Early Postnatal Cardiac Stress Does Not Influence Ventricular Cardiomyocyte Cell-Cycle Withdrawal
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Statement
2.2. Animals and Tissues
2.3. Immunohistochemistry
2.4. Quantification of Cardiomyocyte Cell-Cycle Activity
2.5. In Situ Hybridization
2.6. Picrosiriusred Staining
2.7. Cardiomyocyte Nuclei Isolation and Ploidy Analysis
2.8. RNA-Sequencing
2.9. Analysis of RNA-Seq Data
2.10. Statistics
3. Results
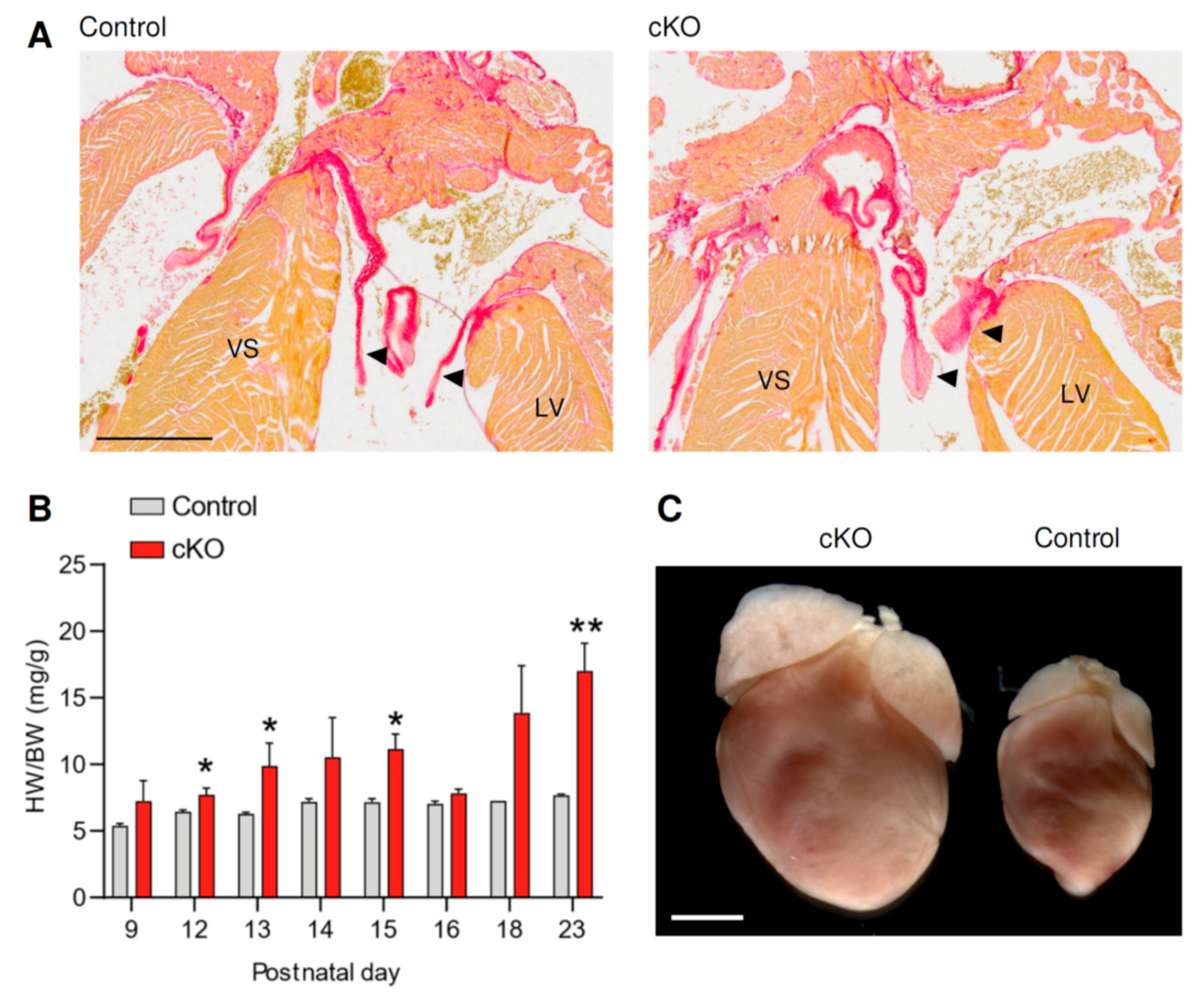
3.1. Fstl1KO/fl; Tie2-Cre Mice Develop Cardiac Stress Soon after Birth
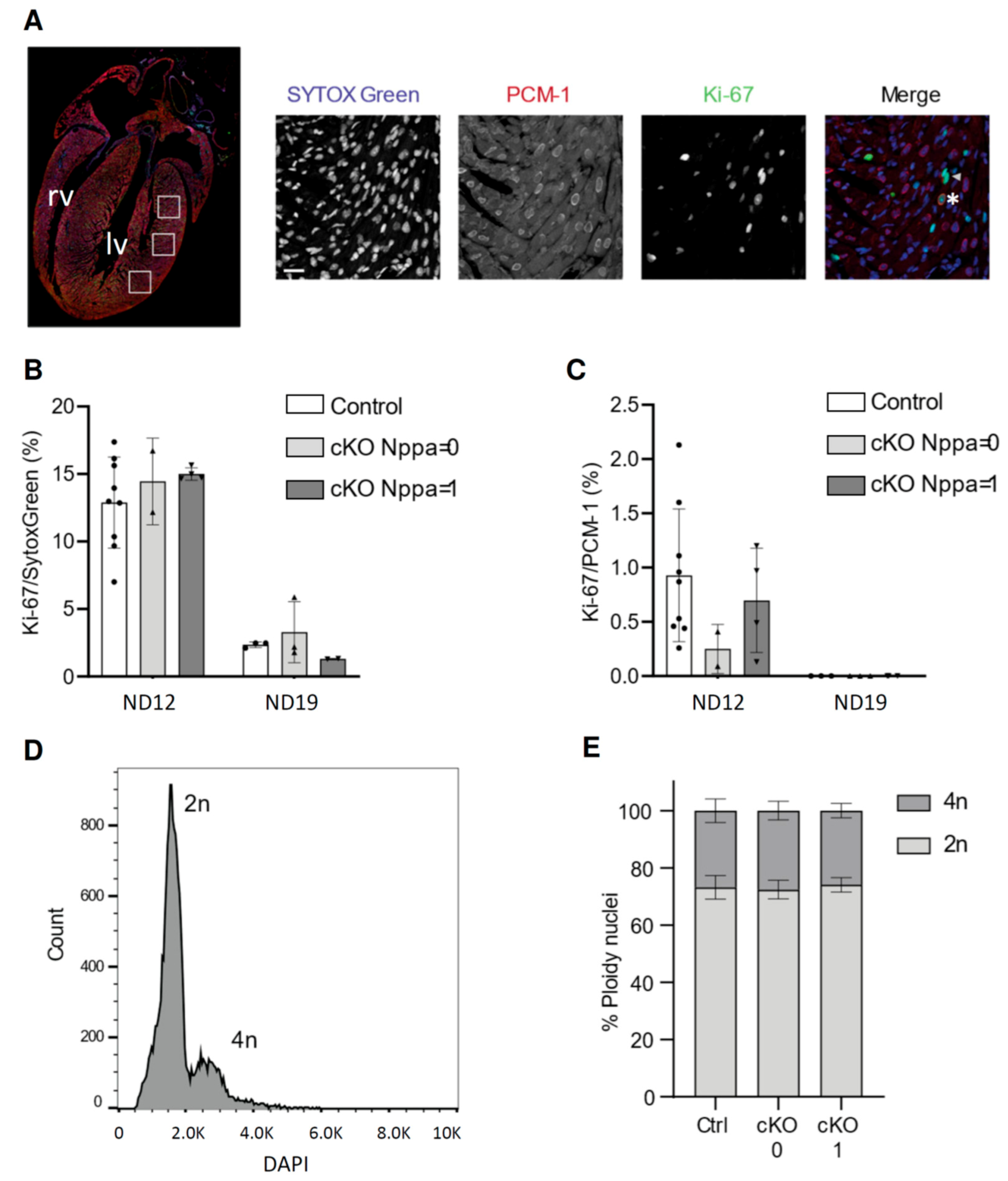
3.2. Cardiac Stress in Pre-Weaning Mice Does Not Affect Cell-Cycle Activity and Ploidy
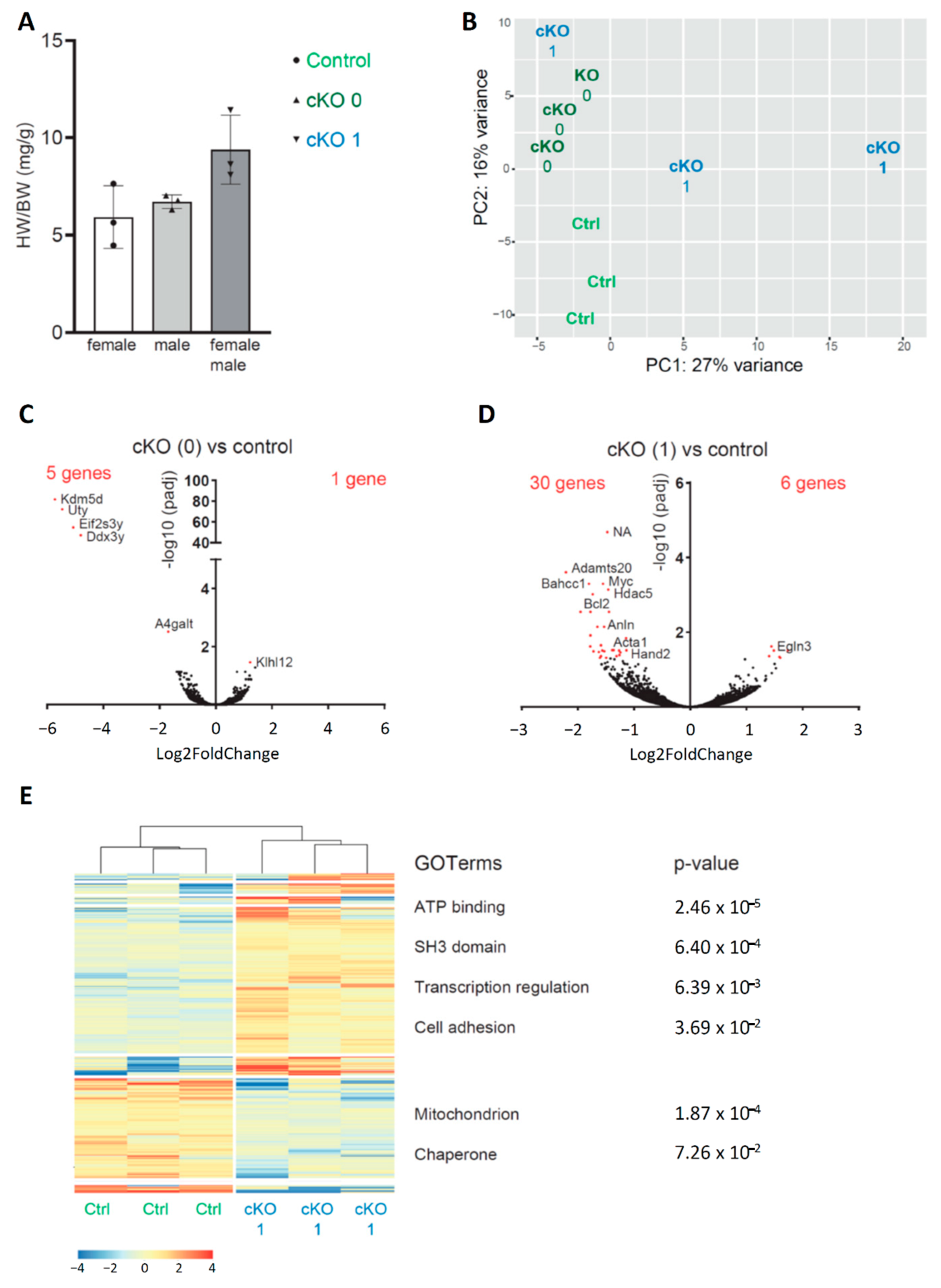
3.3. Neonatal Cardiac Stress Induces Cell-Cycle Gene Expression
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Dolk, H. EUROCAT: 25 years of European surveillance of congenital anomalies. Arch. Dis. Child Fetal. Neonatal. Ed. 2005, 90, F355–F358. [Google Scholar] [CrossRef]
- Schwerzmann, M.; Schwitz, F.; Thomet, C.; Kadner, A.; Pfammatter, J.P.; Wustmann, K. Challenges of congenital heart disease in grown-up patients. Swiss Med. Wkly 2017, 147, w14495. [Google Scholar] [CrossRef] [PubMed]
- Hinton, R.B.; Ware, S.M. Heart Failure in Pediatric Patients With Congenital Heart Disease. Circ. Res. 2017, 120, 978–994. [Google Scholar] [CrossRef]
- Moons, P.; Bovijn, L.; Budts, W.; Belmans, A.; Gewillig, M. Temporal Trends in Survival to Adulthood Among Patients Born With Congenital Heart Disease From 1970 to 1992 in Belgium. Circulation 2010, 122, 2264–2272. [Google Scholar] [CrossRef] [PubMed]
