The Athlete’s Heart and Machine Learning: A Review of Current Implementations and Gaps for Future Research
Abstract
1. Introduction
2. Methods
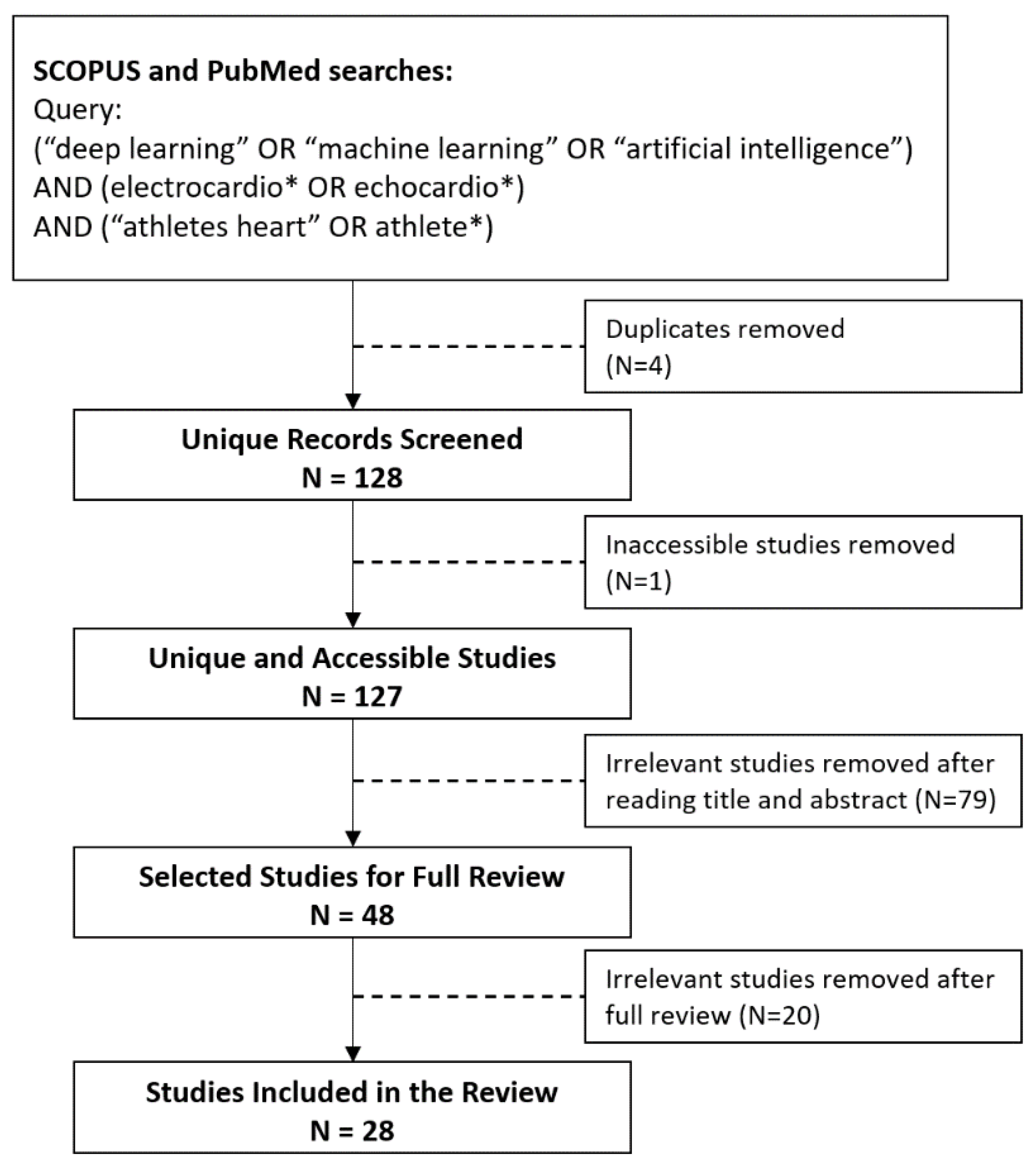
2.1. Search Strategy and Selection Process
2.2. Search Results
3. Results
3.1. Study Subgroups
3.1.1. Predictive Modelling
3.1.2. Reviews
3.1.3. Wearables
3.1.4. Others
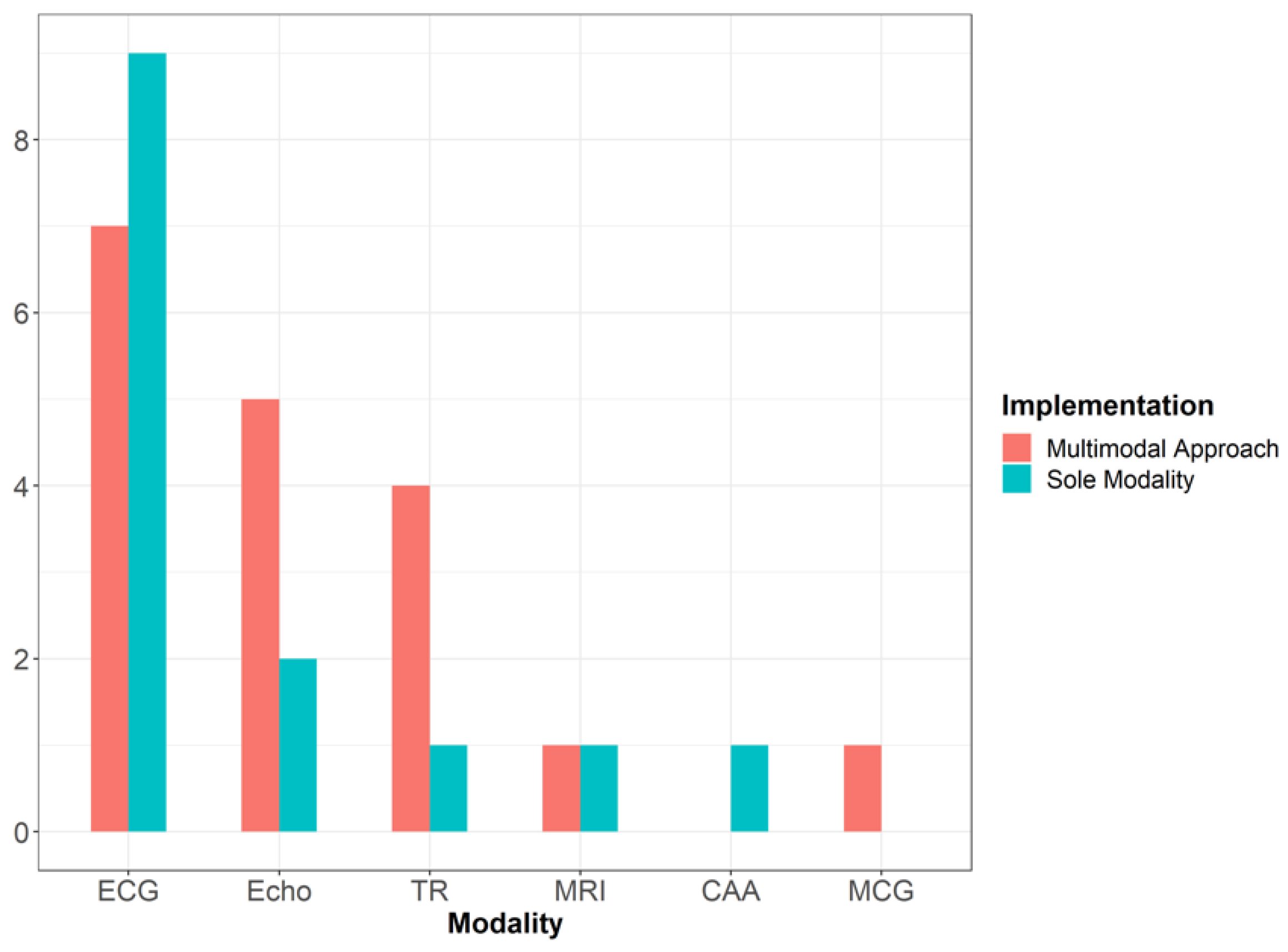
3.2. Data Modalities Used for Athlete’s Heart Assessment
3.3. Machine Learning Approaches Used
4. Discussion
5. Limitations of Current Research
6. Future Research and Impact
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- WHO. The Top 10 Causes of Death. 2020. Available online: https://www.who.int/news-room/fact-sheets/detail/the-top-10-causes-of-death (accessed on 25 November 2021).
