The Landscape of Cell Death Processes with Associated Immunogenic and Fibrogenic Effects in Arrhythmogenic Cardiomyopathy
Abstract
:1. Background
2. Methods
2.1. Collection and Processing of RNA-seq Data
2.2. Signature Gene Sets of Cell Death, Immune Cells, and Pathways
2.3. Differentially Expressed Cell Death Signature Genes
2.4. Comprehensive Analysis of Cell Death and Immune Responses Based on ssGSEA
2.5. Identification of Functional Coexpression Modules by WGCNA
2.6. Additional Methods and Statistics
3. Results
3.1. Differentially Expressed Cell Death Signature Genes
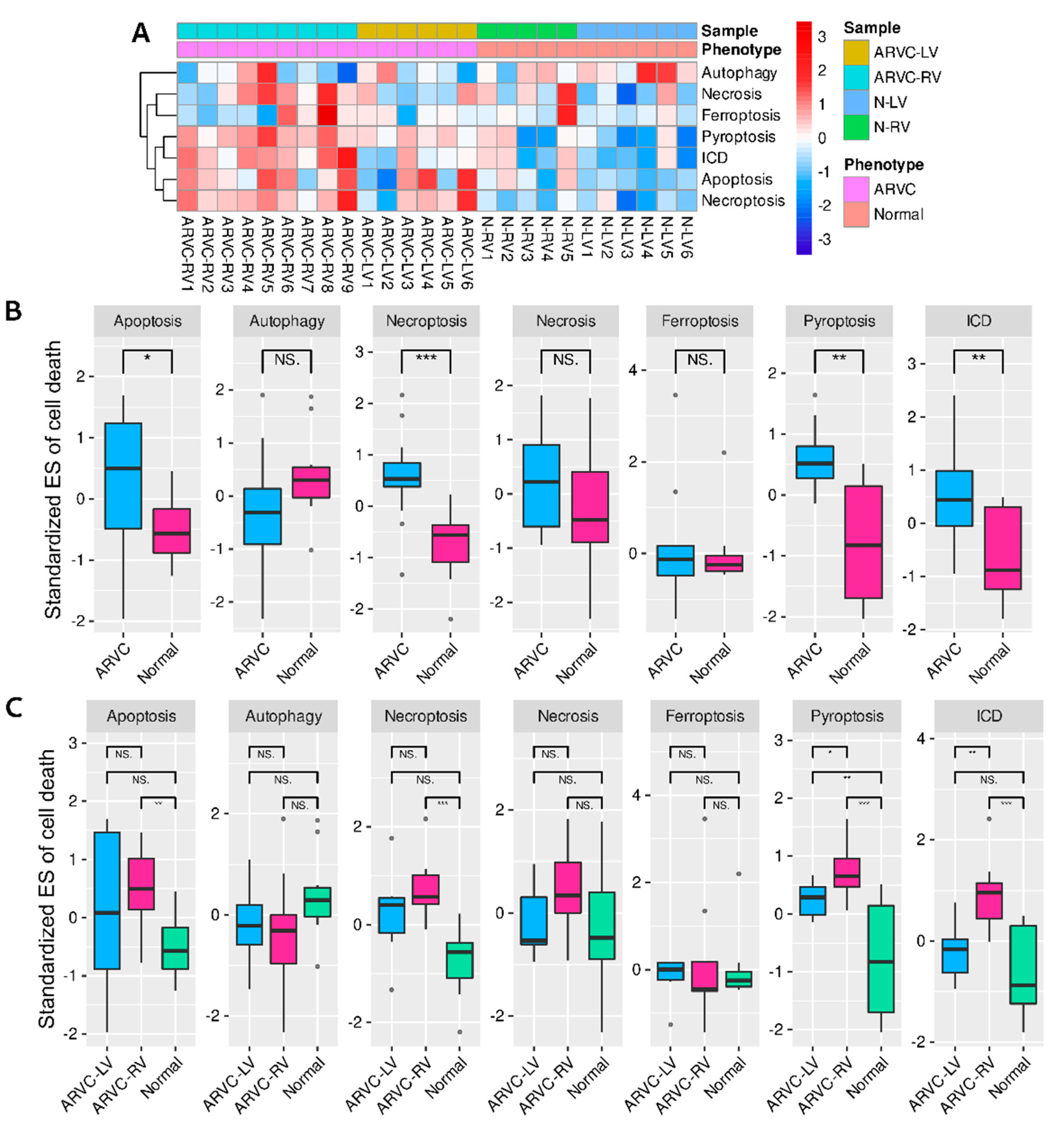
3.2. Comparisons and Correlation Analysis of Cell Death Processes
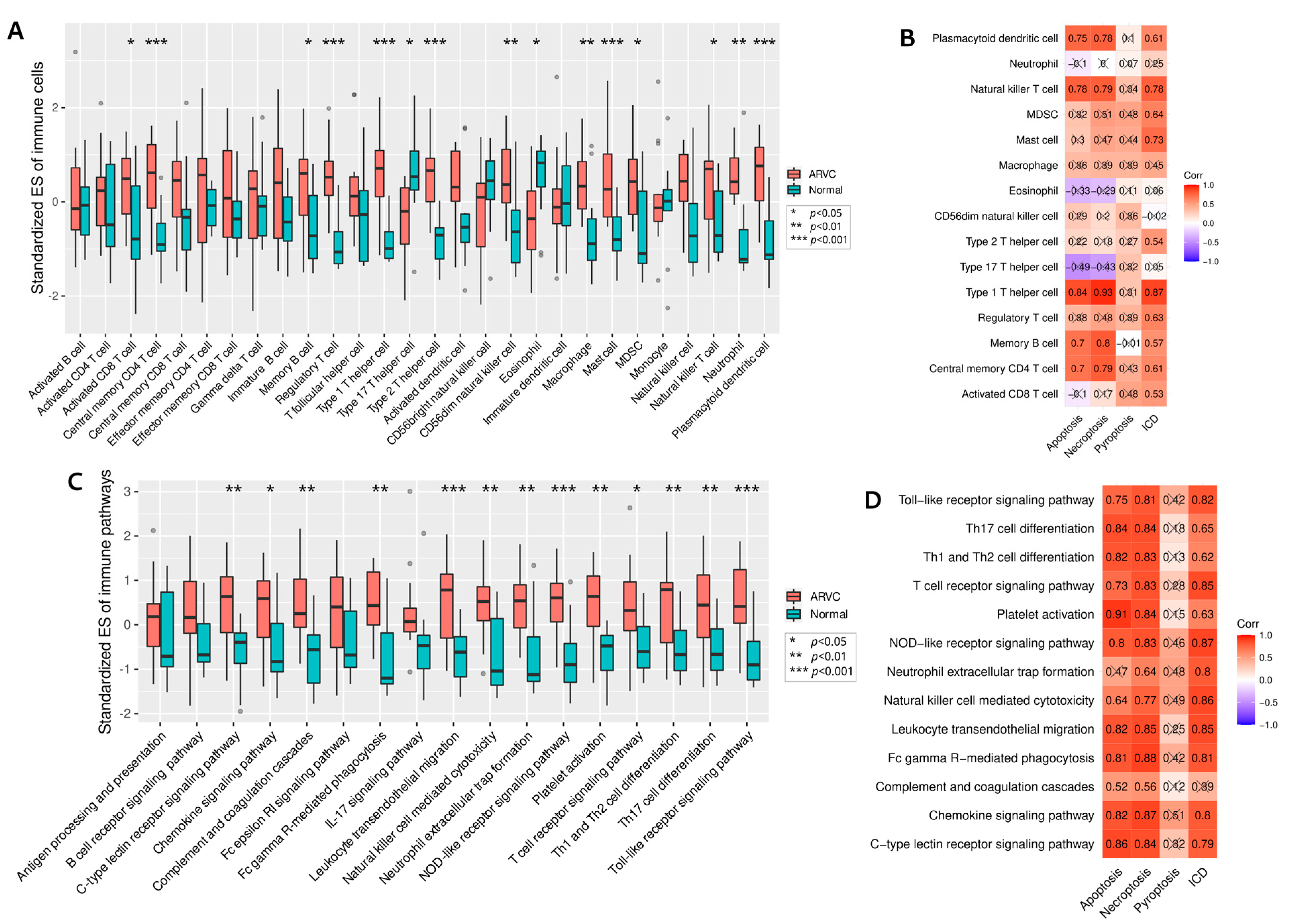
3.3. Relationships between Cell Death and Immune Responses
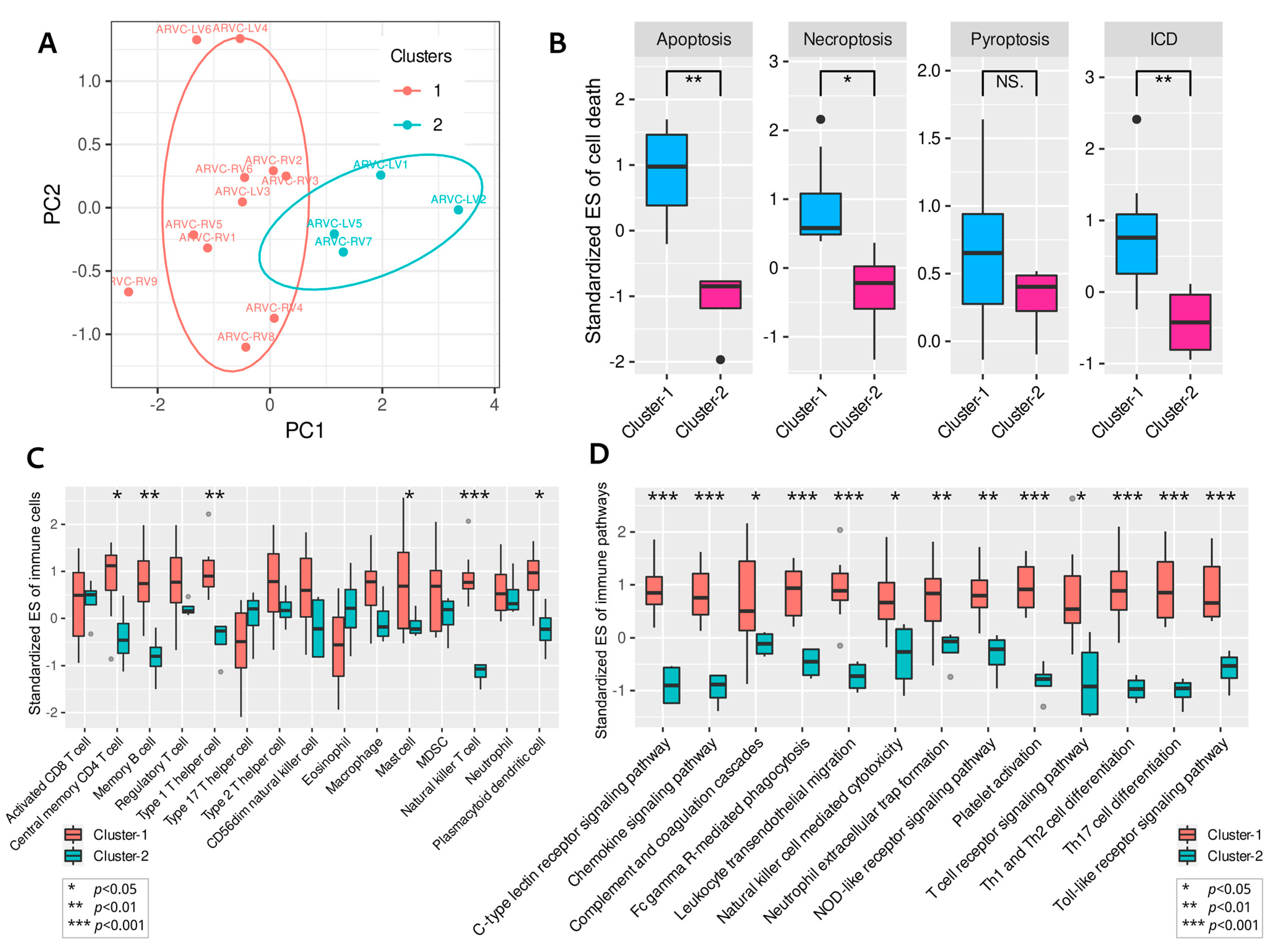
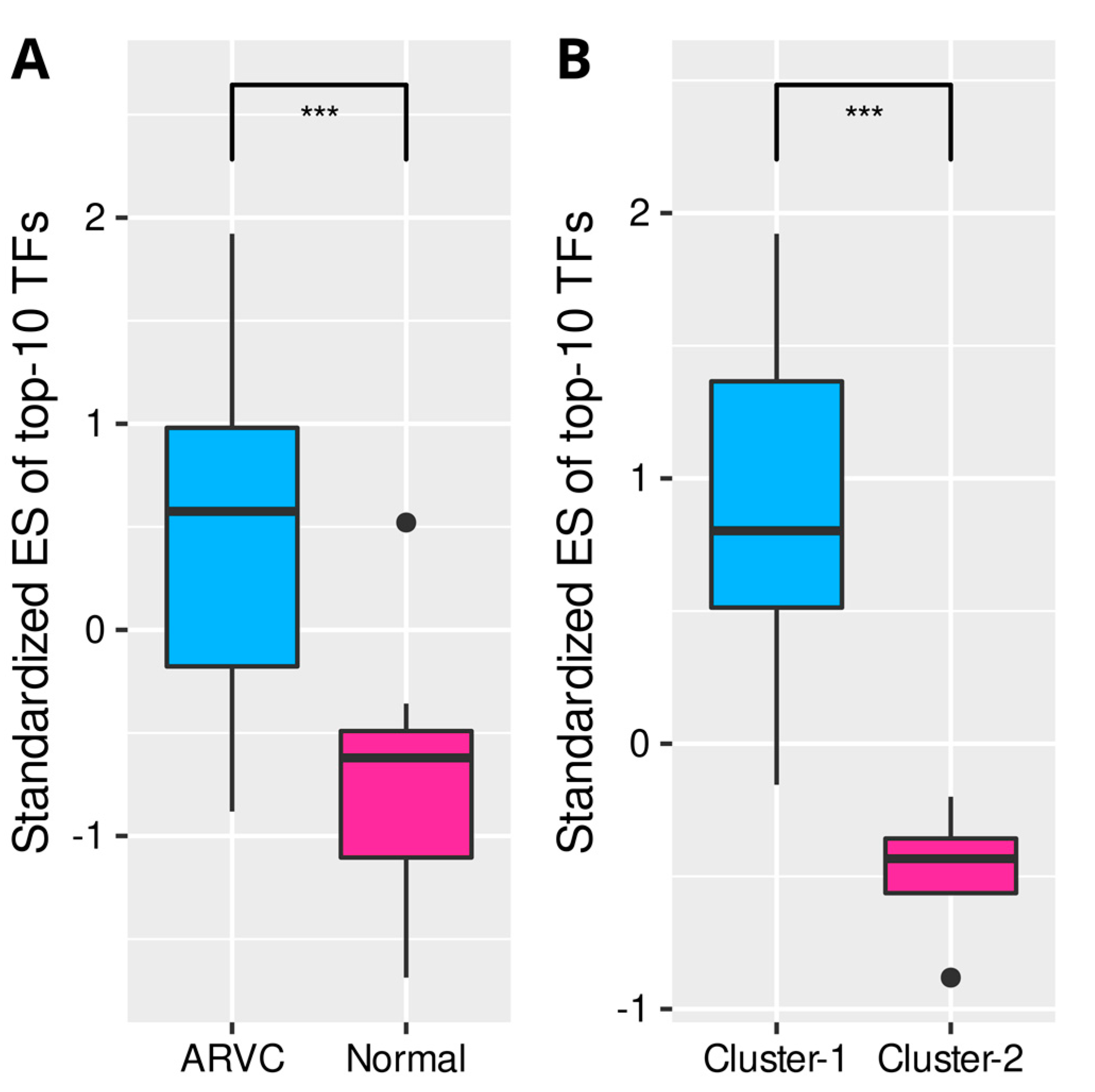
3.4. Clustering of ARVC Samples Based on Cell Death Scores
3.5. Correlations between Cell Death, Immune Responses, and Fibrosis in ARVC
3.6. Identifying the Functional Coexpression Module
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Austin, K.M.; Trembley, M.A.; Chandler, S.F.; Sanders, S.P.; Saffitz, J.E.; Abrams, D.J.; Pu, W.T. Molecular mechanisms of arrhythmogenic cardiomyopathy. Nat. Rev. Cardiol. 2019, 16, 519–537. [Google Scholar] [CrossRef] [PubMed]
- Berte, B.; Denis, A.; Amraoui, S.; Yamashita, S.; Komatsu, Y.; Pillois, X.; Sacher, F.; Mahida, S.; Wielandts, J.-Y.; Sellal, J.-M.; et al. Characterization of the Left-Sided Substrate in Arrhythmogenic Right Ventricular Cardiomyopathy. Circ. Arrhythmia Electrophysiol. 2015, 8, 1403–1412. [Google Scholar] [CrossRef]