- Warnes, C.A. Adult congenital heart disease: The challenges of a lifetime. Eur. Heart J. 2017, 38, 2041–2047. [Google Scholar] [CrossRef]
- Puente, B.N.; Kimura, W.; Muralidhar, S.A.; Moon, J.; Amatruda, J.F.; Phelps, K.L.; Grinsfelder, D.; Rothermel, B.A.; Chen, R.; Garcia, J.A.; et al. The Oxygen-Rich Postnatal Environment Induces Cardiomyocyte Cell-Cycle Arrest through DNA Damage Response. Cell 2014, 157, 565–579. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Wang, X.; Capasso, J.M.; Gerdes, A.M. Rapid transition of cardiac myocytes from hyperplasia to hypertrophy during postnatal development. J. Mol. Cell. Cardiol. 1996, 28, 1737–1746. [Google Scholar] [CrossRef]
- Alkass, K.; Panula, J.; Westman, M.; Wu, T.D.; Guerquin-Kern, J.L.; Bergmann, O. No Evidence for Cardiomyocyte Number Expansion in Preadolescent Mice. Cell 2015, 163, 1026–1036. [Google Scholar] [CrossRef]
- Bergmann, O.; Zdunek, S.; Felker, A.; Salehpour, M.; Alkass, K.; Bernard, S.; Sjostrom, S.L.; Szewczykowska, M.; Jackowska, T.; Dos Remedios, C.; et al. Dynamics of Cell Generation and Turnover in the Human Heart. Cell 2015, 161, 1566–1575. [Google Scholar] [CrossRef]
- Ding, X.; Wang, S.; Wang, Y.; Yang, J.; Bao, N.; Liu, J.; Zhang, Z. Neonatal Heart Responds to Pressure Overload With Differential Alterations in Various Cardiomyocyte Maturation Programs That Accommodate Simultaneous Hypertrophy and Hyperplasia. Front. Cell Dev. Biol. 2020, 8, 596960. [Google Scholar] [CrossRef]
- Kannan, S.; Kwon, C. Regulation of cardiomyocyte maturation during critical perinatal window. J. Physiol. 2020, 598, 2941–2956. [Google Scholar] [CrossRef]
