Targeted Printing of Cells: Evaluation of ADA-PEG Bioinks for Drop on Demand Approaches
Abstract
:1. Introduction
2. Results and Discussion
2.1. Bioink Synthesis and Characterization
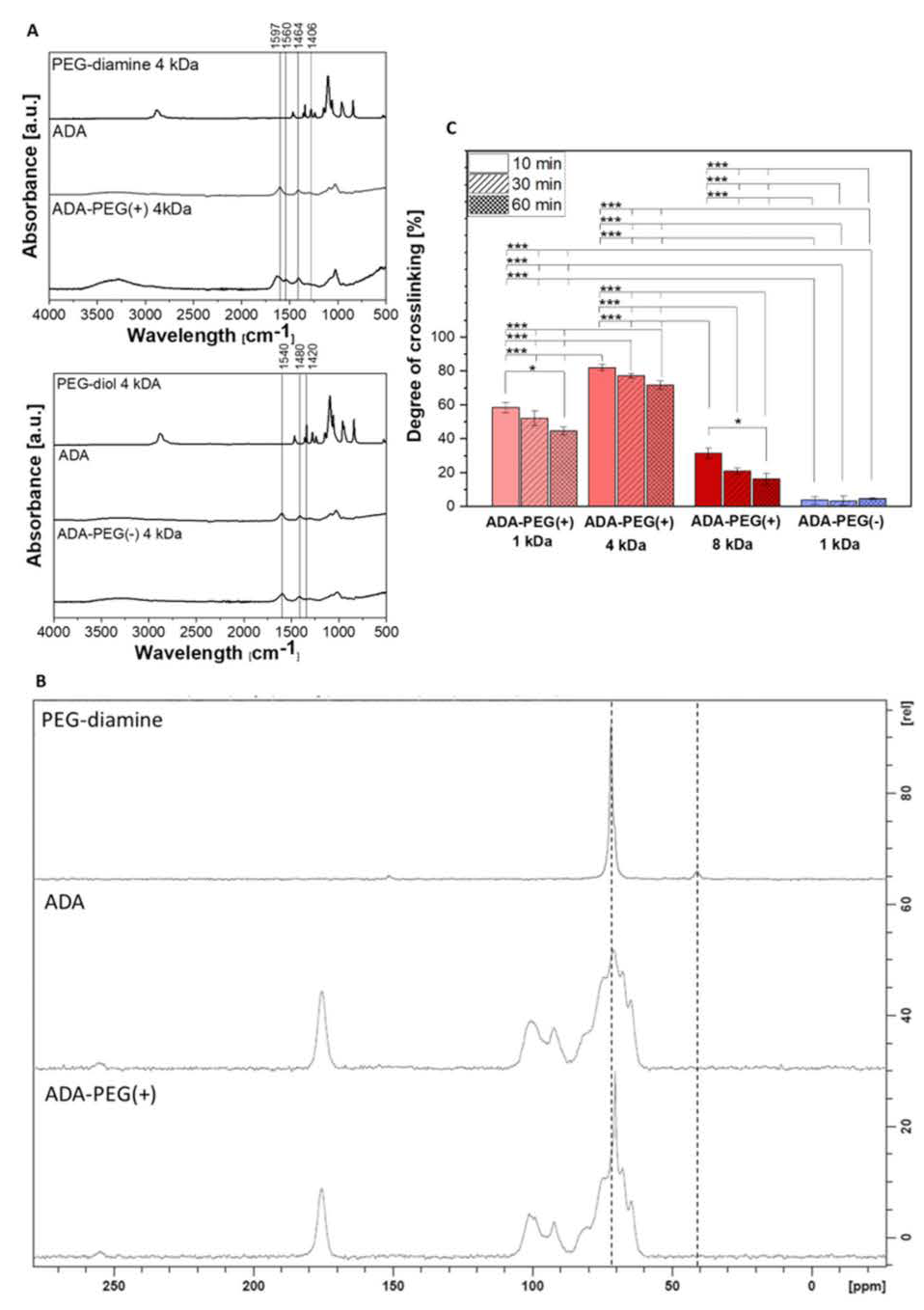
2.1.1. Chemical Composition
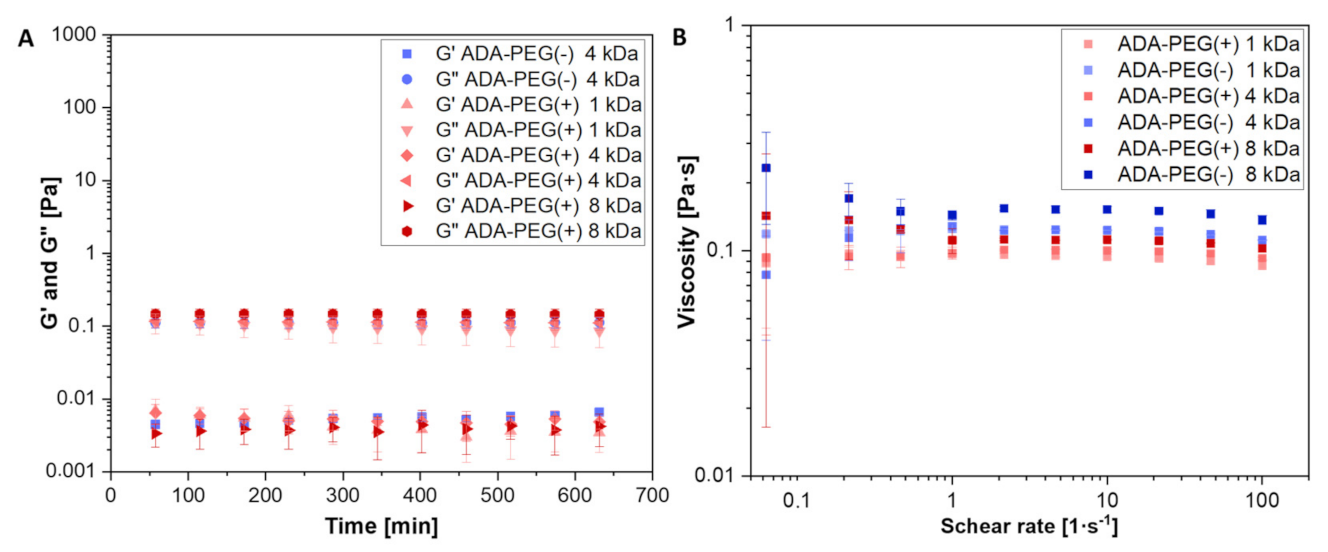
2.1.2. Rheological Characterization
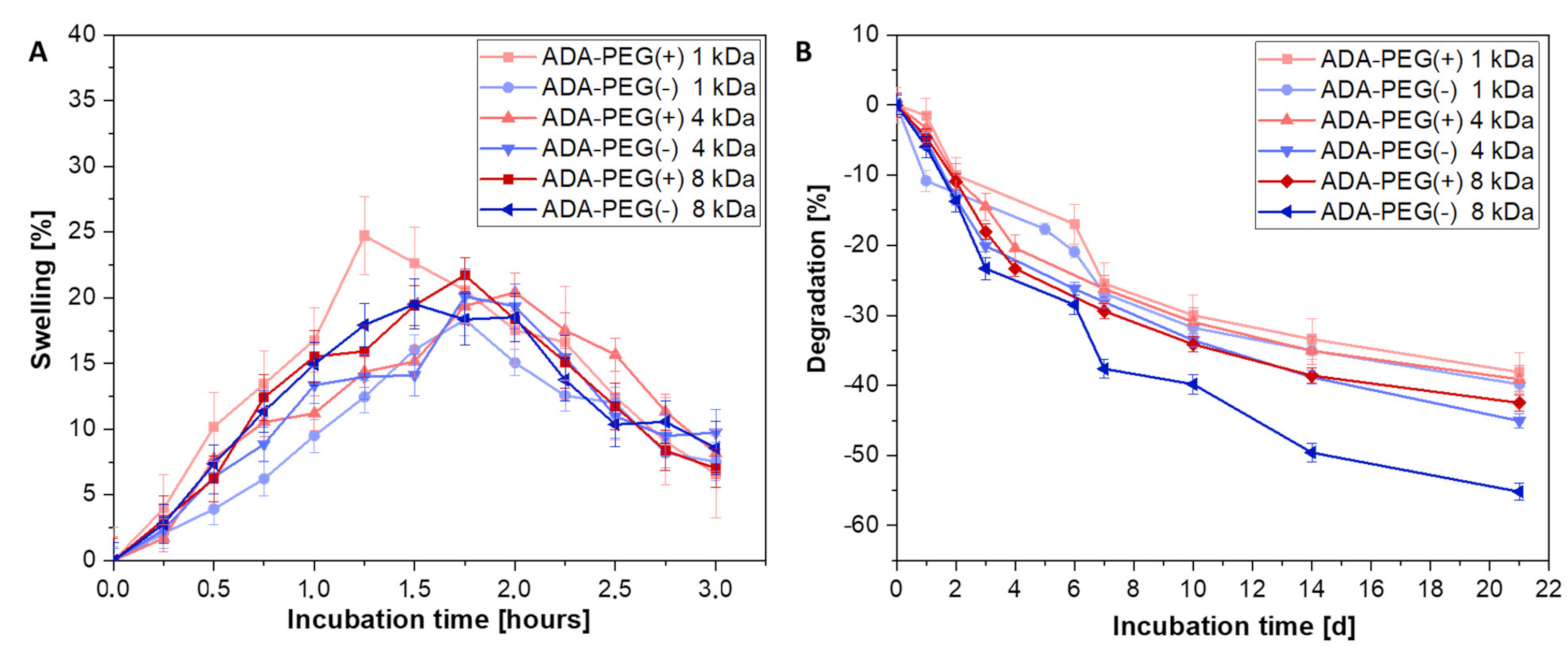
2.1.3. In Vitro Swelling and Degradation Behaviour
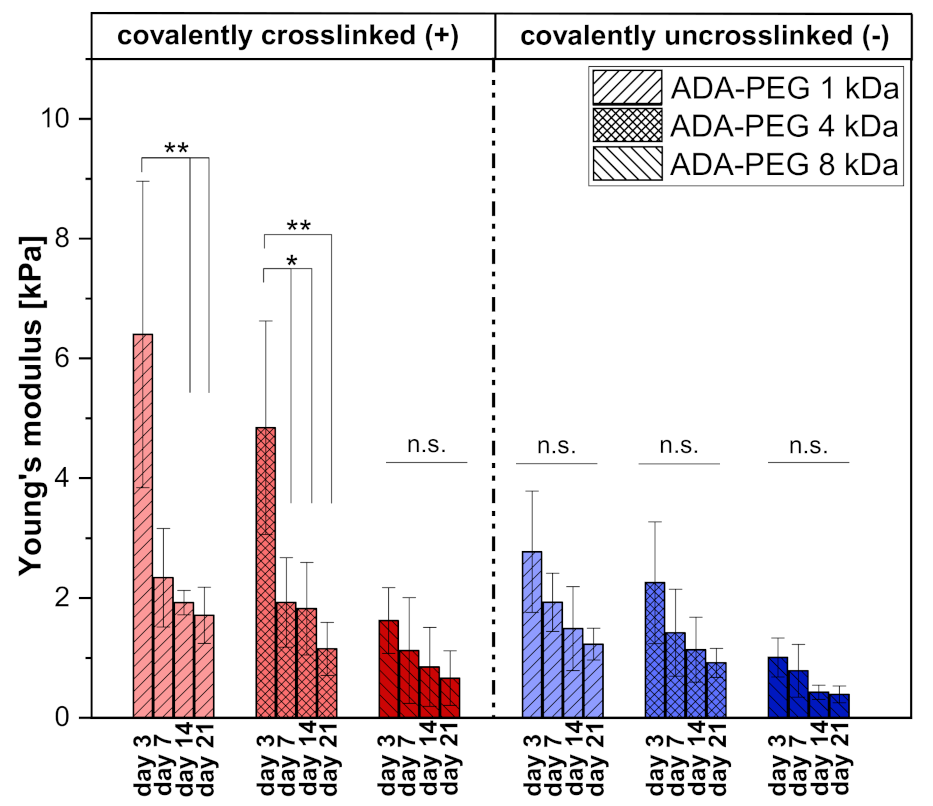
2.1.4. Mechanical Analysis Using Compression Tests
2.2. Evaluation of the Printing Process
2.2.1. Analytical Modulation of the Printing Process
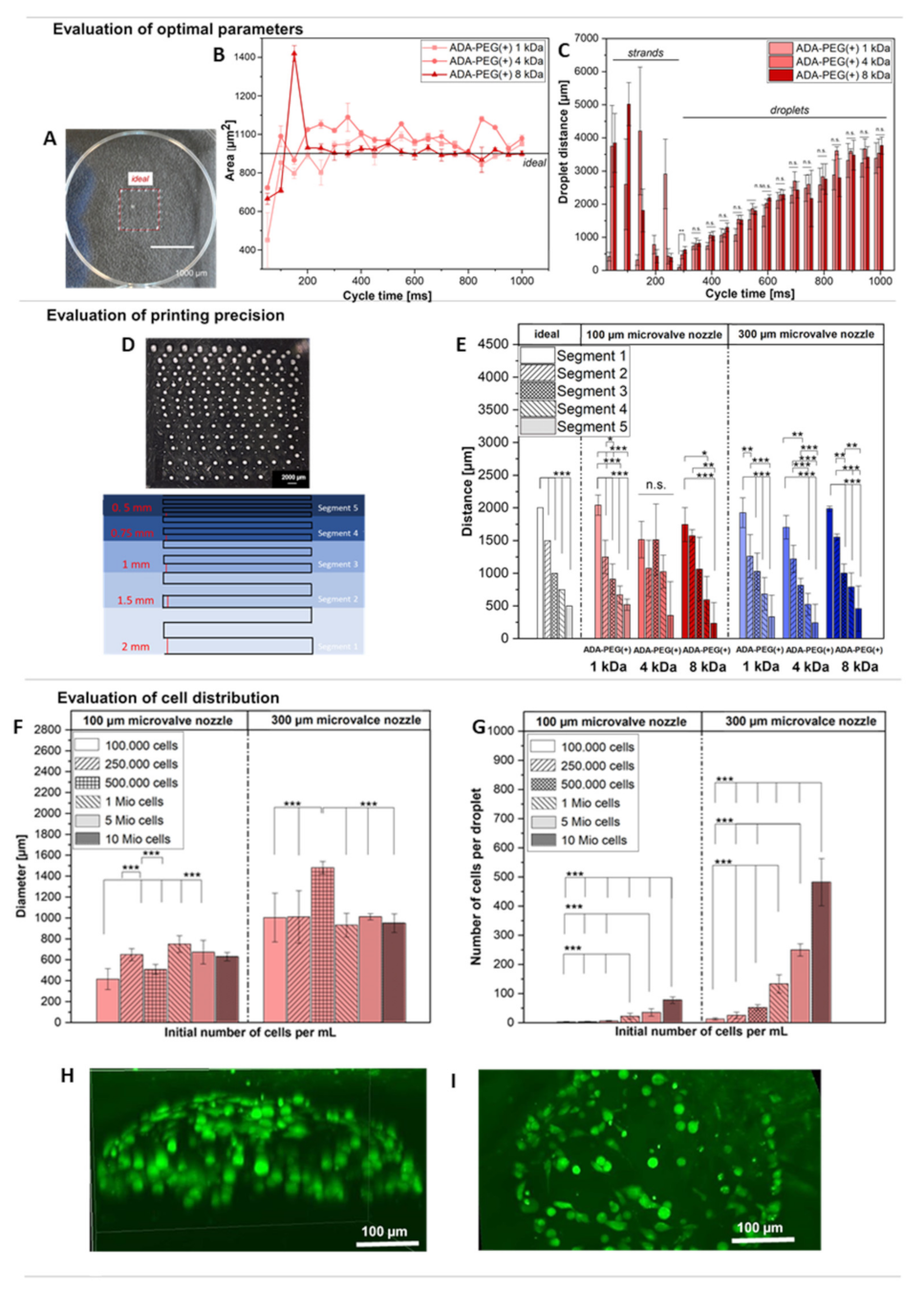
2.2.2. Evaluation of Printing Parameters
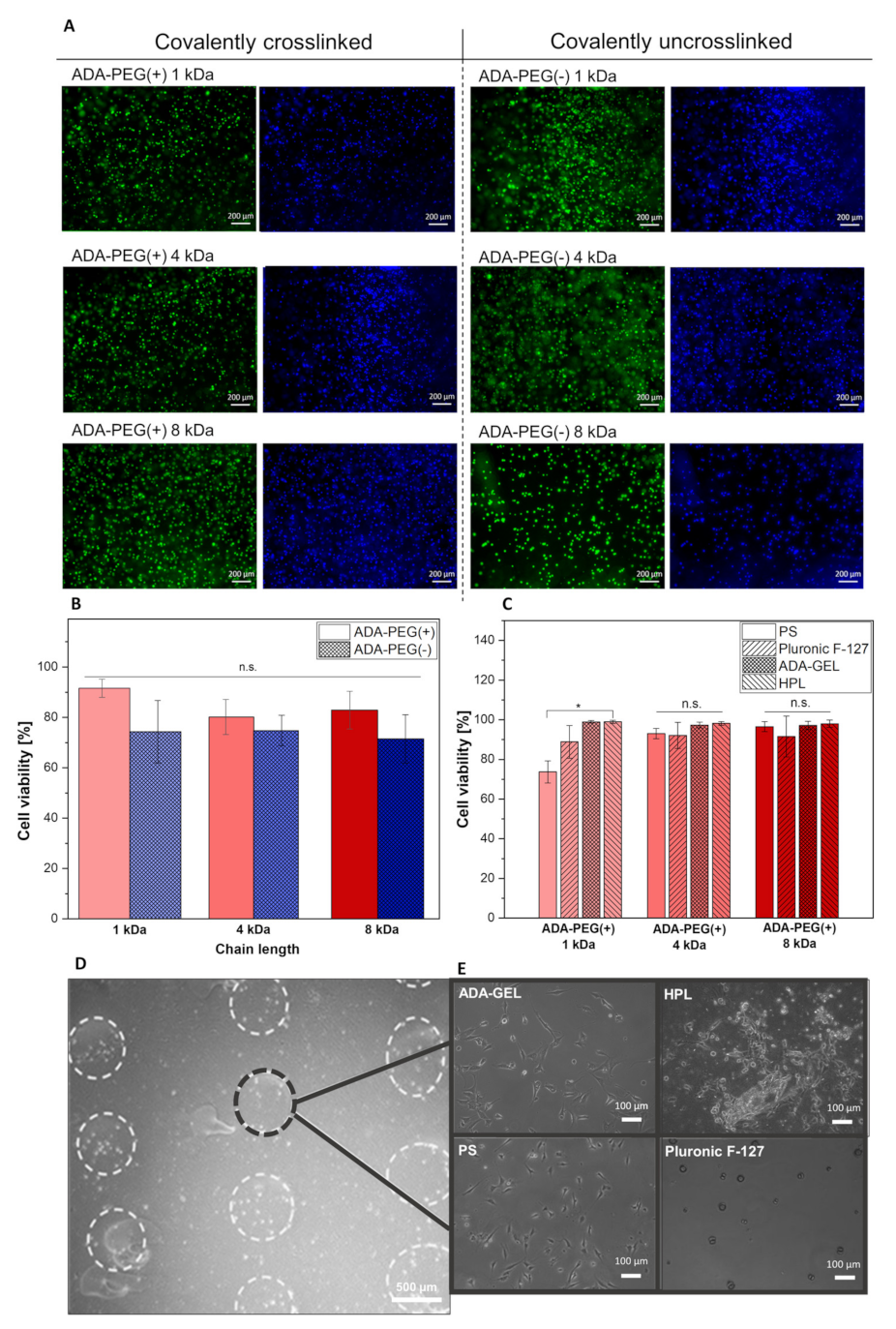
2.2.3. Targeted Cell Printing on Substrates
3. Conclusions
4. Materials and Methods
4.1. Bioink Synthesis
4.1.1. Synthesis of ADA
4.1.2. Synthesis of PEG-Diamine
4.1.3. Preparation of ADA-PEG
4.2. Bioink Characterization
4.2.1. Chemical Composition
4.2.2. Degree of Crosslinking
4.2.3. Rheological Characterization
4.2.4. In Vitro Degradation and Swelling Behaviour
4.2.5. Mechanical Analysis Using Compression Tests
4.3. Preparation of Hydrogels Used as Printing Substrates
4.4. Cytocompatibility
4.5. Evaluation of the Printing Process
4.5.1. Calculation of Shear Stress during Printing
4.5.2. Evaluation of Printing Parameters
4.5.3. Cell Printing
4.6. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Koch, L.; Deiwick, A.; Schlie, S.; Michael, S.; Gruene, M.; Coger, V.; Zychlinski, D.; Schambach, A.; Reimers, K.; Vogt, P.M.; et al. Skin Tissue Generation by Laser Cell Printing. Biotechnol. Bioeng. 2012, 109, 1855–1863. [Google Scholar] [CrossRef] [PubMed]
- Patra, S.; Young, V. A Review of 3D Printing Techniques and the Future in Biofabrication of Bioprinted Tissue. Cell Biochem. Biophys. 2016, 74, 93–98. [Google Scholar] [CrossRef] [PubMed]
- Pati, F.; Gantelius, J.; Svahn, H.A. 3D Bioprinting of Tissue/Organ Models. Angew. Chem. Int. Ed. 2016, 55, 4650–4665. [Google Scholar] [CrossRef]