- Gandjbakhch, E.; Redheuil, A.; Pousset, F.; Charron, P.; Frank, R. Clinical Diagnosis, Imaging, and Genetics of Arrhythmogenic Right Ventricular Cardiomyopathy/Dysplasia: JACC State-of-the-Art Review. J. Am. Coll. Cardiol. 2018, 72, 784–804. [Google Scholar] [CrossRef] [PubMed]
- Nishikawa, T.; Ishiyama, S.; Nagata, M.; Sakomura, Y.; Nakazawa, M.; Momma, K.; Hiroe, M.; Kasajima, T. Programmed cell death in the myocardium of arrhythmogenic right ventricular cardiomyopathy in children and adults. Cardiovasc. Pathol. 1999, 8, 185–189. [Google Scholar] [CrossRef]
- Mattesi, G.; Zorzi, A.; Corrado, D.; Cipriani, A. Natural History of Arrhythmogenic Cardiomyopathy. J. Clin. Med. 2020, 9, 878. [Google Scholar] [CrossRef]
- Asatryan, B.; Asimaki, A.; Landstrom, A.P.; Khanji, M.Y.; Odening, K.E.; Cooper, L.T.; Marchlinski, F.E.; Gelzer, A.R.; Semsarian, C.; Reichlin, T.; et al. Inflammation and Immune Response in Arrhythmogenic Cardiomyopathy: State-of-the-Art Review. Circulation 2021, 144, 1646–1655. [Google Scholar] [CrossRef]
- Galluzzi, L.; Vitale, I.; Aaronson, S.A.; Abrams, J.M.; Adam, D.; Agostinis, P.; Alnemri, E.S.; Altucci, L.; Amelio, I.; Andrews, D.W.; et al. Molecular mechanisms of cell death: Recommendations of the Nomenclature Committee on Cell Death 2018. Cell Death Differ. 2018, 25, 486–541. [Google Scholar] [CrossRef]
- Garg, A.D.; De Ruysscher, D.; Agostinis, P. Immunological metagene signatures derived from immunogenic cancer cell death associate with improved survival of patients with lung, breast or ovarian malignancies: A large-scale meta-analysis. Oncoimmunology 2016, 5, e1069938. [Google Scholar] [CrossRef]