- Prakash, S.; Borreguero, L.J.J.; Sylva, M.; Flores Ruiz, L.; Rezai, F.; Gunst, Q.D.; de la Pompa, J.L.; Ruijter, J.M.; van den Hoff, M.J.B. Deletion of Fstl1 (Follistatin-Like 1) From the Endocardial/Endothelial Lineage Causes Mitral Valve Disease. Arterioscler. Thromb. Vasc. Biol. 2017, 116–130. [Google Scholar] [CrossRef] [PubMed]
- Moorman, A.F.M.; Houweling, A.C.; de Boer, P.A.J.; Christoffels, V.M. Sensitive nonradioactive detection of mRNA in tissue sections: Novel application of the whole-mount in situ hybridization protocol. J. Histochem. Cytochem. 2001, 49, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Van Duijvenboden, K.; de Bakker, D.E.M.; Man, J.C.K.; Janssen, R.; Gunthel, M.; Hill, M.C.; Hooijkaas, I.B.; van der Made, I.; van der Kraak, P.H.; Vink, A.; et al. Conserved NPPB+ Border Zone Switches from MEF2 to AP-1 Driven Gene Program. Circulation 2019. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Huang, d.W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009, 37, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Huang, d.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef]
- Sergeeva, I.A.; Hooijkaas, I.B.; van der Made, I.; Jong, W.M.; Creemers, E.E.; Christoffels, V.M. A transgenic mouse model for the simultaneous monitoring of ANF and BNP gene activity during heart development and disease. Cardiovasc. Res. 2014, 78–86. [Google Scholar] [CrossRef]
- Scholzen, T.; Gerdes, J. The Ki-67 protein: From the known and the unknown. J. Cell Physiol. 2000, 182, 311–322. [Google Scholar] [CrossRef]
- Malek Mohammadi, M.; Abouissa, A.; Azizah, I.; Xie, Y.; Cordero, J.; Shirvani, A.; Gigina, A.; Engelhardt, M.; Trogisch, F.A.; Geffers, R.; et al. Induction of cardiomyocyte proliferation and angiogenesis protects neonatal mice from pressure overload-associated maladaptation. JCI Insight 2019, 5, 1–20. [Google Scholar] [CrossRef]