- British Heart Foundation. Facts and Figures. There Are around 7.6 Million, the Single Biggest Killer Worldwide. 2021. Available online: https://www.bhf.org.uk/what-we-do/news-from-the-bhf/contact-the-press-office/facts-and-figures (accessed on 28 November 2021).
- Wasfy, M.M.; Hutter, A.M.; Weiner, R.B. Sudden Cardiac Death in Athletes. Methodist DeBakey Cardiovasc. J. 2016, 12, 76. [Google Scholar] [CrossRef]
- Kerkhof, D.L.; Lucas, C.; Corrado, G.D. Monitoring Morphologic Changes in Male Rowers Using Limited Portable Echocardiography Performed by a Frontline Physician. J. Ultrasound Med. 2018, 37, 2451–2455. [Google Scholar] [CrossRef]
- Harmon, K.G.; Drezner, J.A.; Wilson, M.G.; Sharma, S. Incidence of sudden cardiac death in athletes: A state-of-the-art review. Br. J. Sports Med. BMJ 2014, 48, 1185–1192. [Google Scholar] [CrossRef]
- Olier, I.; Ortega-Martorell, S.; Pieroni, M.; Lip, G.Y.H. How machine learning is impacting research in atrial fibrillation: Implications for risk prediction and future management. Cardiovasc. Res. 2021, 117, 1700–1717. [Google Scholar] [CrossRef]
- McKinney, S.M.; Sieniek, M.; Godbole, V.; Godwin, J.; Antropova, N.; Ashrafian, H.; Back, T.; Chesus, M.; Corrado, G.S.; Darzi, A.; et al. International evaluation of an AI system for breast cancer screening. Nature 2020, 577, 89–94. Available online: https://www.nature.com/articles/s41586-019-1799-6 (accessed on 25 November 2021). [CrossRef]
- Singh, A.; Thakur, N.; Sharma, A. A review of supervised machine learning algorithms. In Proceedings of the 2016 3rd International Conference on Computing for Sustainable Global Development (INDIACom), New Delhi, India, 16–18 March 2016; pp. 1310–1315. [Google Scholar]
- Katz, D.H.; Deo, R.C.; Aguilar, F.G.; Selvaraj, S.; Martinez, E.E.; Beussink-Nelson, L.; Kim, K.-Y.A.; Peng, J.; Irvin, M.R.; Tiwari, H.; et al. Phenomapping for the Identification of Hypertensive Patients with the Myocardial Substrate for Heart Failure with Preserved Ejection Fraction. J. Cardiovasc. Transl. Res. 2017, 10, 275–284. [Google Scholar] [CrossRef]
- Alizadehsani, R.; Hosseini, M.J.; Khosravi, A.; Khozeimeh, F.; Roshanzamir, M.; Sarrafzadegan, N.; Nahavandi, S. Non-invasive detection of coronary artery disease in high-risk patients based on the stenosis prediction of separate coronary arteries. Comput. Methods Programs Biomed. 2018, 162, 119–127. [Google Scholar] [CrossRef]
- Asselbergs, F.; Meijboom, F.J. Big data analytics in adult congenital heart disease: Why coding matters. Eur. Heart J. 2019, 40, 1078–1080. Available online: https://academic.oup.com/eurheartj/article/40/13/1069/5301309 (accessed on 28 November 2021). [CrossRef]
- Jing, L.; Cerna, A.E.U.; Good, C.W.; Sauers, N.M.; Schneider, G.; Hartzel, D.N.; Leader, J.B.; Kirchner, H.L.; Hu, Y.; Riviello, D.M.; et al. A Machine Learning Approach to Management of Heart Failure Populations. JACC Heart Fail. 2020, 8, 578–587. Available online: https://www.sciencedirect.com/science/article/abs/pii/S2213177920301384 (accessed on 28 November 2021). [CrossRef]