- Wang, X.; Wu, S.; Liu, F.; Ke, D.; Wang, X.; Pan, D.; Xu, W.; Zhou, L.; He, W. An Immunogenic Cell Death-Related Classification Predicts Prognosis and Response to Immunotherapy in Head and Neck Squamous Cell Carcinoma. Front. Immunol. 2021, 12, 781466. [Google Scholar] [CrossRef]
- Ahluwalia, P.; Ahluwalia, M.; Mondal, A.K.; Sahajpal, N.; Kota, V.; Rojiani, M.V.; Rojiani, A.; Kolhe, R. Immunogenomic Gene Signature of Cell-Death Associated Genes with Prognostic Implications in Lung Cancer. Cancers 2021, 13, 155. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed]
- Hu, T.; Zhao, X.; Zhao, Y.; Cheng, J.; Xiong, J.; Lu, C. Identification and Verification of Necroptosis-Related Gene Signature and Associated Regulatory Axis in Breast Cancer. Front. Genet. 2022, 13, 842218. [Google Scholar] [CrossRef] [PubMed]
- Song, W.; Ren, J.; Xiang, R.; Kong, C.; Fu, T. Identification of pyroptosis-related subtypes, the development of a prognosis model, and characterization of tumor microenvironment infiltration in colorectal cancer. OncoImmunology 2021, 10, 1987636. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.Y.; Wang, D.S.; Lin, H.C.; Chen, X.X.; Yang, H.; Zheng, Y.; Li, Y.H. A Novel Ferroptosis-related Gene Signature for Overall Survival Prediction in Patients with Hepatocellular Carcinoma. Int. J. Biol. Sci. 2020, 16, 2430–2441. [Google Scholar] [CrossRef]
- Yue, Q.; Zhang, Y.; Wang, F.; Cao, F.; Duan, X.; Bai, J. Classification of colorectal carcinoma subtypes based on ferroptosis-associated molecular markers. World J. Surg. Oncol. 2022, 20, 117. [Google Scholar] [CrossRef] [PubMed]