- Porrello, E.R.; Mahmoud, A.I.; Simpson, E.; Hill, J.A.; Richardson, J.A.; Olson, E.N.; Sadek, H.A. Transient regenerative potential of the neonatal mouse heart. Science 2011, 331, 1078–1080. [Google Scholar] [CrossRef]
- Zebrowski, D.C.; Jensen, C.H.; Becker, R.; Ferrazzi, F.; Baun, C.; Hvidsten, S.; Sheikh, S.P.; Polizzotti, B.D.; Andersen, D.C.; Engel, F.B. Cardiac injury of the newborn mammalian heart accelerates cardiomyocyte terminal differentiation. Sci. Rep. 2017, 7, 8362. [Google Scholar] [CrossRef]
- Lam, N.T.; Sadek, H.A. Neonatal Heart Regeneration: Comprehensive Literature Review. Circulation 2018, 138, 412–423. [Google Scholar] [CrossRef]
- Quaife-Ryan, G.A.; Sim, C.B.; Ziemann, M.; Kaspi, A.; Rafehi, H.; Ramialison, M.; El-Osta, A.; Hudson, J.E.; Porrello, E.R. Multicellular Transcriptional Analysis of Mammalian Heart Regeneration. Circulation 2017, 136, 1123–1139. [Google Scholar] [CrossRef]
- Porrello, E.R.; Mahmoud, A.I.; Simpson, E.; Johnson, B.A.; Grinsfelder, D.; Canseco, D.; Mammen, P.P.; Rothermel, B.A.; Olson, E.N.; Sadek, H.A. Regulation of neonatal and adult mammalian heart regeneration by the miR-15 family. Proc. Natl. Acad. Sci. USA 2013, 110, 187–192. [Google Scholar] [CrossRef]
- Vivien, C.J.; Hudson, J.E.; Porrello, E.R. Evolution, comparative biology and ontogeny of vertebrate heart regeneration. NPJ Regen. Med. 2016, 1, 16012. [Google Scholar] [CrossRef]