- Agasthi, P.; Ashraf, H.; Pujari, S.H.; Girardo, M.E.; Tseng, A.; Mookadam, F.; Venepally, N.R.; Buras, M.; Khetarpal, B.K.; Allam, M.; et al. Artificial Intelligence Trumps TAVI2-SCORE and CoreValve Score in Predicting 1-Year Mortality Post-Transcatheter Aortic Valve Replacement. Cardiovasc. Revascularizat. Med. 2020, 24, 33–41. [Google Scholar] [CrossRef]
- Adetiba, E.; Iweanya, V.C.; Popoola, S.I.; Adetiba, J.N.; Menon, C. Automated detection of heart defects in athletes based on electrocardiography and artificial neural network. Cogent Eng. 2017, 4, 1411220. [Google Scholar] [CrossRef]
- Claudino, J.G.; de Oliveira Capanema, D.; De Souza, T.V.; Serrão, J.C.; Pereira, A.C.M.; Nassis, G.P. Current Approaches to the Use of Artificial Intelligence for Injury Risk Assessment and Performance Prediction in Team Sports: A Systematic Review. Sports Med. Open 2019, 5, 28. [Google Scholar] [CrossRef]
- Długosz, D.; Królak, A.; Eftestøl, T.; Ørn, S.; Wiktorski, T.; Oskal, K.R.J.; Nygård, M. ECG signal analysis for troponin level assessment and coronary artery disease detection: The NEEDED study 2014. In Proceedings of the 2018 Federated Conference on Computer Science and Information Systems (FedCSIS), Poznan, Poland, 9–12 September 2018; Volume 15, pp. 1065–1068. [Google Scholar]
- Georgijevic, L.; Andric, L. Electrocardiography in pre-participation screening and current guidelines for participation in competitive sports. Srp. Arh. Za Celok. Lek. 2016, 144, 104–110. [Google Scholar] [CrossRef]
- Higgins, J.P.; Ananaba, I.E.; Higgins, C.L. Sudden Cardiac Death in Young Athletes: Preparticipation Screening for Underlying Cardiovascular Abnormalities and Approaches to Prevention. Physician Sportsmed. 2013, 41, 81–93. [Google Scholar] [CrossRef]
- Huang, K.-C.; Lin, C.-E.; Lin, L.-Y.; Hwang, J.-J.; Lin, L.-C. Data-driven clustering supports adaptive remodeling of athlete’s hearts: An echocardiographic study from the Taipei Summer Universiade. J. Formos. Med. Assoc. 2021, 121, 1495–1505. [Google Scholar] [CrossRef]
- Hussain, A.; Zafar, K.; Baig, A.R. Fog-Centric IoT Based Framework for Healthcare Monitoring, Management and Early Warning System. IEEE Access 2021, 9, 74168–74179. [Google Scholar] [CrossRef]
- Laurino, M.; Piarulli, A.; Bedini, R.; Gemignani, A.; Pingitore, A.; L’Abbate, A.; Landi, A.; Piaggi, P.; Menicucci, D. Comparative study of morphological ECG features classificators: An application on athletes undergone to acute physical stress. In Proceedings of the 2011 11th International Conference on Intelligent Systems Design and Applications, Cordoba, Spain, 22–24 November 2011; IEEE: Piscataway, NJ, USA, 2011; pp. 242–246. [Google Scholar]
