Noncoding RNAs as Key Regulators for Cardiac Development and Cardiovascular Diseases
Abstract
:1. Introduction
2. Noncoding RNAs
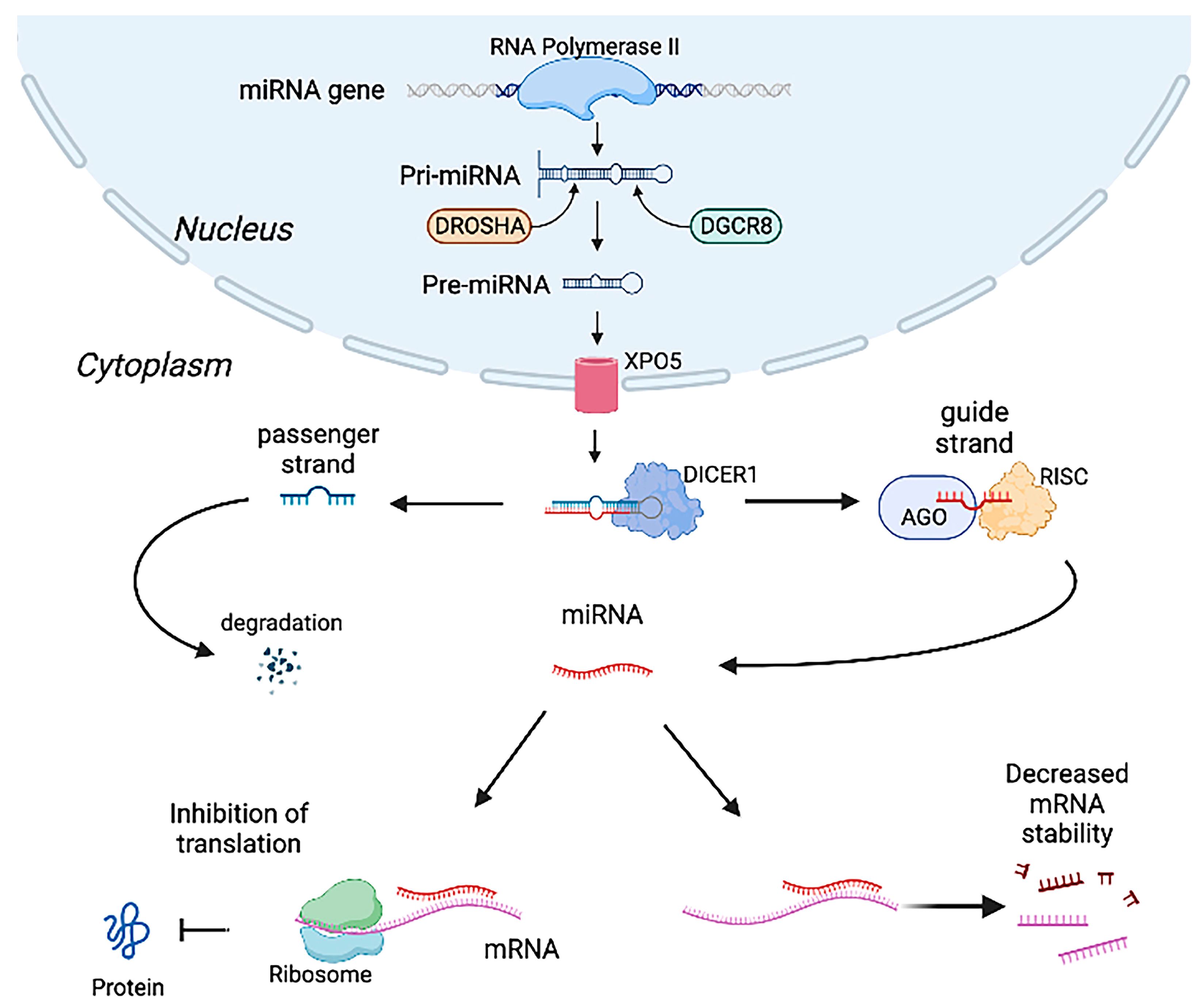
2.1. MiRNAs
2.2. LncRNAs
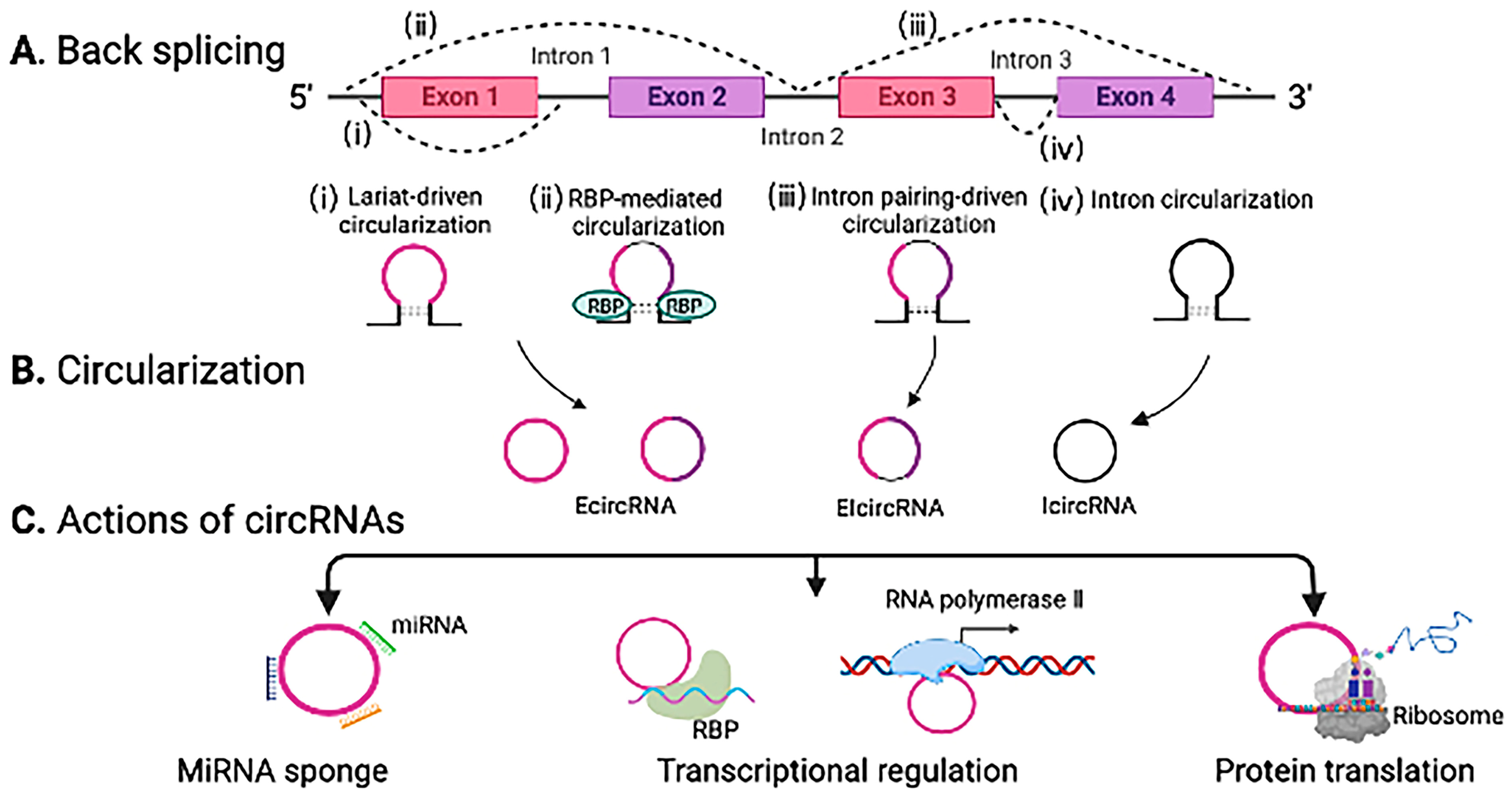
2.3. CircRNAs
3. NcRNAs in Cardiac Development
3.1. Roles of ncRNAs in the Formation of the Heart Tube and Cardiac Morphogenesis
3.2. Roles of ncRNAs in Cardiac Mesoderm Specification
3.3. Roles of ncRNAs in Embryonic CMs and Cardiac Progenitor Cells
4. NcRNAs in Cardiovascular Diseases
4.1. Atherosclerosis
4.2. Cardiac Arrhythmia
4.2.1. Atrial Fibrillation
4.2.2. Bradyarrhythmia
4.2.3. Ventricular Arrhythmias
4.2.4. Tachycardia
4.3. Cardiac Fibrosis
4.4. Cardiac Hypertrophy
4.5. Myocardial Infarction
4.6. Pulmonary Hypertension
5. Summary and Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tsao, C.W.; Aday, A.W.; Almarzooq, Z.I.; Anderson, C.A.; Arora, P.; Avery, C.L.; Baker-Smith, C.M.; Beaton, A.Z.; Boehme, A.K.; Buxton, A.E.; et al. Heart Disease and Stroke Statistics—2023 Update: A Report From the American Heart Association. Circulation 2023, 147, e93–e621. [Google Scholar] [CrossRef] [PubMed]
- Quinn, J.J.; Chang, H.Y. Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 2016, 17, 47–62. [Google Scholar] [CrossRef] [PubMed]
- Jonas, S.; Izaurralde, E. Towards a molecular understanding of microRNA-mediated gene silencing. Nat. Rev. Genet. 2015, 16, 421–433. [Google Scholar] [CrossRef]
- Boon, R.A.; Jaé, N.; Holdt, L.; Dimmeler, S. Long Noncoding RNAs. J. Am. Coll. Cardiol. 2016, 67, 1214–1226. [Google Scholar] [CrossRef]
- Ounzain, S.; Burdet, F.; Ibberson, M.; Pedrazzini, T. Discovery and functional characterization of cardiovascular long noncoding RNAs. J. Mol. Cell. Cardiol. 2015, 89, 17–26. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Chen, H.; Pan, T.; Jiang, C.; Zhao, Z.; Wang, Z.; Zhang, J.; Xu, J.; Li, X. LncRNA ontology: Inferring lncRNA functions based on chromatin states and expression patterns. Oncotarget 2015, 6, 39793–39805. [Google Scholar] [CrossRef] [Green Version]
- Ludwig, N.; Leidinger, P.; Becker, K.; Backes, C.; Fehlmann, T.; Pallasch, C.P.; Rheinheimer, S.; Meder, B.; Stähler, C.; Meese, E.; et al. Distribution of miRNA expression across human tissues. Nucleic Acids Res. 2016, 44, 3865–3877. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Feng, L.; Han, Z.; Li, Y.; Wu, A.; Shao, T.; Ding, N.; Li, L.; Deng, W.; Di, X.; et al. Extensive ceRNA-ceRNA interaction networks mediated by miRNAs regulate development in multiple rhesus tissues. Nucleic Acids Res. 2016, 44, 9438–9451. [Google Scholar] [CrossRef] [Green Version]
- Jiang, C.; Li, Y.; Zhao, Z.; Lu, J.; Chen, H.; Ding, N.; Wang, G.; Xu, J.; Li, X. Identifying and functionally characterizing tissue-specific and ubiquitously expressed human lncRNAs. Oncotarget 2016, 7, 7120–7133. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Liang, C.; Wong, K.-C.; Luo, J.; Zhang, Z. Mirsynergy: Detecting synergistic miRNA regulatory modules by overlapping neighbourhood expansion. Bioinformatics 2014, 30, 2627–2635. [Google Scholar] [CrossRef] [Green Version]
- Wang, D.; Wang, J.; Lu, M.; Song, F.; Cui, Q. Inferring the human microRNA functional similarity and functional network based on microRNA-associated diseases. Bioinformatics 2010, 26, 1644–1650. [Google Scholar] [CrossRef] [Green Version]
- Aonuma, T.; Bayoumi, A.S.; Tang, Y.; Kim, I.-M. A circular RNA regulator quaking: A novel gold mine to be unfolded in doxorubicin-mediated cardiotoxicity. Stem Cell Investig. 2018, 2, 19. [Google Scholar] [CrossRef]
- Archer, K.; Broskova, Z.; Bayoumi, A.S.; Teoh, J.-P.; Davila, A.; Tang, Y.; Su, H.; Kim, I.-M. Long Non-Coding RNAs as Master Regulators in Cardiovascular Diseases. Int. J. Mol. Sci. 2015, 16, 23651–23667. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bayoumi, A.S.; Aonuma, T.; Teoh, J.-P.; Tang, Y.; Kim, I.-M. Circular noncoding RNAs as potential therapies and circulating biomarkers for cardiovascular diseases. Acta Pharmacol. Sin. 2018, 39, 1100–1109. [Google Scholar] [CrossRef] [Green Version]
- Bayoumi, A.S.; Sayed, A.; Broskova, Z.; Teoh, J.-P.; Wilson, J.; Su, H.; Tang, Y.-L.; Kim, I.-M. Crosstalk between Long Noncoding RNAs and MicroRNAs in Health and Disease. Int. J. Mol. Sci. 2016, 17, 356. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moukette, B.; Barupala, N.P.; Aonuma, T.; Sepulveda, M.; Kawaguchi, S.; Kim, I.-M. Interactions between noncoding RNAs as epigenetic regulatory mechanisms in cardiovascular diseases. Methods Cell Biol. 2021, 166, 309–348. [Google Scholar] [CrossRef]
- Krol, J.; Loedige, I.; Filipowicz, W. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 2010, 11, 597–610. [Google Scholar] [CrossRef] [PubMed]
- Orang, A.V.; Safaralizadeh, R.; Kazemzadeh-Bavili, M. Mechanisms of miRNA-Mediated Gene Regulation from Common Downregulation to mRNA-Specific Upregulation. Int. J. Genom. 2014, 2014, 970607. [Google Scholar] [CrossRef] [Green Version]
- van Rooij, E. The Art of MicroRNA Research. Circ. Res. 2011, 108, 219–234. [Google Scholar] [CrossRef] [Green Version]
- De la Mata, M.; Gaidatzis, D.; Vitanescu, M.; Stadler, M.B.; Wentzel, C.; Scheiffele, P.; Filipowicz, W.; Großhans, H. Potent degradation of neuronal mi RNA s induced by highly complementary targets. EMBO Rep. 2015, 16, 500–511. [Google Scholar] [CrossRef] [Green Version]
- Fan, X.; Zhang, Z.; Zheng, L.; Wei, W.; Chen, Z. Long non-coding RNAs in the pathogenesis of heart failure: A literature review. Front. Cardiovasc. Med. 2022, 9, 950284. [Google Scholar] [CrossRef] [PubMed]
- Oo, J.A.; Brandes, R.P.; Leisegang, M.S. Long non-coding RNAs: Novel regulators of cellular physiology and function. Eur. J. Physiol. 2021, 474, 191–204. [Google Scholar] [CrossRef] [PubMed]
- Terashima, M.; Tange, S.; Ishimura, A.; Suzuki, T. MEG3 Long Noncoding RNA Contributes to the Epigenetic Regulation of Epithelial-Mesenchymal Transition in Lung Cancer Cell Lines. J. Biol. Chem. 2017, 292, 82–99. [Google Scholar] [CrossRef] [Green Version]
- Xu, M.; Chen, X.; Lin, K.; Zeng, K.; Liu, X.; Pan, B.; Xu, X.; Xu, T.; Hu, X.; Sun, L.; et al. The long noncoding RNA SNHG1 regulates colorectal cancer cell growth through interactions with EZH2 and miR-154-5p. Mol. Cancer 2018, 17, 141. [Google Scholar] [CrossRef] [Green Version]
- Tang, Y.; Wang, J.; Lian, Y.; Fan, C.; Zhang, P.; Wu, Y.; Li, X.; Xiong, F.; Li, X.; Li, G.; et al. Linking long non-coding RNAs and SWI/SNF complexes to chromatin remodeling in cancer. Mol. Cancer 2017, 16, 42. [Google Scholar] [CrossRef] [Green Version]