- Chaudhary, S.; Chakraborty, E. Hydrogel Based Tissue Engineering and Its Future Applications in Personalized Disease Modeling and Regenerative Therapy. Beni-Suef Univ. J. Basic Appl. Sci. 2022, 11, 3. [Google Scholar] [CrossRef] [PubMed]
- Murphy, S.; Atala, A. 3D Bioprinting of Tissues and Organs. Nat. Biotechnol. 2014, 32, 773–785. [Google Scholar] [CrossRef] [PubMed]
- Jeon, H.; Kang, K.; Park, S.A.; Kim, W.D.; Paik, S.S.; Lee, S.H.; Jeong, J.; Choi, D. Generation of Multilayered 3D Structures of HepG2 Cells Using a Bio-printing Technique. Gut Liver 2017, 11, 121–128. [Google Scholar] [CrossRef] [Green Version]
- Hazur, J.; Detsch, R.; Karakaya, E.; Kaschta, J.; Teßmar, J.; Schneidereit, D.; Friedrich, O.; Schubert, D.W.; Boccaccini, A.R. Improving Alginate Printability for Biofabrication: Establishment of a Universal and Homogeneous Pre-Crosslinking Technique. Biofabrication 2020, 12, 045004. [Google Scholar] [CrossRef] [PubMed]
- Alcinesio, A.; Meacock, O.J.; Allan, R.G.; Monico, C.; Restrepo Schild, V.; Cazimoglu, I.; Cornall, M.T.; Krishna Kumar, R.; Bayley, H. Controlled Packing and Single-Droplet Resolution of 3D-Printed Functional Synthetic Tissues. Nat. Commun. 2020, 11, 2105. [Google Scholar] [CrossRef]
- Kador, K.E.; Grogan, S.P.; Dorthé, E.W.; Venugopalan, P.; Malek, M.F.; Goldberg, J.L.; D’Lima, D.D. Control of Retinal Ganglion Cell Positioning and Neurite Growth: Combining 3D Printing with Radial Electrospun Scaffolds. Tissue Eng. Part A 2016, 22, 286–294. [Google Scholar] [CrossRef] [Green Version]
- Wüst, S.; Müller, R.; Hofmann, S. Controlled Positioning of Cells in Biomaterials-Approaches towards 3D Tissue Printing Functional Biomaterials Controlled Positioning of Cells in Biomaterials-Approaches Towards 3D Tissue Printing. J. Funct. Biomater 2011, 2, 119–154. [Google Scholar] [CrossRef] [Green Version]
- Distler, T.; Solisito, A.A.; Schneidereit, D.; Friedrich, O.; Detsch, R.; Boccaccini, A.R. 3D Printed Oxidized Alginate-Gelatin Bioink Provides Guidance for C2C12 Muscle Precursor Cell Orientation and Differentiation via Shear Stress during Bioprinting. Biofabrication 2020, 12, 045005. [Google Scholar] [CrossRef] [PubMed]
