google Scholar] [CrossRef]
- Hyvärinen, A.; Oja, E. Independent Component Analysis: Algorithms and Applications. Neural Netw. 2000, 13, 411–430. [Google Scholar] [CrossRef]
- Jollife, I.T.; Cadima, J. Principal component analysis: A review and recent developments. Philos. Trans. R. Soc. Math. Phys. Eng. Sci. 2016, 374, 20150202. [Google Scholar] [CrossRef] [PubMed]
- Tharwat, A. Independent component analysis: An introduction. Appl. Comput. Inform. 2018, 17, 222–249. [Google Scholar] [CrossRef]
- Jaadi, Z. Principal Component Analysis (PCA): A Step-by-Step Explanation. Available online: https://builtin.com/data-science/step-step-explanation-principal-component-analysis (accessed on 31 May 2024).
- Toiviainen, M.; Corona, F.; Paaso, J.; Teppola, P. Blind source separation in diffuse reflectance NIR spectroscopy using independent component analysis. J. Chemom. 2010, 24, 514–522. [Google Scholar] [CrossRef]
- Choi, S.; Riken, A.C.; Park, H.M.; Lee, S.Y. Blind Source Separation and Independent Component Analysis: A Review. Neural Inf. Process. Lett. Rev. 2005, 6, 1. [Google Scholar]
- Cao, X.R.; Liu, R.W. General Approach to Blind Source Separation. IEEE Trans. Signal Process. 1996, 44, 562–571. [Google Scholar]
- Lever, J.; Krzywinski, M.; Altman, N. Points of Significance: Principal component analysis. Nat. Methods 2017, 14, 641–642. [Google Scholar] [CrossRef]
- Hyvärinen, A. What Is Independent Component Analysis? Available online: https://www.cs.helsinki.fi/u/ahyvarin/whatisica.shtml (accessed on 31 May 2024).
- De Lathauwer, L.; De Moor, B.; Vandewalle, J. An introduction to independent component analysis. J. Chemom. 2000, 14, 123–149. [Google Scholar] [CrossRef]
- Wiklund, K. The Cocktail Party Problem: Solutions and Applications. Ph.D. Thesis, McMaster University, Hamilton, ON, Canada, 2009. [Google Scholar]
- Brys, G.; Hubert, M.; Rousseeuw, P.J. A robustification of independent component analysis. J. Chemom. 2005, 19, 364–375. [Google Scholar] [CrossRef]
- Westad, F. Independent component analysis and regression applied on sensory data. J. Chemom. 2005, 19, 171–179. [Google Scholar] [CrossRef]
- Bach, F.R.; Jordan, M.I. Finding Clusters in Independent Component Analysis. In Proceedings of the 4th International Symposium on Independent Component Analysis and Blind Signal Separation (ICA2003), Nara, Japan, 1–4 April 2003. [Google Scholar]
- Widom, H. Hankel Matrices. Trans. Am. Math. Soc. 1966, 121, 1–35. [Google Scholar] [CrossRef]
- Yao, F.; Coquery, J.; Lê Cao, K.A. Independent Principal Component Analysis for biologically meaningful dimension reduction of large biological data sets. BMC Bioinform. 2012, 13, 24. [Google Scholar] [CrossRef]
- Zhao, X.; Ye, B. Similarity of signal processing effect between Hankel matrix-based SVD and wavelet transform and its mechanism analysis. Mech. Syst. Signal Process. 2009, 23, 1062–1075. [Google Scholar] [CrossRef]
- Wang, L.; Yang, D.; Chen, Z.; Lesniewski, P.J.; Naidu, R. Application of neural networks with novel independent component analysis methodologies for the simultaneous determination of cadmium, copper, and lead using an ISE array. J. Chemom. 2014, 28, 491–498. [Google Scholar] [CrossRef]
- Golyandina, N. On the choice of parameters in Singular Spectrum Analysis and related subspace-based methods. arXiv 2010, arXiv:1005.4374. [Google Scholar] [CrossRef]
- Broomhead, D.; King, G.P. Extracting qualitative dynamics from experimental data. Phys. D Nonlinear Phenom. 1986, 20, 217–236. [Google Scholar] [CrossRef]
- Zhang, T.; Ramakrishnan, R.; Livny, M. BIRCH: An efficient data clustering method for very large databases. In Proceedings of the 1996 ACM SIGMOD International Conference on Management of Data, SIGMOD’96, New York, NY, USA, 4–6 June 1996; pp. 103–114. [Google Scholar] [CrossRef]
- Ester, M.; Kriegel, H.P.; Sander, J.; Xu, X. A Density-Based Algorithm for Discovering Clusters in Large Spatial Databases with Noise. In Proceedings of the 2nd International Conference on Knowledge Discovery and Data Mining (KDD), Portland, OR, USA, 2–4 August 1996; AAAI Press: Washington, DC, USA, 1996; pp. 226–231. [Google Scholar]
- Schubert, E.; Sander, J.; Ester, M.; Kriegel, H.P.; Xu, X. DBSCAN revisited, revisited: Why and how you should (still) use DBSCAN. ACM Trans. Database Syst. (TODS) 2017, 42, 19. [Google Scholar] [CrossRef]
- Ankerst, M.; Breunig, M.M.; Kriegel, H.P.; Sander, J. OPTICS: Ordering points to identify the clustering structure. ACM SIGMOD Rec. 1999, 28, 49–60. [Google Scholar] [CrossRef]