- Charoentong, P.; Finotello, F.; Angelova, M.; Mayer, C.; Efremova, M.; Rieder, D.; Hackl, H.; Trajanoski, Z. Pan-cancer Immunogenomic Analyses Reveal Genotype-Immunophenotype Relationships and Predictors of Response to Checkpoint Blockade. Cell Rep. 2017, 18, 248–262. [Google Scholar] [CrossRef]
- Jia, Q.; Wu, W.; Wang, Y.; Alexander, P.B.; Sun, C.; Gong, Z.; Cheng, J.-N.; Sun, H.; Guan, Y.; Xia, X.; et al. Local mutational diversity drives intratumoral immune heterogeneity in non-small cell lung cancer. Nat. Commun. 2018, 9, 5361. [Google Scholar] [CrossRef]
- Hänzelmann, S.; Castelo, R.; Guinney, J. GSVA: Gene set variation analysis for microarray and RNA-seq data. BMC Bioinform. 2013, 14, 7. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef]
- Mallat, Z.; Tedgui, A.; Fontaliran, F.; Frank, R.; Durigon, M.; Fontaine, G. Evidence of Apoptosis in Arrhythmogenic Right Ventricular Dysplasia. N. Engl. J. Med. 1996, 335, 1190–1197. [Google Scholar] [CrossRef]
- Kavantzas, N.G.; Lazaris, A.C.; Agapitos, E.V.; Nanas, J.; Davaris, P.S. Histological assessment of apoptotic cell death in cardiomyopathies. Pathology 2000, 32, 176–180. [Google Scholar] [CrossRef]
- Nagata, M.; Hiroe, M.; Ishiyama, S.; Nishikawa, T.; Sakomura, Y.; Kasanuki, H.; Toyosaki, T.; Marumo, F. Apoptotic cell death in arrhythmogenic right ventricular cardiomyopathy: A comparative study with idiopathic sustained ventricular tachycardia. Jpn. Heart J. 2000, 41, 733–741. [Google Scholar] [CrossRef] [PubMed]
- Caspi, O.; Huber, I.; Gepstein, A.; Arbel, G.; Maizels, L.; Boulos, M.; Gepstein, L. Modeling of arrhythmogenic right ventricular cardiomyopathy with human induced pluripotent stem cells. Circ. Cardiovasc. Genet. 2013, 6, 557–568. [Google Scholar] [CrossRef]