- Sarker, B.; Papageorgiou, D.G.; Silva, R.; Zehnder, T.; Gul-E-Noor, F.; Bertmer, M.; Kaschta, J.; Chrissafis, K.; Detsch, R.; Boccaccini, A.R. Fabrication of Alginate–Gelatin Crosslinked Hydrogel Microcapsules and Evaluation of the Microstructure and Physico-Chemical Properties. J. Mater. Chem. B 2014, 2, 1470–1482. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dranseikiene, D.; Schrüfer, S.; Schubert, D.W.; Reakasame, S.; Boccaccini, A.R. Cell-Laden Alginate Dialdehyde–Gelatin Hydrogels Formed in 3D Printed Sacrificial Gel. J. Mater. Sci. Mater. Med. 2020, 31, 1–5. [Google Scholar] [CrossRef]
- Chen, E.P.; Toksoy, Z.; Davis, B.A.; Geibel, J.P. 3D Bioprinting of Vascularized Tissues for in Vitro and in Vivo Applications. Front. Bioeng. Biotechnol. 2021, 9, 664188. [Google Scholar] [CrossRef]
- Albanna, M.; Binder, K.W.; Murphy, V.; Kim, J.; Qasem, A.; Zhao, W.; Tan, J.; El-Amin, I.B.; Dice, D.D.; Marco, J.; et al. In Situ Bioprinting of Autologous Skin Cells Accelerates Wound Healing of Extensive Excisional Full-Thickness Wounds. Sci. Rep. 2019, 9, 1856. [Google Scholar] [CrossRef] [Green Version]
- Grottkau, B.E.; Zhixin, H.; Yonggang, P. A Novel 3D Bioprinter Using Direct-Volumetric Drop-On-Demand Technology for Fabricating Micro-Tissues and Drug-Delivery. Int. J. Mol. Sci. 2020, 21, 3482. [Google Scholar] [CrossRef]
- Saunders, R.E.; Gough, J.E.; Derby, B. Delivery of Human Fibroblast Cells by Piezoelectric Drop-on-Demand Inkjet Printing. Biomaterials 2008, 29, 193–203. [Google Scholar] [CrossRef] [PubMed]
- Boland, T.; Tao, X.; Damon, B.J.; Manley, B.; Kesari, P.; Jalota, S.; Bhaduri, S. Drop-on-Demand Printing of Cells and Materials for Designer Tissue Constructs. Mater. Sci. Eng. C 2007, 27, 372–376. [Google Scholar] [CrossRef]
- Uddin, M.J.; Scoutaris, N.; Klepetsanis, P.; Chowdhry, B.; Prausnitz, M.R.; Douroumis, D. Inkjet Printing of Transdermal Microneedles for the Delivery of Anticancer Agents. Int. J. Pharm. 2015, 494, 593–602. [Google Scholar] [CrossRef] [Green Version]
- Haas, R.; Lohse, S.; Düllmann, C.E.; Eberhardt, K.; Mokry, C.; Runke, J. Development and Characterization of a Drop-on-Demand Inkjet Printing System for Nuclear Target Fabrication. Nucl. Instrum. Methods Phys. Res. Sect. A Accel. Spectrometers Detect. Assoc. Equip. 2017, 874, 43–49. [Google Scholar] [CrossRef]