- Lombardi, G.; Sorbo, A.R.; Guida, G.; La Brocca, L.; Fenici, R.; Brisinda, D. Magnetocardiographic classification and non-invasive electro-anatomical imaging of outflow tract ventricular arrhythmias in recreational sport activity practitioners. J. Electrocardiol. 2018, 51, 433–439. [Google Scholar] [CrossRef]
- Lucas, C.; Kerkhof, D.L.; Briggs, J.E.; Corrado, G.D. The Use of Echocardiograms in Preparticipation Examinations. Curr. Sports Med. Rep. 2017, 16, 77–83. [Google Scholar] [CrossRef][Green Version]
- Młyńczak, M.; Krysztofiak, H. Discovery of Causal Paths in Cardiorespiratory Parameters: A Time-Independent Approach in Elite Athletes. Front. Physiol. 2018, 9, 1455. [Google Scholar] [CrossRef]
- Adetiba, E.; Onosenema, E.N.; Akande, V.; Adetiba, J.N.; Kala, J.R.; Olaloye, F. Development of an ECG Smart Jersey Based on Next Generation Computing for Automated Detection of Heart Defects Among Athletes. In Proceedings of the International Work-Conference on Bioinformatics and Biomedical Engineering, Granada, Spain, 8–10 May 2019; Springer International Publishing: Cham, Switzerland, 2019. [Google Scholar] [CrossRef]
- Narula, S.; Shameer, K.; Omar, A.M.S.; Dudley, J.T.; Sengupta, P.P. Machine-Learning Algorithms to Automate Morphological and Functional Assessments in 2D Echocardiography. J. Am. Coll. Cardiol. 2016, 68, 2287–2295. [Google Scholar] [CrossRef]
- Rahman, Q.A.; Kanagalingam, S.; Pinheiro, A.; Abraham, T.; Shatkay, H. What we found on our way to building a classifier: A critical analysis of the AHA screening questionnaire. In Proceedings of the International Conference on Brain and Health Informatics, Maebashi, Japan, 29–31 October 2013; pp. 225–236. [Google Scholar]
- Rymarczyk, T.; Stanikowski, A.; Nita, P. Wearable sensor array for biopotential measurements. In Proceedings of the 2019 Applications of Electromagnetics in Modern Engineering and Medicine (PTZE), Janów Podlaski, Poland, 9–12 June 2019; Polish Society of Applied Electromagnetics: Warsaw, Poland, 2019; pp. 184–187. [Google Scholar]
- Seshadri, D.R.; Thom, M.L.; Harlow, E.R.; Gabbett, T.J.; Geletka, B.J.; Hsu, J.J.; Drummond, C.K.; Phelan, D.M.; Voos, J.E. Wearable Technology and Analytics as a Complementary Toolkit to Optimize Workload and to Reduce Injury Burden. Front. Sports Act. Living 2021, 2, 630576. [Google Scholar] [CrossRef]
- Van Eetvelde, H.; Mendonça, L.D.; Ley, C.; Seil, R.; Tischer, T. Machine learning methods in sport injury prediction and prevention: A systematic review. J. Exp. Orthop. 2021, 8, 27. [Google Scholar] [CrossRef]
- Vergani, V.; Lazzeroni, D.; Peretto, G. Bridging the gap between hypertrabeculation phenotype, noncompaction phenotype and left ventricular noncompaction cardiomyopathy. J. Cardiovasc. Med. 2020, 21, 192–199. [Google Scholar] [CrossRef]
- Viviers, P.L.; Kirby, J.-A.H.; Viljoen, J.T.; Derman, W. The Diagnostic Utility of Computer-Assisted Auscultation for the Early Detection of Cardiac Murmurs of Structural Origin in the Periodic Health Evaluation. Sports Health 2017, 9, 341–345. [Google Scholar] [CrossRef]