- Pilichou, K.; Remme, C.A.; Basso, C.; Campian, M.E.; Rizzo, S.; Barnett, P.; Scicluna, B.P.; Bauce, B.; Hoff, M.J.V.D.; De Bakker, J.M.; et al. Myocyte necrosis underlies progressive myocardial dystrophy in mouse dsg2-related arrhythmogenic right ventricular cardiomyopathy. J. Exp. Med. 2009, 206, 1787–1802. [Google Scholar]
- Li, D.; Liu, Y.; Maruyama, M.; Zhu, W.; Chen, H.; Zhang, W.; Reuter, S.; Lin, S.-F.; Haneline, L.S.; Field, L.J.; et al. Restrictive loss of plakoglobin in cardiomyocytes leads to arrhythmogenic cardiomyopathy. Hum. Mol. Genet. 2011, 20, 4582–4596. [Google Scholar]
- Snyder, A.G.; Oberst, A. The Antisocial Network: Cross Talk Between Cell Death Programs in Host Defense. Annu. Rev. Immunol. 2021, 39, 77–101. [Google Scholar] [CrossRef]
- Lu, W.; Li, Y.; Dai, Y.; Chen, K. Dominant Myocardial Fibrosis and Complex Immune Microenvironment Jointly Shape the Pathogenesis of Arrhythmogenic Right Ventricular Cardiomyopathy. Front. Cardiovasc. Med. 2022, 9, 900810. [Google Scholar] [CrossRef]
- Stadhouders, R.; Lubberts, E.; Hendriks, R.W. A cellular and molecular view of T helper 17 cell plasticity in autoimmunity. J. Autoimmun. 2018, 87, 1–15. [Google Scholar] [CrossRef]
- Garg, A.D.; Martin, S.; Golab, J.; Agostinis, P. Danger signalling during cancer cell death: Origins, plasticity and regulation. Cell Death Differ. 2014, 21, 26–38. [Google Scholar]
- Talman, V.; Ruskoaho, H. Cardiac fibrosis in myocardial infarction—From repair and remodeling to regeneration. Cell Tissue Res. 2016, 365, 563–581. [Google Scholar] [CrossRef]