- Blaeser, A.; Campos, D.D.F.; Puster, U.; Fischer, H. Mikroventil-Basiertes 3D-Bioprinting-System Zum Additiven Aufbau Mit Lebenden Zellen Beladener Alginat-Strukturen. BioNanoMaterials 2014, 15, s185–s203. [Google Scholar] [CrossRef]
- Ferris, C.; Gilmore, K.; Beirne, S.; McCallum, D.; Wallace, G.; Panhuis, M. Bio-Ink for on-Demand Printing of Living Cells. Biomater. Sci. 2013, 1, 224–230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, Y.J.; Jun, Y.J.; Kim, D.Y.; Yi, H.G.; Chae, S.H.; Kang, J.; Lee, J.; Gao, G.; Kong, J.S.; Jang, J.; et al. A 3D Cell Printed Muscle Construct with Tissue-Derived Bioink for the Treatment of Volumetric Muscle Loss. Biomaterials 2019, 206, 160–169. [Google Scholar] [CrossRef]
- Hölzl, K.; Lin, S.; Tytgat, L.; van Vlierberghe, S.; Gu, L.; Ovsianikov, A. Bioink Properties before, during and after 3D Bioprinting. Biofabrication 2016, 8, 032002. [Google Scholar] [CrossRef] [PubMed]
- Zehnder, T.; Sarker, B.; Boccaccini, A.R.; Detsch, R. Evaluation of an Alginate–Gelatine Crosslinked Hydrogel for Bioplotting. Biofabrication 2015, 7, 025001. [Google Scholar] [CrossRef] [PubMed]
- Genç, H.; Hazur, J.; Karakaya, E.; Dietel, B.; Bider, F.; Groll, J.; Alexiou, C.; Boccaccini, A.R.; Detsch, R.; Cicha, I. Differential Responses to Bioink-Induced Oxidative Stress in Endothelial Cells and Fibroblasts. Int. J. Mol. Sci. 2021, 22, 2358. [Google Scholar] [CrossRef] [PubMed]
- Hernández-González, A.C.; Téllez-Jurado, L.; Rodríguez-Lorenzo, L.M. Preparation of Covalently Bonded Silica-Alginate Hybrid Hydrogels by SCHIFF Base and Sol-Gel Reactions. Carbohydr. Polym. 2021, 267, 118186. [Google Scholar] [CrossRef]
- Rajalekshmi, R.; Kaladevi Shaji, A.; Joseph, R.; Bhatt, A. Scaffold for Liver Tissue Engineering: Exploring the Potential of Fibrin Incorporated Alginate Dialdehyde–Gelatin Hydrogel. Int. J. Biol. Macromol. 2021, 166, 999–1008. [Google Scholar] [CrossRef]