- Aurora, A.B.; Porrello, E.R.; Tan, W.; Mahmoud, A.I.; Hill, J.A.; Bassel-Duby, R.; Sadek, H.A.; Olson, E.N. Macrophages are required for neonatal heart regeneration. J. Clin. Investig. 2014, 124, 1382–1392. [Google Scholar] [CrossRef]
- Engel, F.B.; Schebesta, M.; Keating, M.T. Anillin localization defect in cardiomyocyte binucleation. J. Mol. Cell. Cardiol. 2006, 41, 601–612. [Google Scholar] [CrossRef]
- Sodir, N.M.; Swigart, L.B.; Karnezis, A.N.; Hanahan, D.; Evan, G.I.; Soucek, L. Endogenous Myc maintains the tumor microenvironment. Genes Dev. 2011, 25, 907–916. [Google Scholar] [CrossRef]
- Bywater, M.J.; Burkhart, D.L.; Straube, J.; Sabo, A.; Pendino, V.; Hudson, J.E.; Quaife-Ryan, G.A.; Porrello, E.R.; Rae, J.; Parton, R.G.; et al. Reactivation of Myc transcription in the mouse heart unlocks its proliferative capacity. Nat. Commun. 2020, 11, 1827. [Google Scholar] [CrossRef]
- Limana, F.; Urbanek, K.; Chimenti, S.; Quaini, F.; Leri, A.; Kajstura, J.; Nadal-Ginard, B.; Izumo, S.; Anversa, P. bcl-2 overexpression promotes myocyte proliferation. Proc. Natl. Acad. Sci. USA 2002, 99, 6257–6262. [Google Scholar] [CrossRef]
- Foglia, M.J.; Poss, K.D. Building and re-building the heart by cardiomyocyte proliferation. Development 2016, 143, 729–740. [Google Scholar] [CrossRef]
- Chang, S.; McKinsey, T.A.; Zhang, C.L.; Richardson, J.A.; Hill, J.A.; Olson, E.N. Histone deacetylases 5 and 9 govern responsiveness of the heart to a subset of stress signals and play redundant roles in heart development. Mol. Cell. Biol. 2004, 24, 8467–8476. [Google Scholar] [CrossRef]
- Leone, M.; Magadum, A.; Engel, F.B. Cardiomyocyte proliferation in cardiac development and regeneration: A guide to methodologies and interpretations. Am. J. Physiol. Heart Circ. Physiol. 2015, 309, H1237–H1250. [Google Scholar] [CrossRef]