- Li, L.; Zhao, Q.; Kong, W. Extracellular matrix remodeling and cardiac fibrosis. Matrix Biol. 2018, 68–69, 490–506. [Google Scholar] [CrossRef] [PubMed]
- Frangogiannis, N.G. The extracellular matrix in myocardial injury, repair, and remodeling. J. Clin. Investig. 2017, 127, 1600–1612. [Google Scholar] [CrossRef] [PubMed]
- Ricard-Blum, S. The collagen family. Cold Spring Harb. Perspect. Biol. 2011, 3, a004978. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruoslahti, E. Fibronectin in cell adhesion and invasion. Cancer Metastasis Rev. 1984, 3, 43–51. [Google Scholar] [CrossRef]
- Ali, M.A.M.; Fan, X.; Schulz, R. Cardiac sarcomeric proteins: Novel intracellular targets of matrix metalloproteinase-2 in heart disease. Trends Cardiovasc. Med. 2011, 21, 112–118. [Google Scholar] [CrossRef]
- Guo, D.-C.; Regalado, E.S.; Gong, L.; Duan, X.; Santos-Cortez, R.L.P.; Arnaud, P.; Ren, Z.; Cai, B.; Hostetler, E.M.; Moran, R.; et al. LOX Mutations Predispose to Thoracic Aortic Aneurysms and Dissections. Circ. Res. 2016, 118, 928–934. [Google Scholar] [CrossRef]
- Salmond, R.J.; Filby, A.; Qureshi, I.; Caserta, S.; Zamoyska, R. T-cell receptor proximal signaling via the Src-family kinases, Lck and Fyn, influences T-cell activation, differentiation, and tolerance. Immunol. Rev. 2009, 228, 9–22. [Google Scholar] [CrossRef]
- Jung, S.H.; Yoo, E.H.; Yu, M.J.; Song, H.M.; Kang, H.Y.; Cho, J.Y.; Lee, J.R. ARAP, a Novel Adaptor Protein, Is Required for TCR Signaling and Integrin-Mediated Adhesion. J. Immunol. 2016, 197, 942–952. [Google Scholar] [CrossRef] [Green Version]
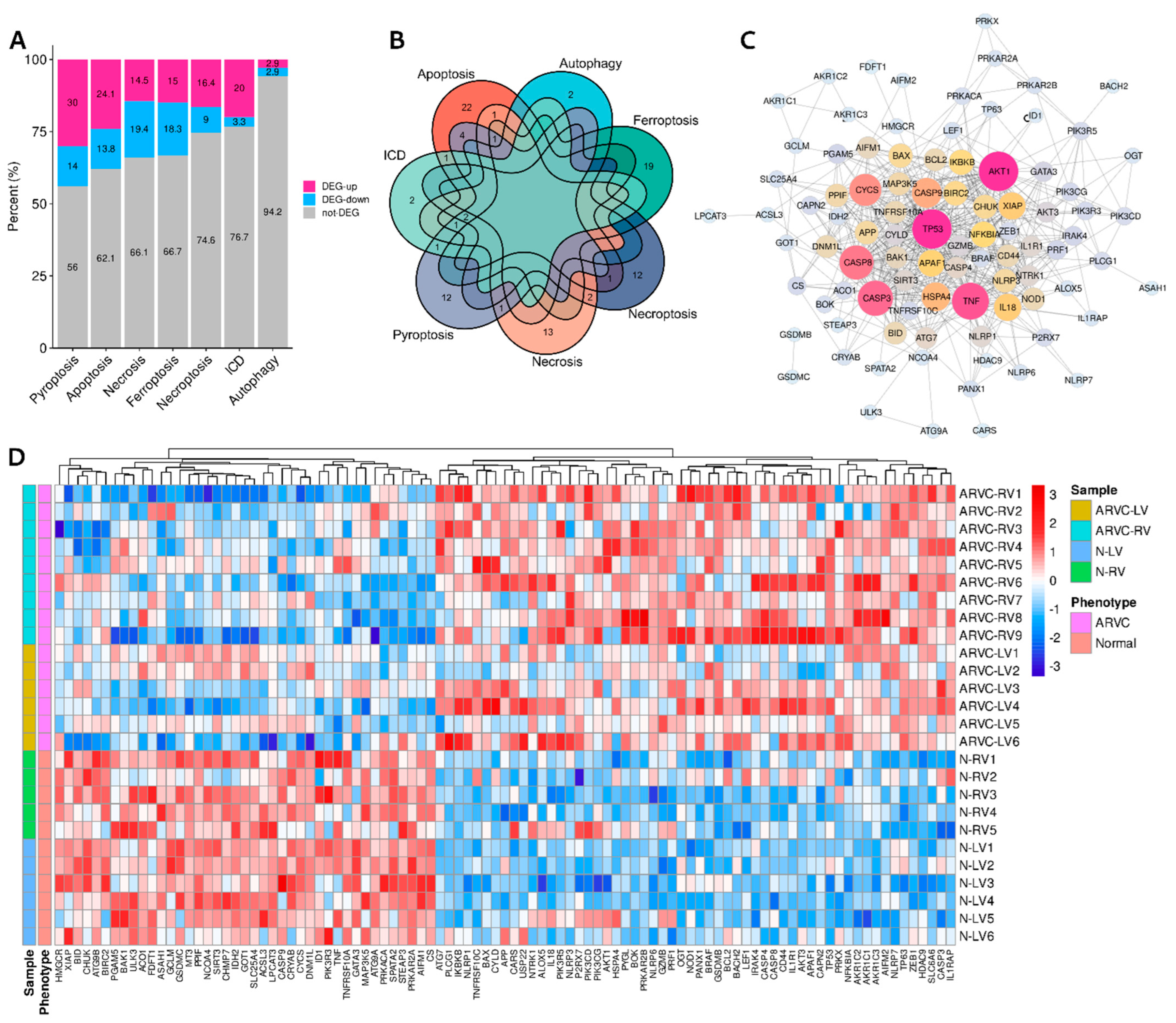

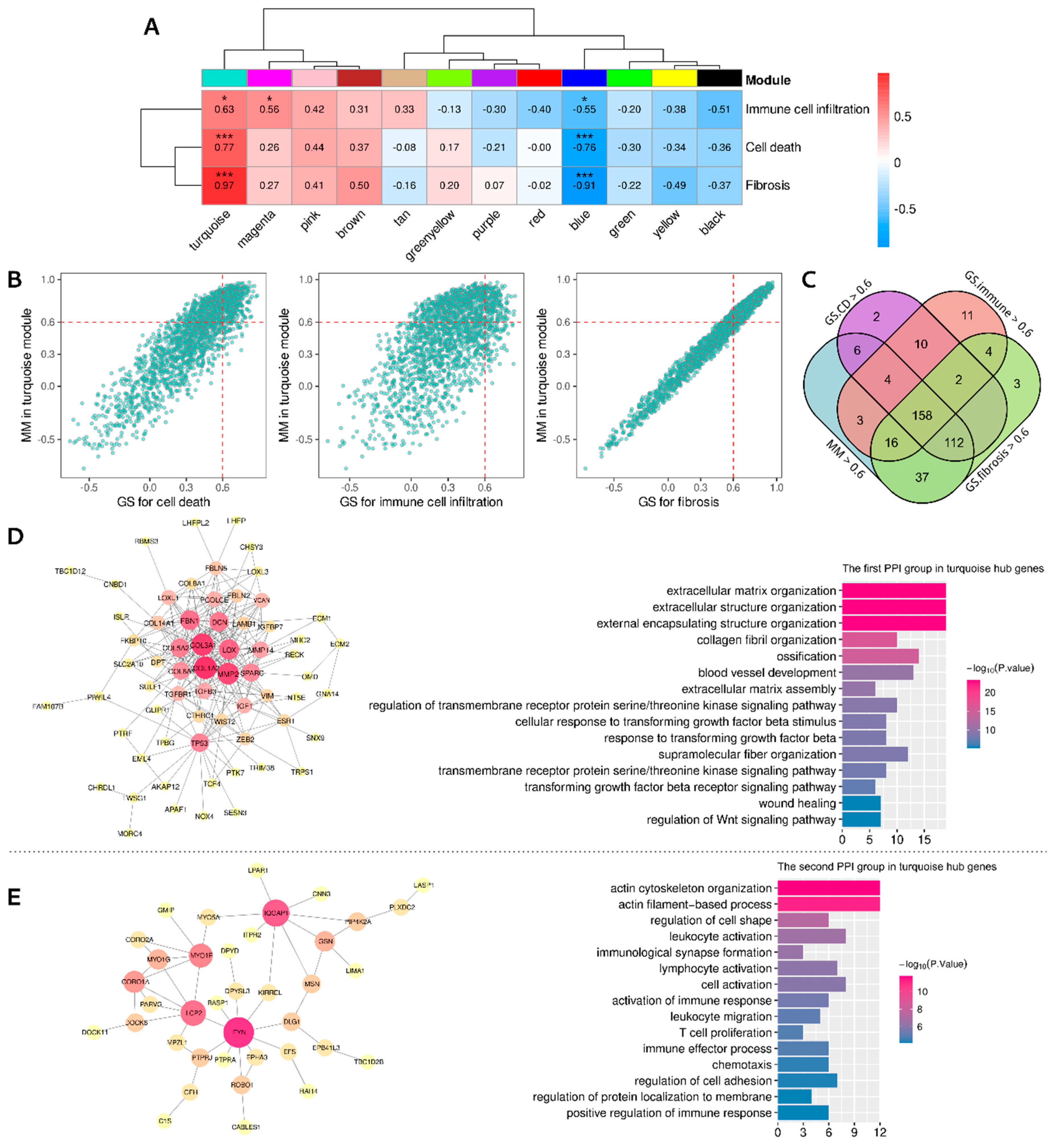