- Mehedi Hasan, M.; Nuruzzaman Khan, M.; Haque, P.; Rahman, M.M. Novel Alginate-Di-Aldehyde Cross-Linked Gelatin/Nano-Hydroxyapatite Bioscaffolds for Soft Tissue Regeneration. Int. J. Biol. Macromol. 2018, 117, 1110–1117. [Google Scholar] [CrossRef]
- Bednarzig, V.; Karakaya, E.; Egaña, A.L.; Teßmar, J.; Boccaccini, A.R.; Detsch, R. Advanced ADA-GEL Bioink for Bioprinted Artificial Cancer Models. Bioprinting 2021, 23, e00145. [Google Scholar] [CrossRef]
- Olsen, D.; Yang, C.; Bodo, M.; Chang, R.; Leigh, S.; Baez, J.; Carmichael, D.; Perälä, M.; Hämäläinen, E.R.; Jarvinen, M.; et al. Recombinant Collagen and Gelatin for Drug Delivery. Adv. Drug Deliv. Rev. 2003, 55, 1547–1567. [Google Scholar] [CrossRef] [PubMed]
- Laurienzo, P.; Malinconico, M.; Motta, A.; Vicinanza, A. Synthesis and Characterization of a Novel Alginate–Poly(Ethylene Glycol) Graft Copolymer. Carbohydr. Polym. 2005, 62, 274–282. [Google Scholar] [CrossRef]
- Jeon, O.; Samorezov, J.E.; Alsberg, E. Single and Dual Crosslinked Oxidized Methacrylated Alginate/PEG Hydrogels for Bioadhesive Applications. Acta Biomater. 2014, 10, 47–55. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holtze, C.; Rowat, A.C.; Agresti, J.J.; Hutchison, J.B.; Angilè, F.E.; Schmitz, C.H.J.; Koster, S.; Duan, H.; Humphry, K.J.; Scanga, R.A.; et al. Biocompatible surfactants for water-in-fluorocarbon emulsions. R. Soc. Chem. 2008, 8, 1632–1639. [Google Scholar] [CrossRef]
- Tirella, A.; Vozzi, F.; de Maria, C.; Vozzi, G.; Sandri, T.; Sassano, D.; Cognolato, L.; Ahluwalia, A. Substrate Stiffness Influences High Resolution Printing of Living Cells with an Ink-Jet System. J. Biosci. Bioeng. 2011, 112, 79–85. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Liu, Y.; Hsu, S. hui Hydrogels Based on Schiff Base Linkages for Biomedical Applications. Molecules 2019, 24, 3005. [Google Scholar] [CrossRef] [Green Version]
- O’Hagan, D. Understanding Organofluorine Chemistry. An Introduction to the C–F Bond. Chem. Soc. Rev. 2008, 37, 308–319. [Google Scholar] [CrossRef]
- Kudernac, T.; Sändig, N.; Landaluce, T.F.; Wees, B.J.; Rudolf, P.; Katsonis, N.; Zerbetto, F.; Feringa, B.L. Intermolecular Repulsion through Interfacial Attraction: Toward Engineering of Polymorphs. J. Am. Chem. Soc. 2009, 131, 15655–15659. [Google Scholar] [CrossRef] [Green Version]