- Xiang, J.-F.; Yin, Q.-F.; Chen, T.; Zhang, Y.; Zhang, X.-O.; Wu, Z.; Zhang, S.; Wang, H.-B.; Ge, J.; Lu, X.; et al. Human colorectal cancer-specific CCAT1-L lncRNA regulates long-range chromatin interactions at the MYC locus. Cell Res. 2014, 24, 513–531. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Y.; Riching, A.S.; Knight, W.E.; Chi, C.; Broadwell, L.J.; Du, Y.; Abdel-Hafiz, M.; Ambardekar, A.V.; Irwin, D.C.; Proenza, C.; et al. Cardiomyocyte-Specific Long Noncoding RNA Regulates Alternative Splicing of the Triadin Gene in the Heart. Circulation 2022, 146, 699–714. [Google Scholar] [CrossRef]
- Ouyang, J.; Zhong, Y.; Zhang, Y.; Yang, L.; Wu, P.; Hou, X.; Xiong, F.; Li, X.; Zhang, S.; Gong, Z.; et al. Long non-coding RNAs are involved in alternative splicing and promote cancer progression. Br. J. Cancer 2021, 126, 1113–1124. [Google Scholar] [CrossRef]
- Han, Y.; Jin, G.; Pan, M.; Fang, Z.; Lu, D.; Cai, W.; Xu, C. Integrated Bioinformatics and Validation of lncRNA-Mediated ceRNA Network in Myocardial Ischemia/Reperfusion Injury. J. Immunol. Res. 2022, 2022, 7260801. [Google Scholar] [CrossRef]
- Imig, J.; Brunschweiger, A.; Brümmer, A.; Guennewig, B.; Mittal, N.; Kishore, S.; Tsikrika, P.; Gerber, A.P.; Zavolan, M.; Hall, J. miR-CLIP capture of a miRNA targetome uncovers a lincRNA H19–miR-106a interaction. Nat. Chem. Biol. 2015, 11, 107–114. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-T.; Kan, C.-H.; Liu, H.; Liu, Y.-H.; Wu, C.-C.; Kuo, Y.-P.; Chang, I.Y.-F.; Chang, K.-P.; Yu, J.-S.; Tan, B.C.-M. Modular scaffolding by lncRNA HOXA10-AS promotes oral cancer progression. Cell Death Dis. 2022, 13, 629. [Google Scholar] [CrossRef] [PubMed]
- Lu, B.-H.; Liu, H.-B.; Guo, S.-X.; Zhang, J.; Li, D.-X.; Chen, Z.-G.; Lin, F.; Zhao, G.-A. Long non-coding RNAs: Modulators of phenotypic transformation in vascular smooth muscle cells. Front. Cardiovasc. Med. 2022, 9, 959955. [Google Scholar] [CrossRef] [PubMed]
- Kung, J.T.Y.; Colognori, D.; Lee, J.T. Long Noncoding RNAs: Past, Present, and Future. Genetics 2013, 193, 651–669. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA circles function as efficient microRNA sponges. Nature 2013, 495, 384–388. [Google Scholar] [CrossRef]
- Ashwal-Fluss, R.; Meyer, M.; Pamudurti, N.R.; Ivanov, A.; Bartok, O.; Hanan, M.; Evantal, N.; Memczak, S.; Rajewsky, N.; Kadener, S. circRNA Biogenesis Competes with Pre-mRNA Splicing. Mol. Cell 2014, 56, 55–66. [Google Scholar] [CrossRef] [Green Version]
- Jiang, T.; Xia, Y.; Lv, J.; Li, B.; Li, Y.; Wang, S.; Xuan, Z.; Xie, L.; Qiu, S.; He, Z.; et al. A novel protein encoded by circMAPK1 inhibits progression of gastric cancer by suppressing activation of MAPK signaling. Mol. Cancer 2021, 20, 66. [Google Scholar] [CrossRef]
- Arráez-Aybar, L.; Turrero-Nogués, A.; Marantos-Gamarra, D. Embryonic Cardiac Morphometry in Carnegie Stages 15–23, from the Complutense University of Madrid Institute of Embryology Human Embryo Collection. Cells Tissues Organs 2008, 187, 211–220. [Google Scholar] [CrossRef]
- Lozano-Velasco, E.; Garcia-Padilla, C.; Muñoz-Gallardo, M.D.M.; Martinez-Amaro, F.J.; Caño-Carrillo, S.; Castillo-Casas, J.M.; Sanchez-Fernandez, C.; Aranega, A.E.; Franco, D. Post-Transcriptional Regulation of Molecular Determinants during Cardiogenesis. Int. J. Mol. Sci. 2022, 23, 2839. [Google Scholar] [CrossRef]
- Kay, M.; Soltani, B.M.; Aghdaei, F.H.; Ansari, H.; Baharvand, H. Hsa-miR-335 regulates cardiac mesoderm and progenitor cell differentiation. Stem Cell Res. Ther. 2019, 10, 191. [Google Scholar] [CrossRef]
- Garcia-Padilla, C.; Garcia-Lopez, V.; Aranega, A.; Franco, D.; Garcia-Martinez, V.; Lopez-Sanchez, C. Inhibition of RhoA and Cdc42 by miR-133a Modulates Retinoic Acid Signalling during Early Development of Posterior Cardiac Tube Segment. Int. J. Mol. Sci. 2022, 23, 4179. [Google Scholar] [CrossRef]
- Kay, M.; Soltani, B.M.; Nemir, M.; Aghagolzadeh, P.; Pezzuto, I.; Chouvardas, P.; Ruberto, F.; Movahedi, F.; Ansari, H.; Baharvand, H.; et al. The conserved long non-coding RNA CARMA regulates cardiomyocyte differentiation. Cardiovasc. Res. 2022, 118, 2339–2353. [Google Scholar] [CrossRef] [PubMed]
- Grote, P.; Herrmann, B.G. The long non-coding RNAFendrrlinks epigenetic control mechanisms to gene regulatory networks in mammalian embryogenesis. RNA Biol. 2013, 10, 1579–1585. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hazra, R.; Brine, L.; Garcia, L.; Benz, B.; Chirathivat, N.; Shen, M.M.; Wilkinson, J.E.; Lyons, S.K.; Spector, D.L. Platr4 is an early embryonic lncRNA that exerts its function downstream on cardiogenic mesodermal lineage commitment. Dev. Cell 2022, 57, 2450–2468.e7. [Google Scholar] [CrossRef] [PubMed]
- Kim, N.-J.; Lee, K.-H.; Son, Y.; Nam, A.-R.; Moon, E.-H.; Pyun, J.-H.; Park, J.; Kang, J.-S.; Lee, Y.J.; Cho, J.-Y. Spatiotemporal expression of long noncoding RNA Moshe modulates heart cell lineage commitment. RNA Biol. 2021, 18, 640–654. [Google Scholar] [CrossRef] [PubMed]
- Lumley, A.I.; Zhang, L.; Ernens, I.; Leszek, P.; Devaux, Y. The Long Noncoding RNA Landscape of Cardiac Regeneration in Zebrafish. Can. J. Cardiol. 2020, 37, 484–492. [Google Scholar] [CrossRef]
- Wamstad, J.A.; Alexander, J.M.; Truty, R.M.; Shrikumar, A.; Li, F.; Eilertson, K.E.; Ding, H.; Wylie, J.N.; Pico, A.R.; Capra, J.A.; et al. Dynamic and Coordinated Epigenetic Regulation of Developmental Transitions in the Cardiac Lineage. Cell 2012, 151, 206–220. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Trager, L.E.; Liu, X.; Hastings, M.H.; Xiao, C.; Guerra, J.; To, S.; Li, G.; Yeri, A.; Rodosthenous, R.; et al. lncExACT1 and DCHS2 Regulate Physiological and Pathological Cardiac Growth. Circulation 2022, 145, 1218–1233. [Google Scholar] [CrossRef]
- Milutinović, A.; Šuput, D.; Zorc-Pleskovič, R. Pathogenesis of atherosclerosis in the tunica intima, media, and adventitia of coronary arteries: An updated review. Bosn. J. Basic Med. Sci. 2020, 20, 21–30. [Google Scholar] [CrossRef]
- Cai, H.; Harrison, D.G. Endothelial Dysfunction in Cardiovascular Diseases: The Role of Oxidant Stress. Circ. Res. 2000, 87, 840–844. [Google Scholar] [CrossRef] [Green Version]
- Cheng, C.-K.; Shang, W.; Liu, J.; Cheang, W.-S.; Wang, Y.; Xiang, L.; Lau, C.-W.; Luo, J.-Y.; Ng, C.-F.; Huang, Y.; et al. Activation of AMPK/miR-181b Axis Alleviates Endothelial Dysfunction and Vascular Inflammation in Diabetic Mice. Antioxidants 2022, 11, 1137. [Google Scholar] [CrossRef]
- Cao, G.; Xuan, X.; Hu, J.; Zhang, R.; Jin, H.; Dong, H. How vascular smooth muscle cell phenotype switching contributes to vascular disease. Cell Commun. Signal. 2022, 20, 180. [Google Scholar] [CrossRef] [PubMed]