- Schubert, E.; Gertz, M. Improving the Cluster Structure Extracted from OPTICS Plots. In Proceedings of the Conference “Lernen, Wissen, Daten, Analysen” (LWDA), Mannheim, Germany, 22–24 August 2018; pp. 318–329. [Google Scholar]
- Comaniciu, D.; Meer, P. Mean shift: A robust approach toward feature space analysis. IEEE Trans. Pattern Anal. Mach. Intell. 2002, 24, 603–619. [Google Scholar] [CrossRef]
- Frey, B.J.; Dueck, D. Clustering by Passing Messages Between Data Points. Science 2007, 315, 972–976. [Google Scholar] [CrossRef] [PubMed]
- Dueck, D. Affinity Propagation: Clustering Data by Passing Messages. Ph.D. Thesis, University of Toronto, Toronto, ON, Canada, 2009. [Google Scholar]
- Kaoungku, N.; Suksut, K.; Chanklan, R.; Kerdprasop, K.; Kerdprasop, N. The silhouette width criterion for clustering and association mining to select image features. Int. J. Mach. Learn. Comput. 2018, 8, 69–73. [Google Scholar] [CrossRef]
- Yuan, C.; Yang, H. Research on K-Value Selection Method of K-Means Clustering Algorithm. J 2019, 2, 226–235. [Google Scholar] [CrossRef]
- Tibshirani, R.; Walther, G.; Hastie, T. Estimating the number of clusters in a data set via the gap statistic. J. R. Stat. Soc. Ser. B (Stat. Methodol.) 2001, 63, 411–423. [Google Scholar] [CrossRef]
- Davies, D.L.; Bouldin, D.W. A Cluster Separation Measure. IEEE Trans. Pattern Anal. Mach. Intell. 1979, PAMI-1, 224–227. [Google Scholar] [CrossRef]
- Caliński, T.; Harabasz, J. A dendrite method for cluster analysis. Commun. Stat. 1974, 3, 1–27. [Google Scholar] [CrossRef]
- Dokmanic, I.; Parhizkar, R.; Ranieri, J.; Vetterli, M. Euclidean Distance Matrices: Essential Theory, Algorithms and Applications. IEEE Signal Process. Mag. 2015, 32, 12–30. [Google Scholar] [CrossRef]
- Singh, A. K-means with Three different Distance Metrics. Int. J. Comput. Appl. 2013, 67, 13–17. [Google Scholar] [CrossRef]
- Lahitani, A.R.; Permanasari, A.E.; Setiawan, N.A. Cosine similarity to determine similarity measure: Study case in online essay assessment. In Proceedings of the 2016 4th International Conference on Cyber and IT Service Management, Bandung, Indonesia, 26–27 April 2016; pp. 1–6. [Google Scholar] [CrossRef]
- Ghorbani, H. Mahalanobis distance and its application for detecting multivariate outliers. Facta Univ. Ser. Math. Inform. 2019, 34, 583. [Google Scholar] [CrossRef]
- Davies, M.E.; James, C.J. Source separation using single channel ICA. Signal Process. 2007, 87, 1819–1832. [Google Scholar] [CrossRef]
- Kachuee, M.; Fazeli, S.; Sarrafzadeh, M. ECG Heartbeat Classification: A Deep Transferable Representation. In Proceedings of the 2018 IEEE International Conference on Healthcare Informatics (ICHI), New York, NY, USA, 4–7 June 2018. [Google Scholar] [CrossRef]
- Setshedi, I.I.; Loveday, P.W.; Long, C.S.; Wilke, D.N. Estimation of rail properties using semi-analytical finite element models and guided wave ultrasound measurements. Ultrasonics 2019, 96, 240–252. [Google Scholar] [CrossRef]
- Setshedi, I.I.; Wilke, D.N.; Loveday, P.W. Feature detection in guided wave ultrasound measurements using simulated spectrograms and generative machine learning. NDT E Int. 2024, 143, 103036. [Google Scholar] [CrossRef]
- Loveday, P.W.; Long, C.S.; Ramatlo, D.A. Ultrasonic guided wave monitoring of an operational rail track. Struct. Health Monit. 2020, 19, 1666–1684. [Google Scholar] [CrossRef]
- Djuwari, D.; Kumar, D.K.; Palaniswami, M. Limitations of ICA for Artefact Removal. Conf. Proc. IEEE Eng. Med. Biol. Soc. 2005, 2005, 4685–4688. [Google Scholar] [CrossRef] [PubMed]






Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stevens, J.; Wilke, D.N.; Setshedi, I.I. Enhancing LS-PIE’s Optimal Latent Dimensional Identification: Latent Expansion and Latent Condensation. Math. Comput. Appl. 2024, 29, 65. https://doi.org/10.3390/mca29040065
Stevens J, Wilke DN, Setshedi II. Enhancing LS-PIE’s Optimal Latent Dimensional Identification: Latent Expansion and Latent Condensation. Mathematical and Computational Applications. 2024; 29(4):65. https://doi.org/10.3390/mca29040065
Chicago/Turabian StyleStevens, Jesse, Daniel N. Wilke, and Isaac I. Setshedi. 2024. "Enhancing LS-PIE’s Optimal Latent Dimensional Identification: Latent Expansion and Latent Condensation" Mathematical and Computational Applications 29, no. 4: 65. https://doi.org/10.3390/mca29040065
APA StyleStevens, J., Wilke, D. N., & Setshedi, I. I. (2024). Enhancing LS-PIE’s Optimal Latent Dimensional Identification: Latent Expansion and Latent Condensation. Mathematical and Computational Applications, 29(4), 65. https://doi.org/10.3390/mca29040065