- Chopin-Doroteo, M.; Mandujano-Tinoco, E.A.; Krötzsch, E. Tailoring of the Rheological Properties of Bioinks to Improve Bioprinting and Bioassembly for Tissue Replacement. Biochim. Biophys. Acta Gen. Subj. 2021, 1865, 129782. [Google Scholar] [CrossRef]
- Ghosh, K.; Shu, X.Z.; Mou, R.; Lombardi, J.; Prestwich, G.D.; Rafailovich, M.H.; Clark, R.A.F. Rheological Characterization of in Situ Cross-Linkable Hyaluronan Hydrogels. Biomacromolecules 2005, 6, 2857–2865. [Google Scholar] [CrossRef]
- Mahendra, A.; James, H.P.; Jadhav, S. PEG-Grafted Phospholipids in Vesicles: Effect of PEG Chain Length and Concentration on Mechanical Properties. Chem. Phys. Lipids 2019, 218, 47–56. [Google Scholar] [CrossRef] [PubMed]
- Cha, C.; Jeong, J.H.; Shim, J.; Kong, H. Tuning the Dependency between Stiffness and Permeability of a Cell Encapsulating Hydrogel with Hydrophilic Pendant Chains. Acta Biomater. 2011, 7, 3719–3728. [Google Scholar] [CrossRef] [PubMed]
- Emami, Z.; Ehsani, M.; Zandi, M.; Foudazi, R. Controlling Alginate Oxidation Conditions for Making Alginate-Gelatin Hydrogels. Carbohydr. Polym. 2018, 198, 509–517. [Google Scholar] [CrossRef] [PubMed]
- Blaeser, A.; Duarte Campos, D.F.; Puster, U.; Richtering, W.; Stevens, M.M.; Fischer, H. Controlling Shear Stress in 3D Bioprinting Is a Key Factor to Balance Printing Resolution and Stem Cell Integrity. Adv. Healthc. Mater. 2016, 5, 326–333. [Google Scholar] [CrossRef] [PubMed]
- Cai, F.-F.; Heid, S.; Boccaccini, A.R. Potential of Laponite® Incorporated Oxidized Alginate–Gelatin (ADA-GEL) Composite Hydrogels for Extrusion-Based 3D Printing. J. Biomed. Mater. Res. Part B Appl. Biomater. 2021, 109, 1090–1104. [Google Scholar] [CrossRef]
- Dani, S.; Ahlfeld, T.; Albrecht, F.; Duin, S.; Kluger, P.; Lode, A.; Gelinsky, M. Homogeneous and Reproducible Mixing of Highly Viscous Biomaterial Inks and Cell Suspensions to Create Bioinks. Gels 2021, 7, 227. [Google Scholar] [CrossRef]