- Durham, A.L.; Speer, M.Y.; Scatena, M.; Giachelli, C.M.; Shanahan, C.M. Role of smooth muscle cells in vascular calcification: Implications in atherosclerosis and arterial stiffness. Cardiovasc. Res. 2018, 114, 590–600. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clarke, M.C.; Littlewood, T.D.; Figg, N.; Maguire, J.J.; Davenport, A.P.; Goddard, M.; Bennett, M.R. Chronic Apoptosis of Vascular Smooth Muscle Cells Accelerates Atherosclerosis and Promotes Calcification and Medial Degeneration. Circ. Res. 2008, 102, 1529–1538. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Tang, Y.; Yan, J. LncRNA-XIST Promotes Proliferation and Migration in ox-LDL Stimulated Vascular Smooth Muscle Cells through miR-539-5p/SPP1 Axis. Oxidative Med. Cell. Longev. 2022, 2022, 9911982. [Google Scholar] [CrossRef] [PubMed]
- You, L.; Zheng, Y.; Yang, J.; Hou, Q.; Wang, L.; Zhang, Y.; Zhao, C.; Xie, R. LncRNA MDRL Mitigates Atherosclerosis through miR-361/SQSTM1/NLRP3 Signaling. Mediat. Inflamm. 2022, 2022, 5463505. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Zhu, Y.; Zhu, J.; Xie, Y.; Wu, R.; Zhong, J.; Qiu, Z.; Jiang, L. circ_0086296 induced atherosclerotic lesions via the IFIT1/STAT1 feedback loop by sponging miR-576-3p. Cell. Mol. Biol. Lett. 2022, 27, 80. [Google Scholar] [CrossRef]
- Lin, J.; Liu, C.; Xu, J.; Li, S.; Dai, D.; Zhang, L.; Yonghui, P. Circ_0021155 can participate in the phenotypic transformation of human vascular smooth muscle cells via the miR-4459/TRPM7 axis. Biochem. Biophys. Res. Commun. 2022, 630, 133–142. [Google Scholar] [CrossRef]
- Lin, J.; Zhou, S.; Zhao, T.; Ju, T.; Zhang, L. TRPM7 channel regulates ox-LDL-induced proliferation and migration of vascular smooth muscle cells via MEK-ERK pathways. FEBS Lett. 2016, 590, 520–532. [Google Scholar] [CrossRef] [Green Version]
- Sheng, Y.; Yang, Z.; Feng, Z.; Wang, Y.; Ji, N. MicroRNA-499-5p promotes vascular smooth muscle cell proliferation and migration via inhibiting SOX6. Physiol. Genom. 2023, 55, 67–74. [Google Scholar] [CrossRef]
- Zhang, P.; Luo, J.; Wu, T.; Wang, X.; Yang, F.; Yu, Y.; Lu, L.; Yu, H. MiR-32-5p/AIDA Mediates OxLDL-Induced Endothelial Injury and Inflammation. Int. Heart J. 2022, 63, 928–938. [Google Scholar] [CrossRef]
- Li, H.; Song, D.; Liu, Q.; Li, L.; Sun, X.; Guo, J.; Li, D.; Li, P. miR-351 promotes atherosclerosis in diabetes by inhibiting the ITGB3/PIK3R1/Akt pathway and induces endothelial cell injury and lipid accumulation. Mol. Med. 2022, 28, 120. [Google Scholar] [CrossRef]
- Wang, W.; Tang, W.; Shan, E.; Zhang, L.; Chen, S.; Yu, C.; Gao, Y. MiR-130a-5p contributed to the progression of endothelial cell injury by regulating FAS. Eur. J. Histochem. 2022, 66, 3342. [Google Scholar] [CrossRef] [PubMed]
- Deng, Z.; Li, L. Effect of miR-663 on atherosclerosis by regulating the proliferation of vascular smooth muscle cells in lipid plaques. Vascular 2022. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Wang, X. miR-320a Targeting RGS5 Aggravates Atherosclerosis by Promoting Migration and Proliferation of ox-LDL–Stimulated Vascular Smooth Muscle Cells. J. Cardiovasc. Pharmacol. 2022, 80, 110–117. [Google Scholar] [CrossRef]
- Jiang, H.; Gong, R.; Wu, Y. miR-129-5p inhibits oxidized low-density lipoprotein-induced A7r5 cell viability and migration by targeting HMGB1 and the PI3k/Akt signaling pathway. Exp. Ther. Med. 2022, 23, 243. [Google Scholar] [CrossRef] [PubMed]
- Ren, X.; Zhu, H.; Deng, K.; Ning, X.; Li, L.; Liu, D.; Yang, B.; Shen, C.; Wang, X.; Wu, N.; et al. Long Noncoding RNA TPRG1-AS1 Suppresses Migration of Vascular Smooth Muscle Cells and Attenuates Atherogenesis via Interacting With MYH9 Protein. Arter. Thromb. Vasc. Biol. 2022, 42, 1378–1397. [Google Scholar] [CrossRef]
- Wu, J.-J.; Jin, J.; Li, Y.-H.; Wang, C.; Bai, J.; Jiang, Q.-J.; He, T.-X.; Nie, S.-J.; Li, D.-J.; Qu, L.-F. LncRNA FGF7-5 and lncRNA GLRX3 together inhibits the formation of carotid plaque via regulating the miR-2681-5p/ERCC4 axis in atherosclerosis. Cell Cycle 2023, 22, 165–182. [Google Scholar] [CrossRef]
- Gao, F.; Wang, X.; Luo, Z.; Hu, G.; Ma, M.; Liang, Y.; Xu, B.; Lin, X. LncRNA HOXA11-AS promotes vascular endothelial cell injury in atherosclerosis by regulating the miR-515-5p/ROCK1 axis. ESC Heart Fail. 2022, 9, 2259–2271. [Google Scholar] [CrossRef]
- Tang, F.; Zhang, S.; Wang, H.; Xu, S.; Yang, S.; Zhu, X.; Zeng, H.; Yang, Y. lncRNA H19 Promotes Ox-LDL-Induced Dysfunction of Human Aortic Endothelial Cells through the miR-152/VEGFA Axis. J. Healthc. Eng. 2022, 2022, 3795060. [Google Scholar] [CrossRef]
- An, F.; Yin, Y.; Ju, W. Long noncoding RNA DANCR expression and its predictive value in patients with atherosclerosis. Bioengineered 2022, 13, 6919–6928. [Google Scholar] [CrossRef]
- Zhang, T.; Feng, C.; Zhang, X.; Sun, B.; Bian, Y. Abnormal expression of long non-coding RNA rhabdomyosarcoma 2-associated transcript (RMST) participates in the pathological mechanism of atherosclerosis by regulating miR-224-3p. Bioengineered 2022, 13, 2648–2657. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Zong, W.; Liu, L.; Wu, J.; Pang, J. Knockdown of long non-coding RNA plasmacytoma variant translocation 1 relieves ox-LDL-induced endothelial cell injury through regulating microRNA-30c-5p in atherosclerosis. Bioengineered 2022, 13, 2791–2802. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Zheng, G.; Xie, H.; Guo, Y.; Zeng, H.; Fu, Y.; Liu, X. Study on the Mechanism of circRNA-0024103 Reducing Endothelial Cell Injury by Regulating miR-363/MMP-10. Contrast Media Mol. Imaging 2022, 2022, 1709325. [Google Scholar] [CrossRef] [PubMed]
- Mei, R.; Wu, M.; Ren, F. Knockdown of circ_0002194 protects against ox-LDL-induced cell damages via the regulation of miR-637/PACS2 axis in human vascular endothelial Cells. Interact. Cardiovasc. Thorac. Surg. 2022, 35, ivac210. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Li, L.; Ye, B.; Chen, W.; Zheng, G.; Xie, H.; Guo, Y. Knockdown of hsa_circ_0005699 attenuates inflammation and apoptosis induced by ox-LDL in human umbilical vein endothelial cells through regulation of the miR-450b-5p/NFKB1 axis. Mol. Med. Rep. 2022, 26, 290. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Zhou, X. CircRNA-PTPRA Knockdown Inhibits Atherosclerosis Progression by Repressing ox-LDL-Induced Endothelial Cell Injury via Sponging of miR-671-5p. Biochem. Genet. 2023, 61, 187–201. [Google Scholar] [CrossRef]
- Xiu, J.; Yang, Z.; Sui, Y.; Zhang, L.; Zhou, Y. CircNMD3 relieves endothelial cell injury induced by oxidatively modified low-density lipoprotein through regulating miR-498/ BMP and activin membrane-bound inhibitor (BAMBI) axis. Bioengineered 2022, 13, 12558–12571. [Google Scholar] [CrossRef]
- Wang, Y.; Chen, X.; Lu, Z.; Lai, C. Circ_0093887 regulated ox-LDL induced human aortic endothelial cells viability, apoptosis, and inflammation through modulating miR-758-3p/BAMBI axis in atherosclerosis. Clin. Hemorheol. Microcirc. 2022, 81, 343–358. [Google Scholar] [CrossRef]
- Ma, J.; Liu, J.; Li, T.; Ren, J. Hsa_circ_0030042 Facilitates the Proliferation and Migration of Vascular Smooth Muscle Cells via the miR-514a-3p/FOXO1 Axis. J. Endovasc. Ther. 2022, 29, 611–622. [Google Scholar] [CrossRef]