- Dockerill, C.; Lapidaire, W.; Lewandowski, A.J.; Leeson, P. Cardiac remodelling and exercise: What happens with ultra-endurance exercise? Eur. J. Prev. Cardiol. 2020, 27, 1464–1466. [Google Scholar] [CrossRef]
- Barbieri, D.; Chawla, N.; Zaccagni, L.; Grgurinović, T.; Šarac, J.; Čoklo, M.; Missoni, S. Predicting Cardiovascular Risk in Athletes: Resampling Improves Classification Performance. Int. J. Environ. Res. Public Health 2020, 17, 7923. [Google Scholar] [CrossRef]
- Beavers, D.L.; Chung, E.H. Wearables in Sports Cardiology. Clin. Sports Med. 2022, 41, 405–423. Available online: https://linkinghub.elsevier.com/retrieve/pii/S0278591922000060 (accessed on 11 July 2022). [CrossRef]
- Bernardino, G.; Benkarim, O.; la Garza, M.S.-D.; Prat-Gonzàlez, S.; Sepulveda-Martinez, A.; Crispi, F.; Sitges, M.; Butakoff, C.; De Craene, M.; Bijnens, B.; et al. Handling confounding variables in statistical shape analysis—Application to cardiac remodelling. Med. Image Anal. 2020, 65, 101792. [Google Scholar] [CrossRef]
- Castillo-Atoche, A.; Caamal-Herrera, K.; Atoche-Enseñat, R.; Estrada-López, J.J.; Vázquez-Castillo, J.; Castillo-Atoche, A.C.; Palma-Marrufo, O.; Espinoza-Ruiz, A. Energy Efficient Framework for a AIoT Cardiac Arrhythmia Detection System Wearable during Sport. Appl. Sci. 2022, 12, 2716. [Google Scholar] [CrossRef]
- Chang, A.C. Primary Prevention of Sudden Cardiac Death of the Young Athlete: The Controversy About the Screening Electrocardiogram and Its Innovative Artificial Intelligence Solution. Pediatr. Cardiol. 2012, 33, 428–433. [Google Scholar] [CrossRef]
- Chatzakis, I.; Vassilakis, K.; Lionis, C.; Germanakis, I. Electronic health record with computerized decision support tools for the purposes of a pediatric cardiovascular heart disease screening program in Crete. Comput. Methods Programs Biomed. 2018, 159, 159–166. Available online: https://reader.elsevier.com/reader/sd/pii/S0169260717310829?token=84C38F2B617ADF4F313FF41CB8931F87EF613D8408E265AC70973B0637D45B24CF4ADF6ADE6942DA9DC0CBDAB4214746&originRegion=eu-west-1&originCreation=20211125174057 (accessed on 28 November 2021). [CrossRef] [PubMed]
- Christ, P.; Rückert, U. Identification of athletes during walking and jogging based on gait and electrocardiographic patterns. Commun. Comput. Inf. Sci. 2014, 452, 240–257. [Google Scholar]
- Liu, X.; Faes, L.; Kale, A.U.; Wagner, S.K.; Fu, D.J.; Bruynseels, A.; Mahendiran, T.; Moraes, G.; Shamdas, M.; Kern, C.; et al. A comparison of deep learning performance against health-care professionals in detecting diseases from medical imaging: A systematic review and meta-analysis. Lancet Digit. Health 2019, 1, e271–e297. [Google Scholar] [CrossRef]
- Marr, B. The Big Risks of Big Data in Sports. Forbes. 2017. Available online: https://www.forbes.com/sites/bernardmarr/2017/04/28/the-big-risks-of-big-data-in-sports/%3E (accessed on 25 November 2021).