- Ding, W.; Zhou, J.; Zeng, Y.; Wang, Y.-N.; Shi, B. ARTICLE IN PRESS G Model Preparation of Oxidized Sodium Alginate with Different Molecular Weights and Its Application for Crosslinking Collagen Fiber. Carbohydr. Polym. 2016. [Google Scholar] [CrossRef]
- Monavari, M.; Homaeigohar, S.; Fuentes-Chandía, M.; Nawaz, Q.; Monavari, M.; Venkatraman, A.; Boccaccini, A.R. 3D Printing of Alginate Dialdehyde-Gelatin (ADA-GEL) Hydrogels Incorporating Phytotherapeutic Icariin Loaded Mesoporous SiO2-CaO Nanoparticles for Bone Tissue Engineering. Mater. Sci. Eng. C 2021, 131, 112470. [Google Scholar] [CrossRef]
- Soltan, N.; Ning, L.; Mohabatpour, F.; Papagerakis, P.; Chen, X. Printability and Cell Viability in Bioprinting Alginate Dialdehyde-Gelatin Scaffolds. ACS Biomater. Sci. Eng. 2019, 5, 2976–2987. [Google Scholar] [CrossRef]
- Distler, T.; Polley, C.; Shi, F.; Schneidereit, D.; Ashton, M.D.; Friedrich, O.; Kolb, J.F.; Hardy, J.G.; Detsch, R.; Seitz, H.; et al. Electrically Conductive and 3D-Printable Oxidized Alginate-Gelatin Polypyrrole:PSS Hydrogels for Tissue Engineering. Adv. Healthc. Mater. 2021, 10, 2001876. [Google Scholar] [CrossRef]
- Goktas, M.; Cinar, G.; Orujalipoor, I.; Ide, S.; Tekinay, A.B.; Guler, M.O. Self-Assembled Peptide Amphiphile Nanofibers and PEG Composite Hydrogels as Tunable ECM Mimetic Microenvironment. Biomacromolecules 2015, 16, 1247–1258. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Kao, W.J. Synthesis of Polyethylene Glycol (PEG) Derivatives and PEGylated−Peptide Biopolymer Conjugates. Biomacromolecules 2003, 4, 1055–1067. [Google Scholar] [CrossRef] [PubMed]
- Iijima, M.; Ulkoski, D.; Sakuma, S.; Matsukuma, D.; Nishiyama, N.; Otsuka, H.; Scholz, C. Synthesis of PEGylated Poly(Amino Acid) Pentablock Copolymers and Their Self-Assembly. Polym. Int. 2016, 65, 1132–1141. [Google Scholar] [CrossRef]
- Danjou, P.E.; Wallyn, D.; Cazier-Dennin, F.; Delattre, F. Ultrasound-Promoted Tosylation of Oligo(Ethylene Glycols). Ultrason. Sonochem. 2012, 19, 1201–1204. [Google Scholar] [CrossRef]
- Liu, C.; Yuan, J.; Luo, X.; Chen, M.; Chen, Z.; Zhao, Y.; Li, X. Folate-Decorated and Reduction-Sensitive Micelles Assembled from Amphiphilic Polymer-Camptothecin Conjugates for Intracellular Drug Delivery. Mol. Pharm. 2014, 11, 4258–4269. [Google Scholar] [CrossRef]