- Hou, X.; Dai, H.; Zheng, Y. Circular RNA hsa_circ_0008896 accelerates atherosclerosis by promoting the proliferation, migration and invasion of vascular smooth muscle cells via hsa-miR-633/CDC20B (cell division cycle 20B) axis. Bioengineered 2022, 13, 5987–5998. [Google Scholar] [CrossRef]
- Li, Y.; Wang, B. Circular RNA circCHFR downregulation protects against oxidized low-density lipoprotein-induced endothelial injury via regulation of microRNA-15b-5p/growth arrest and DNA damage inducible gamma. Bioengineered 2022, 13, 4481–4492. [Google Scholar] [CrossRef] [PubMed]
- Miao, R.; Qi, C.; Fu, Y.; Wang, Y.; Lang, Y.; Liu, W.; Zhang, Y.; Zhang, Z.; Liu, A.; Chai, H.; et al. Silencing of circARHGAP12 inhibits the progression of atherosclerosis via miR-630/EZH2/TIMP2 signal axis. J. Cell. Physiol. 2022, 237, 1057–1069. [Google Scholar] [CrossRef]
- Xie, L.; Huang, G.; Gao, M.; Huang, J.; Li, H.; Xia, H.; Xiang, X.; Wu, S.; Ruan, Y. Identification of Atrial Fibrillation-Related lncRNA Based on Bioinformatic Analysis. Dis. Markers 2022, 2022, 8307975. [Google Scholar] [CrossRef] [PubMed]
- Ye, Q.; Liu, Q.; Ma, X.; Bai, S.; Chen, P.; Zhao, Y.; Bai, C.; Liu, Y.; Liu, K.; Xin, M.; et al. MicroRNA-146b-5p promotes atrial fibrosis in atrial fibrillation by repressing TIMP4. J. Cell. Mol. Med. 2021, 25, 10543–10553. [Google Scholar] [CrossRef]
- Liu, D.; Yang, M.; Yao, Y.; He, S.; Wang, Y.; Cao, Z.; Chen, H.; Fu, Y.; Liu, H.; Zhao, Q. Cardiac Fibroblasts Promote Ferroptosis in Atrial Fibrillation by Secreting Exo-miR-23a-3p Targeting SLC7A11. Oxidative Med. Cell. Longev. 2022, 2022, 3961495. [Google Scholar] [CrossRef]
- Wiedmann, F.; Kraft, M.; Kallenberger, S.; Büscher, A.; Paasche, A.; Blochberger, P.L.; Seeger, T.; Jávorszky, N.; Warnecke, G.; Arif, R.; et al. MicroRNAs Regulate TASK-1 and Are Linked to Myocardial Dilatation in Atrial Fibrillation. J. Am. Heart Assoc. 2022, 11, e023472. [Google Scholar] [CrossRef]
- Barstow, C.; McDivitt, J.D. Cardiovascular Disease Update: Bradyarrhythmias. FP Essent. 2017, 454, 18–23. [Google Scholar] [PubMed]
- Yanni, J.; D’Souza, A.; Wang, Y.; Li, N.; Hansen, B.J.; Zakharkin, S.O.; Smith, M.; Hayward, C.; Whitson, B.A.; Mohler, P.J.; et al. Silencing miR-370-3p rescues funny current and sinus node function in heart failure. Sci. Rep. 2020, 10, 11279. [Google Scholar] [CrossRef] [PubMed]
- Aminu, A.J.; Petkova, M.; Atkinson, A.J.; Yanni, J.; Morris, A.D.; Simms, R.T.; Chen, W.; Yin, Z.; Kuniewicz, M.; Holda, M.K.; et al. Further insights into the molecular complexity of the human sinus node—The role of ‘novel’ transcription factors and microRNAs. Prog. Biophys. Mol. Biol. 2021, 166, 86–104. [Google Scholar] [CrossRef]
- Li, P.; Wang, K.; Yin, J.; Qi, L.; Hu, H.; Yang, P.; Shi, Y.; Li, Y.; Feng, M.; Lyu, H.; et al. lncRNA LOC100911717-targeting GAP43-mediated sympathetic remodeling after myocardial infarction in rats. Front. Cardiovasc. Med. 2022, 9, 1019435. [Google Scholar] [CrossRef]
- Liang, Y.; Wang, B.; Huang, H.; Wang, M.; Wu, Q.; Zhao, Y.; He, Y. Silenced SOX2-OT alleviates ventricular arrhythmia associated with heart failure by inhibiting NLRP3 expression via regulating miR-2355-3p. Immun. Inflamm. Dis. 2020, 9, 255–264. [Google Scholar] [CrossRef]
- Shi, Y.; Qiao, L.; Han, F.; Xie, X.; Wang, W. MiR-1231 regulates L-calcium in ventricular arrhythmia in chronic heart failure. Minerva Med. 2021, 112, 305–306. [Google Scholar] [CrossRef] [PubMed]
- Djalinac, N.; Kolesnik, E.; Maechler, H.; Scheruebel-Posch, S.; Pelzmann, B.; Rainer, P.P.; Foessl, I.; Wallner, M.; Scherr, D.; Heinemann, A.; et al. miR-1183 Is a Key Marker of Remodeling upon Stretch and Tachycardia in Human Myocardium. Int. J. Mol. Sci. 2022, 23, 6962. [Google Scholar] [CrossRef] [PubMed]
- Park, H.; Park, H.; Park, J. Circulating microRNA-423 attenuates the phosphorylation of calcium handling proteins in atrial fibrillation. Mol. Med. Rep. 2022, 25, 186. [Google Scholar] [CrossRef]
- Xu, Y.; Li, W.; Wan, K.; Liang, Y.; Jiang, X.; Wang, J.; Mui, D.; Li, Y.; Tang, S.; Guo, J.; et al. Myocardial Tissue Reverse Remodeling After Guideline-Directed Medical Therapy in Idiopathic Dilated Cardiomyopathy. Circ. Heart Fail. 2021, 14, e007944. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Tang, Y.; Cai, X.; Gong, J. Long Noncoding RNAs Testis Development Related Gene 1 Aggravates Transforming Growth Factor-β1–Induced Fibrogenesis and Inflammatory Response of Cardiac Fibroblasts Via miR-605-3p/Tumor Necrosis Factor Receptor Superfamily-21 Axis. J. Cardiovasc. Pharmacol. 2022, 79, 296–303. [Google Scholar] [CrossRef] [PubMed]
- Quaife, N.M.; Chothani, S.; Schulz, J.F.; Lindberg, E.L.; Vanezis, K.; Adami, E.; O’Fee, K.; Greiner, J.; Litviňuková, M.; van Heesch, S.; et al. LINC01013 Is a Determinant of Fibroblast Activation and Encodes a Novel Fibroblast-Activating Micropeptide. J. Cardiovasc. Transl. Res. 2022, 16, 77–85. [Google Scholar] [CrossRef] [PubMed]
- Chignon, A.; Argaud, D.; Boulanger, M.-C.; Mkannez, G.; Bon-Baret, V.; Li, Z.; Thériault, S.; Bossé, Y.; Mathieu, P. Genome-wide chromatin contacts of super-enhancer-associated lncRNA identify LINC01013 as a regulator of fibrosis in the aortic valve. PLoS Genet. 2022, 18, e1010010. [Google Scholar] [CrossRef]
- Feng, B.; Liu, J.; Wang, E.; Su, Z.; Chakrabarti, S. Endothelial derived miRNA-9 mediated cardiac fibrosis in diabetes and its regulation by ZFAS1. PLoS ONE 2022, 17, e0276076. [Google Scholar] [CrossRef]
- Peng, T.; Liu, M.; Hu, L.; Guo, D.; Di Wang, D.; Qi, B.; Ren, G.; Hu, C.; Zhang, F.; Chun, H.J.; et al. LncRNA Airn alleviates diabetic cardiac fibrosis by inhibiting activation of cardiac fibroblasts via a m6A-IMP2-p53 axis. Biol. Direct 2022, 17, 32. [Google Scholar] [CrossRef]
- Zhou, J.; Tian, G.; Quan, Y.; Kong, Q.; Huang, F.; Li, J.; Wu, W.; Tang, Y.; Zhou, Z.; Liu, X. The long non-coding RNA THBS1-AS1 promotes cardiac fibroblast activation in cardiac fibrosis by regulating TGFBR1. J. Clin. Investig. 2023, 8, e160745. [Google Scholar] [CrossRef]
- Tan, W.; Wang, K.; Yang, X.; Wang, N.; Jiang, T.-B. LncRNA HOTAIR promotes myocardial fibrosis in atrial fibrillation through binding with PTBP1 to increase the stability of Wnt5a. Int. J. Cardiol. 2022, 369, 21–28. [Google Scholar] [CrossRef]
- Frey, N.; Olson, E. Cardiac Hypertrophy: The Good, the Bad, and the Ugly. Annu. Rev. Physiol. 2003, 65, 45–79. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, M.; Sadoshima, J. Mechanisms of physiological and pathological cardiac hypertrophy. Nat. Rev. Cardiol. 2018, 15, 387–407. [Google Scholar] [CrossRef] [PubMed]
- Tu, S.; Wang, X.-Y.; Zeng, L.-X.; Shen, Z.-J.; Zhang, Z.-H. LncRNA TINCR improves cardiac hypertrophy by regulating the miR-211-3p-VEGFB-SDF-1α-CXCR4 pathway. Lab. Investig. 2022, 102, 253–262. [Google Scholar] [CrossRef]