- Soonpaa, M.H.; Zebrowski, D.C.; Platt, C.; Rosenzweig, A.; Engel, F.B.; Field, L.J. Cardiomyocyte Cell-Cycle Activity during Preadolescence. Cell 2015, 163, 781–782. [Google Scholar] [CrossRef]
- Velayutham, N.; Agnew, E.J.; Yutzey, K.E. Postnatal Cardiac Development and Regenerative Potential in Large Mammals. Pediatric Cardiol. 2019, 40, 1345–1358. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Günthel, M.; van Duijvenboden, K.; Jeremiasse, J.; van den Hoff, M.J.B.; Christoffels, V.M. Early Postnatal Cardiac Stress Does Not Influence Ventricular Cardiomyocyte Cell-Cycle Withdrawal. J. Cardiovasc. Dev. Dis. 2021, 8, 38. https://doi.org/10.3390/jcdd8040038
Günthel M, van Duijvenboden K, Jeremiasse J, van den Hoff MJB, Christoffels VM. Early Postnatal Cardiac Stress Does Not Influence Ventricular Cardiomyocyte Cell-Cycle Withdrawal. Journal of Cardiovascular Development and Disease. 2021; 8(4):38. https://doi.org/10.3390/jcdd8040038
Chicago/Turabian StyleGünthel, Marie, Karel van Duijvenboden, Jorn Jeremiasse, Maurice J. B. van den Hoff, and Vincent M. Christoffels. 2021. "Early Postnatal Cardiac Stress Does Not Influence Ventricular Cardiomyocyte Cell-Cycle Withdrawal" Journal of Cardiovascular Development and Disease 8, no. 4: 38. https://doi.org/10.3390/jcdd8040038
APA StyleGünthel, M., van Duijvenboden, K., Jeremiasse, J., van den Hoff, M. J. B., & Christoffels, V. M. (2021). Early Postnatal Cardiac Stress Does Not Influence Ventricular Cardiomyocyte Cell-Cycle Withdrawal. Journal of Cardiovascular Development and Disease, 8(4), 38. https://doi.org/10.3390/jcdd8040038