| Cell Death Category | Up-Regulated in ARVC Samples | Down-Regulated in ARVC Samples |
|---|---|---|
| Immunogenic cell death (n = 30) | PRF1, IL1R1, P2RX7, CASP8, NLRP3, BAX (n = 6) | TNF (n = 1) |
| Apoptosis (n = 87) | TNFRSF10C, NTRK1, NFKBIA, IL1R1, PIK3R5, PRKAR2B, BCL2, IL1RAP, CASP8, IKBKB, PIK3CG, AKT3, PIK3CD, CASP3, BAX, APAF1, IRAK4, TP53, AKT1, PRKX, CAPN2 (n = 21) | TNF, PRKACA, BID, AIFM1, CYCS, PIK3R3, TNFRSF10A, CASP9, PRKAR2A, BIRC2, CHUK, XIAP (n = 12) |
| Necroptosis (n = 67) | LEF1, HDAC9, PANX1, BACH2, BCL2, CASP8, CYLD, BRAF, HSPA4, USP22, APP (n = 11) | TNF, IDH2, SPATA2, GATA3, ID1, SIRT3 (n = 6) |
| Pyroptosis (n = 50) | NLRP6, NLRP7, GZMB, TP63, NLRP1, GSDMB, IL18, CASP8, NLRP3, CASP3, BAX, CASP4, NOD1, PLCG1, TP53 (n = 15) | GSDMC, TNF, PRKACA, CYCS, CASP9, BAK1, CHMP7 (n = 7) |
| Necrosis (n = 62) | NLRP6, SLC6A6, BOK, PYGL, CASP8, BAX, TP53, CYLD, OGT (n = 9) | ATG9B, MT3, TNF, SPATA2, PPIF, SLC25A4, MAP3K5, PGAM5, BIRC2, ATG9A, ASAH1, DNM1L (n = 12) |
| Ferroptosis (n = 60) | AKR1C2, AKR1C1, CD44, AKR1C3, ALOX5, ZEB1, AIFM2, TP53, CARS (n = 9) | LPCAT3, CS, STEAP3, GOT1, CRYAB, NCOA4, GCLM, ACSL3, ACO1, FDFT1, HMGCR (n = 11) |
| Autophagy (n = 35) | ATG7 (n = 1) | ULK3 (n = 1) |
| Key TF | Description | p-Value | FDR | List of Overlapped Genes |
|---|---|---|---|---|
| HIF1A | Hypoxia inducible factor 1, alpha subunit | 0.000005 | 0.0002 | MMP2, RECK, LOX, VIM, TGFB3, NT5E, TWIST2 |
| TFAP2C | Transcription factor AP-2 gamma | 0.00006 | 0.001 | MMP2, ESR1, ECM1 |
| TP53 | Tumor protein p53 | 0.0004 | 0.006 | TP53, CABLES1, VCAN, TRIM22, ESR1, APAF1, MMP2 |