- Moody, G.B.; Mark, R.G. The impact of the MIT-BIH Arrhythmia Database. IEEE Eng. Med. Biol. Mag. 2001, 20, 45–50. [Google Scholar] [CrossRef]
- Lugovaya, T. Biometric Human Identification Based on Electrocardiogram. Master’s Thesis, Electrotechnical University ‘LETI’, Saint-Petersburg, Russia, 2005. [Google Scholar]
- Greenwald, S.D.; Patil, R.S.; Mark, R.G. Improved Detection and Classification of Arrhythmias in Noise-Corrupted Electrocardiograms Using Contextual Information; IEEE: Piscataway, NJ, USA, 1990. [Google Scholar]
- Moody, G.B.; Mark, R.G. A new method for detecting atrial fibrillation using R-R intervals. Comput. Cardiol. 1983, 227–230. [Google Scholar]
- Laguna, P.; Mark, R.G.; Goldberg, A.; Moody, G.B. Database for evaluation of algorithms for measurement of QT and other waveform intervals in the ECG. Comput. Cardiol. 1997, 24, 673–676. [Google Scholar]
- Jager, F.; Taddei, A.; Moody, G.B.; Emdin, M.; Antolič, G.; Dorn, R.; Smrdel, A.; Marchesi, C.; Mark, R.G. Long-term ST database: A reference for the development and evaluation of automated ischaemia detectors and for the study of the dynamics of myocardial ischaemia. Med. Biol. Eng. Comput. 2003, 41, 172–182. [Google Scholar] [CrossRef]
- Goldberger, A.L.; Amaral, L.A.; Glass, L.; Hausdorff, J.M.; Ivanov, P.C.; Mark, R.G.; Mietus, J.E.; Moody, G.B.; Peng, C.K.; Stanley, H.E. PhysioBank, PhysioToolkit, and PhysioNet: Components of a new research resource for complex physiologic signals. Circulation 2000, 101, e215–e220. Available online: https://pubmed.ncbi.nlm.nih.gov/10851218/ (accessed on 13 December 2021). [CrossRef]
- Makimoto, H.; Höckmann, M.; Lin, T.; Glöckner, D.; Gerguri, S.; Clasen, L.; Schmidt, J.; Assadi-Schmidt, A.; Bejinariu, A.; Müller, P.; et al. Performance of a convolutional neural network derived from an ECG database in recognizing myocardial infarction. Sci. Rep. 2020, 10, 8445. Available online: https://www.nature.com/articles/s41598-020-65105-x (accessed on 25 November 2021). [CrossRef]
- Kwon, J.-M.; Kim, K.-H.; Medina-Inojosa, J.; Jeon, K.-H.; Park, J.; Oh, B.-H. Artificial intelligence for early prediction of pulmonary hypertension using electrocardiography. J. Heart Lung Transplant. 2020, 39, 805–814. [Google Scholar] [CrossRef]
- Erdenebayar, U.; Kim, H.; Park, J.-U.; Kang, D.; Lee, K.-J. Automatic Prediction of Atrial Fibrillation Based on Convolutional Neural Network Using a Short-term Normal Electrocardiogram Signal. J. Korean Med. Sci. 2019, 34, e64. Available online: https://jkms.org/DOIx.php?id=10.3346/jkms.2019.34.e64 (accessed on 25 November 2021). [CrossRef] [PubMed]
| Criteria | Term Location | |
|---|---|---|
| A | “deep learning” OR “machine learning” OR “artificial intelligence” | Anywhere within the manuscript |
| B | electrocardio* OR echocardio* | Anywhere within the manuscript |
| C | “athletes heart” OR “athlete*” | Title, Abstract, or Keywords |
| Group | Criteria | |
|---|---|---|
| 1 | Predictive Modelling | Main aim is to use some methodology to create a model or framework that can be used to classify data |
| 2 | Review | Consolidate existing literature in some way to construct practical guidelines or conduct a systematic review, etc. |
| 3 | Wearables | Main aim is the discussion or development of wearable technology for use as either a solely data collection enterprise or to conduct automatic analysis |
| 4 | Others | Does not fit the above criteria |
| Study | Sample Size (N) | Type of Method | Problem Addressed | Performance Metrics Stated |
|---|---|---|---|---|
| Adetiba et al. [14] | 40 | ANN | Automatic heart defect detection for athletes | Accuracy = 0.9 |