- Wang, E.R.; Jarrah, A.; Benard, L.; Chen, J.; Schwarzkopf, M.; Hadri, L.; Tarzami, S.T. Deletion of CXCR4 in cardiomyocytes exacerbates cardiac dysfunction following isoproterenol administration. Gene Ther. 2014, 21, 496–506. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Sha, Z.; Zhu, X.; Xu, W.; Yuan, W.; Yang, T.; Jin, B.; Yan, Y.; Chen, R.; Wang, S.; et al. Targeting miR-30d reverses pathological cardiac hypertrophy. Ebiomedicine 2022, 81, 104108. [Google Scholar] [CrossRef]
- Lin, X.; Zhang, L.; Zhang, W.; Lei, X.; Lu, Q.; Ma, A. Circular RNA circ_0001006 aggravates cardiac hypertrophy via miR-214-3p/PAK6 axis. Aging 2022, 14, 2210–2220. [Google Scholar] [CrossRef]
- Yuan, J.; Yuan, G. miR-212 Promotes Cardiomyocyte Hypertrophy through Regulating Transcription Factor 7 Like 2. Mediat. Inflamm. 2022, 2022, 5187218. [Google Scholar] [CrossRef]
- Chang, W.-T.; Shih, J.-Y.; Lin, Y.-W.; Huang, T.-L.; Chen, Z.-C.; Chen, C.-L.; Chu, J.-S.; Liu, P.Y. miR-21 upregulation exacerbates pressure overload-induced cardiac hypertrophy in aged hearts. Aging 2022, 14, 5925–5945. [Google Scholar] [CrossRef]
- Silva, T.D.O.; Lino, C.A.; Miranda, J.B.; Balbino-Silva, C.S.; Lunardon, G.; Lima, V.M.; Jensen, L.; Donato, J.; Irigoyen, M.C.; Barreto-Chaves, M.L.M.; et al. The miRNA-143-3p–Sox6–Myh7 pathway is altered in obesogenic diet-induced cardiac hypertrophy. Exp. Physiol. 2022, 107, 892–905. [Google Scholar] [CrossRef]
- Li, D.; Shen, M.; Deng, X.; Bai, Y. MicroRNA miR-27a-3p accelerates cardiac hypertrophy by targeting neuro-oncological ventral antigen 1. Bioengineered 2022, 13, 8982–8993. [Google Scholar] [CrossRef]
- Gaddam, R.R.; Kim, Y.; Jacobs, J.S.; Yoon, J.; Li, Q.; Cai, A.; Shankaiahgari, H.; London, B.; Irani, K.; Vikram, A. The microRNA-204-5p inhibits APJ signalling and confers resistance to cardiac hypertrophy and dysfunction. Clin. Transl. Med. 2022, 12, e693. [Google Scholar] [CrossRef] [PubMed]
- Jia, G.; Liang, C.; Li, W.; Dai, H. MiR-410-3p facilitates Angiotensin II–induced cardiac hypertrophy by targeting Smad7. Bioengineered 2021, 13, 119–127. [Google Scholar] [CrossRef] [PubMed]
- Bi, X.; Zhang, Y.; Yu, Y.; Yuan, J.; Xu, S.; Liu, F.; Ye, J.; Liu, P. MiRNA-339-5p promotes isoproterenol-induced cardiomyocyte hypertrophy by targeting VCP to activate the mTOR signaling. Cell Biol. Int. 2021, 46, 288–299. [Google Scholar] [CrossRef]
- Li, G.; Shao, Y.; Guo, H.C.; Zhi, Y.; Qiao, B.; Ma, K.; Du, J.; Lai, Y.Q.; Li, Y. MicroRNA-27b-3p down-regulates FGF1 and aggravates pathological cardiac remodelling. Cardiovasc. Res. 2021, 118, 2139–2151. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.-Y.; Yue, L.-J.; Luo, Y.-C.; Lu, J.; Wu, G.-D.; Sheng, S.-Q.; Shi, Y.-Q.; Dong, Z.-X. SUMOylation of SIRT1 activating PGC-1α/PPARα pathway mediates the protective effect of LncRNA-MHRT in cardiac hypertrophy. Eur. J. Pharmacol. 2022, 930, 175155. [Google Scholar] [CrossRef]
- Chen, F.; Li, W.; Zhang, D.; Fu, Y.; Yuan, W.; Luo, G.; Liu, F.; Luo, J. MALAT1 regulates hypertrophy of cardiomyocytes by modulating the miR-181a/HMGB2 pathway. Eur. J. Histochem. 2022, 66, 3426. [Google Scholar] [CrossRef]
- Zhu, C.; Wang, M.; Yu, X.; Shui, X.; Tang, L.; Chen, Z.; Xiong, Z. lncRNA NBR2 attenuates angiotensin II-induced myocardial hypertrophy through repressing ER stress via activating LKB1/AMPK/Sirt1 pathway. Bioengineered 2022, 13, 13667–13679. [Google Scholar] [CrossRef]
- Yang, Y.; Mbikyo, M.B.; Zhang, J.; Zhang, Y.; Zhang, N.; Li, Z. The lncRNA MIAT regulates CPT-1a mediated cardiac hypertrophy through m6A RNA methylation reading protein Ythdf2. Cell Death Discov. 2022, 8, 167. [Google Scholar] [CrossRef]
- Wang, H.; Lian, X.; Gao, W.; Gu, J.; Shi, H.; Ma, Y.; Li, Y.; Fan, Y.; Wang, Q.; Wang, L. Long noncoding RNA H19 suppresses cardiac hypertrophy through the MicroRNA-145-3p/SMAD4 axis. Bioengineered 2022, 13, 3826–3839. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Li, J.; Wang, X.; Zeng, Z.; Zhang, H.; Zou, Z.; Huang, N.; Sun, X. LincRNA RMRP Regulates Phenylephrine-induced Cardiomyocyte Hypertrophy by Means of Targeting miR-1. J. Cardiovasc. Pharmacol. 2022, 80, 709–717. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Wang, Y.; An, Y.; Wang, J.; Gao, Y. The Particular Expression Profiles of Circular RNA in Peripheral Blood of Myocardial Infarction Patients by RNA Sequencing. Front. Cardiovasc. Med. 2022, 9, 810257. [Google Scholar] [CrossRef]
- Yang, X.; Dai, R.; Qin, Z.; Cai, R.; Xu, Y.; Su, Q. LncRNA MALAT1 functions as a biomarker of no-reflow phenomenon in ST-segment elevation myocardial infarction patients receiving primary percutaneous coronary intervention. Sci. Rep. 2022, 12, 3294. [Google Scholar] [CrossRef] [PubMed]
- Elbaz, M.; Faccini, J.; Laperche, C.; Grazide, M.-H.; Ruidavets, J.-B.; Vindis, C. MiR-223 and MiR-186 Are Associated with Long-Term Mortality after Myocardial Infarction. Biomolecules 2022, 12, 1243. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Jin, J.; Liu, E.; Zhang, J. A novel circulating biomarker lnc-MALAT1 for acute myocardial infarction: Its relationship with disease risk, features, cytokines, and major adverse cardiovascular events. J. Clin. Lab. Anal. 2022, 36, e24771. [Google Scholar] [CrossRef]
- Liu, X.-M.; Zhang, Z.; Zhong, J.; Li, N.; Wang, T.; Wang, L.; Zhang, Q. Long non-coding RNA MALAT1 modulates myocardial ischemia-reperfusion injury through the PI3K/Akt/eNOS pathway by sponging miRNA-133a-3p to target IGF1R expression. Eur. J. Pharmacol. 2022, 916, 174719. [Google Scholar] [CrossRef]
- Nugroho, A.B.; Stafford, N.; Zi, M.; Prehar, S.; Potter, R.; Kwon, D.; Kohar, Y.S.; Triastuti, E.; Bui, T.A.; Cartwright, E.J.; et al. Micro RNA-411 Expression Improves Cardiac Phenotype Following Myocardial Infarction in Mice. JACC Basic Transl. Sci. 2022, 7, 859–875. [Google Scholar] [CrossRef]
- Aonuma, T.; Moukette, B.; Kawaguchi, S.; Barupala, N.P.; Sepúlveda, M.N.; Frick, K.; Tang, Y.; Guglin, M.; Raman, S.V.; Cai, C.; et al. MiR-150 Attenuates Maladaptive Cardiac Remodeling Mediated by Long Noncoding RNA MIAT and Directly Represses Profibrotic Hoxa4. Circ. Hear. Fail. 2022, 15, e008686. [Google Scholar] [CrossRef]
- Hayasaka, T.; Takehara, N.; Aonuma, T.; Kano, K.; Horiuchi, K.; Nakagawa, N.; Tanaka, H.; Kawabe, J.-I.; Hasebe, N. Sarcopenia-derived exosomal micro-RNA 16-5p disturbs cardio-repair via a pro-apoptotic mechanism in myocardial infarction in mice. Sci. Rep. 2021, 11, 19163. [Google Scholar] [CrossRef]
- Zeng, H.; Hu, F.; Duan, Y.; Li, H.; Wang, Y. Expression of lncRNA APF in Peripheral Blood of Patients with Acute Myocardial Infarction Caused by Coronary Heart Disease and its Clinical Significance. Int. Heart J. 2022, 63, 21–434. [Google Scholar] [CrossRef] [PubMed]