| VHL | Von Hippel-Lindau tumor suppressor, E3 ubiquitin protein ligase | 0.0005 | 0.006 | SPARC, TCF4, TP53 |
| ETV4 | Ets variant 4 | 0.0007 | 0.006 | MMP14, VIM, MMP2 |
| STAT3 | Signal transducer and activator of transcription 3 | 0.001 | 0.007 | MMP2, NOX4, AKAP12, TWIST2, TP53, MMP14 |
| HDAC2 | Histone deacetylase 2 | 0.001 | 0.007 | COL1A2, IGF1, APAF1 |
| LEF1 | Lymphoid enhancer-binding factor 1 | 0.002 | 0.007 | ESR1, TCF4, NT5E |
| NCOR1 | Nuclear receptor corepressor 1 | 0.001 | 0.007 | IGF1, ESR1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lu, W.; Rao, Y.; Li, Y.; Dai, Y.; Chen, K. The Landscape of Cell Death Processes with Associated Immunogenic and Fibrogenic Effects in Arrhythmogenic Cardiomyopathy. J. Cardiovasc. Dev. Dis. 2022, 9, 301. https://doi.org/10.3390/jcdd9090301
Lu W, Rao Y, Li Y, Dai Y, Chen K. The Landscape of Cell Death Processes with Associated Immunogenic and Fibrogenic Effects in Arrhythmogenic Cardiomyopathy. Journal of Cardiovascular Development and Disease. 2022; 9(9):301. https://doi.org/10.3390/jcdd9090301
Chicago/Turabian StyleLu, Wenzhao, Yanfang Rao, Yao Li, Yan Dai, and Keping Chen. 2022. "The Landscape of Cell Death Processes with Associated Immunogenic and Fibrogenic Effects in Arrhythmogenic Cardiomyopathy" Journal of Cardiovascular Development and Disease 9, no. 9: 301. https://doi.org/10.3390/jcdd9090301
APA StyleLu, W., Rao, Y., Li, Y., Dai, Y., & Chen, K. (2022). The Landscape of Cell Death Processes with Associated Immunogenic and Fibrogenic Effects in Arrhythmogenic Cardiomyopathy. Journal of Cardiovascular Development and Disease, 9(9), 301. https://doi.org/10.3390/jcdd9090301