| Adetiba et al. [25] | 40 | ANN | Develop a wearable ECG that can be worn by athletes to help automatically detect defects | Accuracy = 1 |
| Barbieri et al. [34] | 26,002 | Decision trees Logistic regression | Classify whether an athlete is at cardiovascular risk or not | AUC = 0.78 |
| Bernardino et al. [36] | - | Logistic regression Principal component analysis Statistical shape analysis | Highlight areas of the heart that undergo cardiac remodelling due to endurance exercise | - |
| Castillo-Atroche et al. [37] | 56,542 samples from 487 patients | CNN | Automatically predict arrhythmias in athletes in real time | Accuracy = 0.939 |
| Christ and Rückert [40] | 22 and 9 | ANN Random forest Support vector machine | Predict whether a patient was an athlete or not based on ECG readings | Accuracy = 0.981 |
| Długosz et al. [16] | 160 | Decision tree Logistic regression | (1) Use ECGs to estimate the level of cardiac troponin (cTnI) in amateur athletes (2) Detect coronary artery disease (CAD) in athletes | AUC = 0.91 |
| Huang et al. [19] | 598 | Agglomerative hierarchical Clustering Multiple regression analysis | (1) Identify athlete groups with similar characteristics (2) Investigate the validity of sport-specific adaption for evaluating athlete’s hearts | - |
| Hussain at al [20] | 7200 data points from 4 athletes | LSTM | (1) Predict and athlete’s health state (2) Predict the activity being performed by an athlete | (1) Accuracy = 0.97 (2) Accuracy = 0.83 |
| Laurino et al. [21] | 14 and 12 | ANN K nearest neighbours Naïve Bayes Support vector machines | Classifying heart states in athletes between those at rest and those in stressful conditions | Accuracy = 0.86 |
| Lombardi et al. [22] | 26 | Linear discriminant analysis | Determine whether patients with idiopathic ventricular arrhythmias with left bundle branch block and inferior axis morphology arrhythmia originated from the aortic sinus cusps or the right ventricular outflow tract | Accuracy = 0.947 |
| Narula et al. [26] | 139 | ANN Random forest Support vector machine | Discriminate between hypertrophic cardiomyopathy from physiological hypertrophy in athletes | AUC = 0.795 |
| Rahmen et al. [27] | 470 | Naïve Bayes Random forest Support vector machines | Predict whether an athlete’s heart is normal or not | Accuracy 0.742 and 0.553 for experiments 1 and 2, respectively |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bellfield, R.A.A.; Ortega-Martorell, S.; Lip, G.Y.H.; Oxborough, D.; Olier, I. The Athlete’s Heart and Machine Learning: A Review of Current Implementations and Gaps for Future Research. J. Cardiovasc. Dev. Dis. 2022, 9, 382. https://doi.org/10.3390/jcdd9110382
Bellfield RAA, Ortega-Martorell S, Lip GYH, Oxborough D, Olier I. The Athlete’s Heart and Machine Learning: A Review of Current Implementations and Gaps for Future Research. Journal of Cardiovascular Development and Disease. 2022; 9(11):382. https://doi.org/10.3390/jcdd9110382
Chicago/Turabian StyleBellfield, Ryan A. A., Sandra Ortega-Martorell, Gregory Y. H. Lip, David Oxborough, and Ivan Olier. 2022. "The Athlete’s Heart and Machine Learning: A Review of Current Implementations and Gaps for Future Research" Journal of Cardiovascular Development and Disease 9, no. 11: 382. https://doi.org/10.3390/jcdd9110382
APA StyleBellfield, R. A. A., Ortega-Martorell, S., Lip, G. Y. H., Oxborough, D., & Olier, I. (2022). The Athlete’s Heart and Machine Learning: A Review of Current Implementations and Gaps for Future Research. Journal of Cardiovascular Development and Disease, 9(11), 382. https://doi.org/10.3390/jcdd9110382