- Bampatsias, D.; Mavroeidis, I.; Tual-Chalot, S.; Vlachogiannis, N.I.; Bonini, F.; Sachse, M.; Mavraganis, G.; Mareti, A.; Kritsioti, C.; Laina, A.; et al. Beta-Secretase-1 Antisense RNA Is Associated with Vascular Ageing and Atherosclerotic Cardiovascular Disease. Thromb. Haemost. 2022, 122, 1932–1942. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Chen, R.; Wang, L.; Lu, Z.; Li, Y.; Tang, D. Molecular mechanism of CAIF inhibiting myocardial infarction by sponging miR-488 and regulating AVEN expression. Mol. Med. Rep. 2022, 26, 270. [Google Scholar] [CrossRef]
- Zhang, H.; Kou, X.; Xiao, D.; Yu, Z. Long non-coding RNA lincRNA-erythroid prosurvival attenuates inflammation by enhancing myosin heavy chain 6 stability through recruitment of heterogeneous nuclear ribonucleoprotein L in myocardial infarction. Bioengineered 2022, 13, 14426–14437. [Google Scholar] [CrossRef] [PubMed]
- Gao, F.; Wang, X.; Fan, T.; Luo, Z.; Ma, M.; Hu, G.; Li, Y.; Liang, Y.; Lin, X.; Xu, B. LncRNA LINC00461 exacerbates myocardial ischemia–reperfusion injury via microRNA-185-3p/Myd88. Mol. Med. 2022, 28, 33. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Luo, J.-Y.; Liu, F.; Zhang, X.-H.; Luo, F.; Yang, Y.-N.; Li, X.-M. Long noncoding RNA MALAT1 polymorphism predicts MACCEs in patients with myocardial infarction. BMC Cardiovasc. Disord. 2022, 22, 152. [Google Scholar] [CrossRef]
- Liu, W.; Lin, W.; Yu, L. Long non-coding RNA muscleblind like splicing regulator 1 antisense RNA 1 (LncRNA MBNL1-AS1) promotes the progression of acute myocardial infarction by regulating the microRNA-132-3p/SRY-related high-mobility-group box 4 (SOX4) axis. Bioengineered 2022, 13, 1424–1435. [Google Scholar] [CrossRef]
- Chen, K.; Xi, M.; Huang, Q.; Wu, H.; Lu, G.; Song, S.; Shi, W. Long non-coding RNA MCM3AP antisense RNA 1 silencing upregulates microRNA-24-3p to accelerate proliferation and migration of vascular endothelial cells in myocardial infarction rats by reducing EIF4G2. Cell Cycle 2022, 21, 674–684. [Google Scholar] [CrossRef]
- Zhou, J. LncRNA MIAT promotes hypoxia-induced H9C2 cell pyroptosis via binding toSF1to inhibitCGRPtranscription. Exp. Physiol. 2021, 107, 58–67. [Google Scholar] [CrossRef]
- Wang, X.; Ren, L.; Chen, S.; Tao, Y.; Zhao, D.; Wu, C. Long non-coding RNA MIR4435-2HG/microRNA-125a-5p axis is involved in myocardial ischemic injuries. Bioengineered 2022, 13, 10707–10720. [Google Scholar] [CrossRef]
- Yao, J.; Ma, R.; Wang, C.; Zhao, G. LncRNA-HOTAIR Inhibits H9c2 Apoptosis After Acute Myocardial Infarction via miR-206/FN1 Axis. Biochem. Genet. 2022, 60, 1781–1792. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Xue, J.-Y.; Chen, S.; Wang, C.; Sun, P.; Fu, S.; Li, Y.; Zhao, P.; Tian, J.; Du, G.-Q. LncRNA PVT1 is a novel mediator promoting the angiogenesis response associated with collateral artery formation. Int. J. Biochem. Cell Biol. 2022, 151, 106294. [Google Scholar] [CrossRef]
- Zhan, J.; Yin, Q.; Zhao, P.; Hong, L. Role and mechanism of the lncRNA SNHG1/miR-450b-5p/IGF1 axis in the regulation of myocardial ischemia reperfusion injury. Mol. Med. Rep. 2022, 25, 176. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.; Liao, W.; Chen, W.; Lai, D.; Tang, Q.; Li, Y. Circulating long non-coding RNA TTTY15 and HULC serve as potential novel biomarkers for predicting acute myocardial infarction. BMC Cardiovasc. Disord. 2022, 22, 86. [Google Scholar] [CrossRef] [PubMed]
- Corrigendum to: 2022 ESC/ERS Guidelines for the diagnosis and treatment of pulmonary hypertension: Developed by the task force for the diagnosis and treatment of pulmonary hypertension of the European Society of Cardiology (ESC) and the European Respiratory Society (ERS). Endorsed by the International Society for Heart and Lung Transplantation (ISHLT) and the European Reference Network on rare respiratory diseases (ERN-LUNG). Eur. Heart J. 2023, 43, 3618–3731. [CrossRef]
- Ali, K.; Schimmel, K.; Zhao, L.; Chen, C.-K.; Dua, K.; Nicolls, M.R.; Spiekerkoetter, E. The role of circular RNAs in pulmonary hypertension. Eur. Respir. J. 2022, 60, 2200012. [Google Scholar] [CrossRef]
- Li, Y.; Cai, H.; Wei, J.; Zhu, L.; Yao, Y.; Xie, M.; Song, L.; Zhang, C.; Huang, X.; Wang, L. Dihydroartemisinin Attenuates Hypoxic Pulmonary Hypertension via the Downregulation of miR-335 Targeting Vangl2. DNA Cell Biol. 2022, 41, 750–767. [Google Scholar] [CrossRef]
- Cai, H.; Fan, S.; Cai, L.M.; Zhu, L.M.; Zhao, Z.M.; Li, Y.; Yao, Y.M.; Huang, X.; Wang, L. Dihydroartemisinin Attenuates Hypoxia-Induced Pulmonary Hypertension Through the ELAVL2/miR-503/PI3K/AKT Axis. J. Cardiovasc. Pharmacol. 2022, 80, 95–109. [Google Scholar] [CrossRef]
- Wang, Q.; Chai, L.; Zhang, Q.; Wang, J.; Liu, J.; Chen, H.; Wang, Y.; Chen, Y.; Shen, N.; Xie, X.; et al. Induction of GLI1 by miR-27b-3p/FBXW7/KLF5 pathway contributes to pulmonary arterial hypertension. J. Mol. Cell. Cardiol. 2022, 171, 16–29. [Google Scholar] [CrossRef]
- Sun, Y.; Jiang, R.; Hu, X.; Gong, S.; Wang, L.; Wu, W.; Li, J.; Kang, X.; Xia, S.; Liu, J.; et al. CircGSAP alleviates pulmonary microvascular endothelial cells dysfunction in pulmonary hypertension via regulating miR-27a-3p/BMPR2 axis. Respir. Res. 2022, 23, 322. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, D.; Long, X.-X.; Fang, Q.-C.; Jia, W.-P.; Li, H.-T. The role of FGF21 in the pathogenesis of cardiovascular disease. Chin. Med. J. 2021, 134, 2931–2943. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhang, Y.; Su, L.; Cai, L.; Zhang, C.; Zhang, J.; Sun, J.; Chai, M.; Cai, M.; Wu, Q.; et al. FGF21 alleviates pulmonary hypertension by inhibiting mTORC1/EIF4EBP1 pathway via H19. J. Cell. Mol. Med. 2022, 26, 3005–3021. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Hei, B.; Hao, W.; Lin, S.; Wang, Y.; Liu, X.; Meng, X.; Guan, Z. Clinical value of lncRNA SOX2-OT in pulmonary arterial hypertension and its role in pulmonary artery smooth muscle cell proliferation, migration, apoptosis, and inflammatory. Hear. Lung 2022, 55, 16–23. [Google Scholar] [CrossRef]
- Yen, T.-A.; Huang, H.-C.; Wu, E.-T.; Chou, H.-W.; Chou, H.-C.; Chen, C.-Y.; Huang, S.-C.; Chen, Y.-S.; Lu, F.; Wu, M.-H.; et al. Microrna-486-5P Regulates Human Pulmonary Artery Smooth Muscle Cell Migration via Endothelin-1. Int. J. Mol. Sci. 2022, 23, 10400. [Google Scholar] [CrossRef]
- Wang, M.; Su, L.; Sun, J.; Cai, L.; Li, X.; Zhu, X.; Song, L.; Li, J.; Tong, S.; He, Q.; et al. FGF21 attenuates pulmonary arterial hypertension via downregulation of miR-130, which targets PPARγ. J. Cell. Mol. Med. 2022, 26, 1034–1049. [Google Scholar] [CrossRef] [PubMed]
- Goel, K.; Egersdorf, N.; Gill, A.; Cao, D.; Collum, S.D.; Jyothula, S.S.; Huang, H.J.; Sauler, M.; Lee, P.J.; Majka, S.; et al. Characterization of pulmonary vascular remodeling and MicroRNA-126-targets in COPD-pulmonary hypertension. Respir. Res. 2022, 23, 349. [Google Scholar] [CrossRef]
- Hu, X.; Wang, Q.; Zhao, H.; Wu, W.; Zhao, Q.; Jiang, R.; Liu, J.; Wang, L.; Yuan, P. Role of miR-21-5p/ FilGAP axis in estradiol alleviating the progression of monocrotaline-induced pulmonary hypertension. Anim. Model. Exp. Med. 2022, 5, 217–226. [Google Scholar] [CrossRef]
- Luo, X.; Hang, C.; Zhang, Z.; Le, K.; Ying, Y.; Lv, Y.; Yan, L.; Huang, Y.; Ye, L.; Xu, X.; et al. PVECs-Derived Exosomal microRNAs Regulate PASMCs via FoxM1 Signaling in IUGR-induced Pulmonary Hypertension. J. Am. Heart Assoc. 2022, 11, e027177. [Google Scholar] [CrossRef]
- Deng, L.; Chen, J.; Chen, B.; Wang, T.; Yang, L.; Liao, J.; Yi, J.; Chen, Y.; Wang, J.; Linneman, J.; et al. LncPTSR Triggers Vascular Remodeling in Pulmonary Hypertension by Regulating [Ca2+]I in Pulmonary Arterial Smooth Muscle Cells. Am. J. Respir. Cell Mol. Biol. 2022, 66, 524–538. [Google Scholar] [CrossRef]
- Jing, X.; Wu, S.; Liu, Y.; Wang, H.; Huang, Q. Circular RNA Sirtuin1 represses pulmonary artery smooth muscle cell proliferation, migration and autophagy to ameliorate pulmonary hypertension via targeting microRNA-145-5p/protein kinase-B3 axis. Bioengineered 2022, 13, 8759–8771. [Google Scholar] [CrossRef]

| NcRNA | Expression in AS | Experimental Models | Mechanisms of Action | Roles | Reference |
|---|---|---|---|---|---|
| MiR-499-5p | ↑ | Oxidized low density lipoprotein (ox-LDL)-treated mouse aortic vascular smooth muscle cells (MAVSMCs) and apolipoprotein E knockout (ApoE−/−) mice | Regulation of SOX6 | Proliferation and migration of smooth muscle cells | [59] |
| MiR-32-5p | ↑ | Ox-LDL-treated human umbilical vein endothelial cells (HUVECs) | Regulation of AIDA | Inflammation | [60] |
| MiR-351 | ↑ | Ox-LDL-treated mouse aortic endothelial cells and miR-351 knockout mice | ITGB3/PIK3R1/AKT pathway | Apoptosis, lipid accumulation, and oxidative stress | [61] |
| MiR-130a-5p | ↓ | Ox-LDL-treated HUVECs | Regulation of FAS | Apoptosis, proliferation, and migration | [62] |
| MiR-663 | ↑ | Ox-LDL-treated VSMCs and ApoE−/− mice | Regulation of HMGA2 | Inflammation and proliferation | [63] |
| MiR-320a | ↑ | Ox-LDL-treated VSMCs | Regulation of RGS5 | Promoting migration and proliferation and reducing apoptosis | [64] |
| MiR-129-5p | ↓ | Ox-LDL-treated A7r5 cells | HMGB1/PI3K/AKT pathway | Reduction in migration | [65] |
| LncRNA TPRG1-AS1 | ↑ | Human aortic smooth muscle cells and ApoE knockout mice | Regulation of MYH9 | Migration and neointimal formation | [66] |
| LncRNA FGF7-5 and lncRNA GLRX3 | ↑ | Carotid plaque of atherosclerotic patients and ox-LDL-treated HUVECs | MiR-2681-5p/ERCC4 pathway | Reduction in atherosclerosis-induced apoptosis | [67] |
| LncRNA HOXA11-AS | ↓ | Ox-LDL-treated HUVECs and ApoE knockout mice | MiR-515-5p/ROCK1 pathway | Proliferation, apoptosis, and dephosphorylation of eNOS | [68] |
| LncRNA H19 | ↑ | Ox-LDL-treated human aortic endothelial cells (HAECs) | MiR-152/VEGFA pathway | Proliferation, migration, and tube formation | [69] |
| LncRNA DANCR | ↑ | Human serum and VSMCs | Regulation of miR-335-5p | Proliferative abilities and migration capacities | [70] |
| LncRNA RMST | ↑ | Human serum and ox-LDL-treated HUVECs | Regulation of miR-224-3p | Inflammation | [71] |
| LncRNA PVT1 | ↑ | Human serum and ox-LDL-treated HUVECs | Regulation of miR-30c-5p | Proliferation, apoptosis, and inflammation | [72] |
| LncRNA XIST | ↑ | Ox-LDL-treated VSMCs | MiR-539-5p/SPP1 pathway | Proliferation and migration | [54] |
| LncRNA MDRL | ↓ | MAVSMCs and LDLR knockout mice with high-fat diet | MiR-361/SQSTM1/NLRP3 pathway | Attenuation of apoptosis and inflammation | [55] |
| Circ_0021155 | ↑ | Ox-LDL-treated VSMCs | MiR-4459/TPRM7 pathway | Proliferation, migration, and phenotypic transformation | [57] |
| Circ_0086296 | ↑ | Human carotid plaque, ox-LDL-treated HUVECs, and ApoE knockout mice | MiR-576-3p/IFIT1/STAT1 pathway | Proliferation, migration, and inflammation | [56] |
| Circ_0024103 | ↑ | Ox-LDL-treated HUVECs | MiR-363/MMP-10 pathway | Migration, tube formation, and apoptosis | [73] |
| Circ_0002194 | ↑ | Ox-LDL-treated HUVECs | MiR-637/PACS2 pathway | Apoptosis and oxidative stress | [74] |
| Circ_0005699 | ↑ | Ox-LDL-treated HUVECs and ApoE knockout mice | MiR-450b-5p/NFKB1 pathway | Apoptosis and inflammation | [75] |
| Circ_PTPRA | ↑ | Human serum and ox-LDL-treated HUVECs | Regulation of miR-671-5p | Apoptosis and inflammation | [76] |
| Circ_NMD3 | ↓ | Ox-LDL-treated HUVECs | MiR-498/BAMBI pathway | Attenuation of proliferation and apoptosis | [77] |
| Circ_0093887 | ↓ | Ox-LDL-treated HAECs | MiR-758-3p/BAMBI pathway | Apoptosis and inflammation | [78] |
| Hsa_circ_0030042 | ↑ | TNF-α-treated VSMCs | MiR-514a-3p/FOXO1 pathway | Proliferation, migration, and apoptosis | [79] |
| Hsa_circ_0008896 | ↑ | Ox-LDL-treated VSMCs | MiR-633/CDC20B pathway | Proliferation and migration | [80] |
| Circ_CHFR | ↑ | Human serum and ox-LDL-treated HUVECs | MiR-15b-5p/GADD45G pathway | Apoptosis and inflammation | [81] |
| Circ_ARHGAP12 | ↑ | Ox-LDL-treated MAVSMCs and ApoE knockout mice | MiR-630/EZH2/TIMP2 pathway | Regulation of AS progression | [82] |
| NcRNA | Expression in CH | Experimental Models | Mechanisms of Action | Roles | Reference |
|---|---|---|---|---|---|
| MiR-212 | ↑ | PE- and Ang II-treated NRVCs and a rat model of abdominal aortic constriction | Regulation of TCF7L1 a | Cardiac hypertrophy | [109] |
| MiR-21 | ↑ | Ang II-treated NRVCs and miR-21 knockout mice | S100a8/NF-κB/NFAT pathway | Cardiac hypertrophy | [110] |
| MiR-143-3p | ↑ | Female mice with obesity- induced cardiac hypertrophy | Sox6/Myh7 pathway | Obesity-induced cardiac hypertrophy | [111] |
| MiR-30d | ↓ | PE- and Ang II-treated hypertrophy in NRVCs and ISO-treated rats | MAP4K4/GRP78 a/NFAT pathway | Attenuation of pathological hypertrophic changes | [107] |
| MiR-27a-3p | ↑ | Human plasma and Ang II-treated H9C2 and mice | Regulation of NOVA1 | Cardiac hypertrophy | [112] |
| MiR-204-5p | ↑ | Stretch-induced H9C2 and NRVCs, and TAC in miR-204 knockout mice | APJ signaling pathway | Attenuation of cardiac hypertrophy | [113] |
| MiR-410-3p | ↑ | Ang II-treated NRVCs | Regulation of Smad7 | Cardiac hypertrophy | [114] |
| MiR-339-5p | ↑ | ISO-induced NRVCs | VCP/mTOR/S6K pathway | Cardiac hypertrophy | [115] |
| MiR-27b-3p | ↑ | MiR-27b knockout mice subjected to TAC and AngII | Regulation of FGF1 | Cardiac hypertrophy | [116] |
| LncRNA MHRT | ↓ | Ang II-treated NRVCs and TAC-induced mice | SUMOylation of SIRT1 activating PGC1-α/PPAR-α pathway | Attenuation of cardiac hypertrophy | [117] |
| LncRNA MALAT1 | ↓ | Ang II-treated NRVCs | MiR-181a/HMGB2 axis | Attenuation of cardiac hypertrophy | [118] |
| LncRNA NBR2 | ↓ | Human plasma and Ang II- treated AC16 | LKB1/AMPK/SIRT1 pathway | Attenuation of cardiac hypertrophy | [119] |
| LncRNA MIAT | ↑ | Ang II-treated NRVCs and TAC-induced mice | YTHDF2/PPARα/CPT-1a pathway | Cardiac hypertrophy | [120] |
| LncRNA TINCR | ↓ | Ang II-treated H9C2 and TAC-induced mice | MiR-211-3p/VEGFB/SDF-1α/CXCR4 pathway | Attenuation of cardiac hypertrophy | [105] |
| LncRNA H19 | ↓ | Human serum and ISO-treated mice | MiR-145-3p/SMAD4 pathway | Attenuation of cardiac hypertrophy | [121] |
| LncRNA RMRP | ↑ | Human cardiac hypertrophic tissues and PE-treated cardiomyocytes | Regulation of miR-1 | Cardiac hypertrophy | [122] |
| Circ_0001006 | ↑ | Ang II-treated NRVCs and TAC-induced mice | MiR-214-3p/PAK6 axis | Cardiac hypertrophy | [108] |
| NcRNA | Expression in MI | Experimental Models | Mechanisms of Action | Roles | Reference |
|---|---|---|---|---|---|
| CircTMEM165, circUBAC2, circZNF609, circANKRD12, and circSLC8A1 | ↑ | Blood of human MI patients and H2O2-induced oxidative stress model in AC-16 cells | Several miRs | Apoptosis and diagnostic biomarker | [123] |
| LncRNA APF | ↑ | Blood of human MI patients | Targeting autophagy via miR-188-3p | Diagnostic biomarker | [131] |
| LncRNA BACE1-AS | ↑ | Blood mononuclear cells derived from CAD patients | Undetermined | Diagnostic biomarker | [132] |
| LncRNA CAIF | ↓ | Mouse MI model and H2O2- induced oxidative stress model in mouse cardiomyocytes | CAIF/miR-488-5p/AVEN axis | Apoptosis | [133] |
| LncRNA EPS | ↓ | Mouse MI model and HL-1 cells treated in oxygen and glucose deprivation (OGD) | Maintaining MYH6 stability through recruitment of HNRNPL | Inflammation and apoptosis | [134] |
| LncRNA LINC00461 | ↑ | Mouse I/R injury model | MiR-185-3p/Myd88 axis | Apoptosis | [135] |
| LncRNA MALAT1 | ↑ | Blood of human MI patients | Undetermined | Prognostic biomarker | [136] |
| LncRNA MALAT1 | ↑ | Monocytes of human MI patients | Undetermined | Diagnostic biomarker | [126] |
| LncRNA MALAT1 | ↑ | Plasma of human STEMI patients | Sponging miR-30e, miR-126, and miR-155 | Biomarker to diagnose no flow | [124] |
| LncRNA MALAT1 | ↑ | Mouse I/R injury model and H9c2 and HL-1 cells subjected to hypoxia/reoxygenation | PI3K/AKT/eNOS signaling via miR-133a-3p | Apoptosis | [127] |
| LncRNA MBNL1-AS1 | ↑ | Rat MI models and H9c2 cells treated by hypoxia | MiR-132-3p/SOX4 axis | Apoptosis | [137] |
| LncRNA MCM3AP-AS1 | ↑ | Rat MI models and knockdown in vascular ECs (VECs) | MiR-24-3p/EIF4G2 pathway | Promoting proliferation and migration of VECs | [138] |
| LncRNA MIAT | ↑ | H9C2 cells subjected to hypoxia | SF1/CGRP pathway | Pyroptosis | [139] |
| LncRNA MIR4435-2HG | ↑ | Human MI patients, mouse I/R model, and H2O2-induced oxidative stress model | MiR-125a-5p/MTFP1 pathway | Apoptosis | [140] |
| LncRNA HOTAIR | ↑ | Mouse MI model and cardiomyocytes subjected to hypoxia/reoxygenation | MiR-206/FN1 axis | Apoptosis | [141] |
| LncRNA PVT1 | ↑ | Plasma of patients with good coronary collateral circulation, HUVECs, and mouse hind limb ischemia and MI models | PVT1/miR-15b-5p/AKT3 axis | Angiogenesis | [142] |
| LncRNA SNHG1 | ↓ | AC-16 cells subjected to hypoxia/reoxygenation | MiR-450b-5p/IGF1 axis | Apoptosis | [143] |
| LncRNA TTTY15 and LncRNA HULC | ↑ | Plasma of human MI patients | Undetermined | Biomarkers to diagnose AMI | [144] |
| MiR-150 | ↓ | Mouse MI model and HCFs | MIAT/miR-150/HOXA4 pathway | Blunting CF activation | [129] |
| MiR-223 and miR-186 | ↑ | Blood of human MI patients | Undetermined | Prognostic biomarkers | [125] |
| MiR-411 | - | Mouse MI model and NRVCs | Hippo/YAP pathway | Cardiomyocyte proliferation and survival | [128] |
| NcRNA | Expression in PH | Experimental Models | Mechanisms of Action | Roles | Reference |
|---|---|---|---|---|---|
| MiR-335 | ↑ | DHA-treated hypoxic PH (HPH) mice and PASMCs | Regulation of Vangl2 | PASMC proliferation in PH | [147] |
| MiR-503 | ↓ | DHA-treated HPH mice and PASMCs | ELAVL2/miR-503/PI3K/AKT pathway | Reduced PASMC proliferation in PH | [148] |
| MiR-486-5p | ↓ | Hypoxia-induced human primary PASMCs | Smad2/3 pathway | hPASMC proliferation and migration in PAH | [154] |
| MiR-130 | ↓ | PASMCs and FGF21-treated mice | FGF21/PPARγ axis | Inhibited PASMC proliferation and migration in PAH | [155] |
| MiR-126 | ↓ | Primary HLMVECs | Regulation of ADAM9 | Angiogenesis and pulmonary vascular remodeling in COPD-PH | [156] |
| MiR-27b-3p | ↑ | Monocrotaline (MCT)- induced PAH rats and rat PASMCs | FBXW7/KLF5/GLI1 pathway | PASMC proliferation and migration in PAH | [149] |
| MiR-21-5p | ↑ | MCT-induced PAH rats | Regulation of FilGAP | PASMC proliferation in PH | [157] |
| MiR-214-3p, miR-326-3p, and miR-125b-2-3p | ↓ | IUGR-induced PH rats | Regulation of FoxM1 | PASMC proliferation and migration in PH | [158] |
| LncRNA H19 | ↓ | FGF21-treated HPH mice | MTORC1/EIF4EBP1 pathway | Reduced PASMC proliferation in PH | [152] |
| LncPTSR | ↓ | Rat PASMCs and HPAH rats | Regulation of PMCA4 and intracellular Ca2+ | Reduced PASMC proliferation in PAH | [159] |
| LncRNA SOX2-OT | ↓ | Hypoxia-induced human PASMCs (hPASMCs) | MiR-455-3p/SUMO1 pathway | Attenuating hPASMC proliferation, migration, antiapoptosis, and inflammation in PAH | [153] |
| CircGSAP | ↓ | MCT-induced PH rats and CO2-treated human PMECs | MiR-27a-3p/BMPR2 pathway | Reduced PMEC proliferation, migration, and increased cell mortality in IPAH | [150] |
| CircSIRT1 | ↑ | MCT-induced PH rats and PASMCs | MiR-145-5p/AKT3 axis | PASMC proliferation and migration in PH | [160] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kawaguchi, S.; Moukette, B.; Hayasaka, T.; Haskell, A.K.; Mah, J.; Sepúlveda, M.N.; Tang, Y.; Kim, I.-m. Noncoding RNAs as Key Regulators for Cardiac Development and Cardiovascular Diseases. J. Cardiovasc. Dev. Dis. 2023, 10, 166. https://doi.org/10.3390/jcdd10040166
Kawaguchi S, Moukette B, Hayasaka T, Haskell AK, Mah J, Sepúlveda MN, Tang Y, Kim I-m. Noncoding RNAs as Key Regulators for Cardiac Development and Cardiovascular Diseases. Journal of Cardiovascular Development and Disease. 2023; 10(4):166. https://doi.org/10.3390/jcdd10040166
Chicago/Turabian StyleKawaguchi, Satoshi, Bruno Moukette, Taiki Hayasaka, Angela K. Haskell, Jessica Mah, Marisa N. Sepúlveda, Yaoliang Tang, and Il-man Kim. 2023. "Noncoding RNAs as Key Regulators for Cardiac Development and Cardiovascular Diseases" Journal of Cardiovascular Development and Disease 10, no. 4: 166. https://doi.org/10.3390/jcdd10040166
APA StyleKawaguchi, S., Moukette, B., Hayasaka, T., Haskell, A. K., Mah, J., Sepúlveda, M. N., Tang, Y., & Kim, I.-m. (2023). Noncoding RNAs as Key Regulators for Cardiac Development and Cardiovascular Diseases. Journal of Cardiovascular Development and Disease, 10(4), 166. https://doi.org/10.3390/jcdd10040166