- Mollica, G.; Ziarelli, F.; Lack, S.; Brunel, F.; Viel, S. Characterization of Insoluble Calcium Alginates by Solid-State NMR. Carbohydr. Polym. 2012, 87, 383–391. [Google Scholar] [CrossRef]
- Nguyen, T.P.; Lee, B.T. Fabrication of Oxidized Alginate-Gelatin-BCP Hydrogels and Evaluation of the Microstructure, Material Properties and Biocompatibility for Bone Tissue Regeneration. J. Biomater. Appl. 2012, 27, 311–321. [Google Scholar] [CrossRef]

| ADA-PEG(+) 1 kDa | ADA-PEG(+) 4 kDa | ADA-PEG(+) 8 kDa | |
|---|---|---|---|
| Consistency factor k (mPa·s) | 20.90 | 26.79 | 43.41 |
| Flow exponent n | 0.96 | 0.98 | 0.97 |
| Drop ejection speed (m·s−1) | 7.35 | 6.61 | 5.94 |
| Average nozzle shear stress (kPa) | 1.3 | 1.9 | 2.3 |
| Label of Ink | ADA Type | PEG Type |
|---|---|---|
| ADA-PEG(+) 1 kDa | %DO = 13% | PEG-diamine 1 kDa |
| ADA-PEG(+) 4 kDa | %DO = 13% | PEG-diamine 4 kDa |
| ADA-PEG(+) 8 kDa | %DO = 13% | PEG-diamine 8 kDa |
| ADA-PEG(-) 1 kDa | %DO = 13% | PEG-diol 1 kDa |
| ADA-PEG(-) 4 kDa | %DO = 13% | PEG-diol 4 kDa |
| ADA-PEG(-) 8 kDa | %DO = 13% | PEG-diol 8 kDa |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Karakaya, E.; Bider, F.; Frank, A.; Teßmar, J.; Schöbel, L.; Forster, L.; Schrüfer, S.; Schmidt, H.-W.; Schubert, D.W.; Blaeser, A.; et al. Targeted Printing of Cells: Evaluation of ADA-PEG Bioinks for Drop on Demand Approaches. Gels 2022, 8, 206. https://doi.org/10.3390/gels8040206
Karakaya E, Bider F, Frank A, Teßmar J, Schöbel L, Forster L, Schrüfer S, Schmidt H-W, Schubert DW, Blaeser A, et al. Targeted Printing of Cells: Evaluation of ADA-PEG Bioinks for Drop on Demand Approaches. Gels. 2022; 8(4):206. https://doi.org/10.3390/gels8040206
Chicago/Turabian StyleKarakaya, Emine, Faina Bider, Andreas Frank, Jörg Teßmar, Lisa Schöbel, Leonard Forster, Stefan Schrüfer, Hans-Werner Schmidt, Dirk Wolfram Schubert, Andreas Blaeser, and et al. 2022. "Targeted Printing of Cells: Evaluation of ADA-PEG Bioinks for Drop on Demand Approaches" Gels 8, no. 4: 206. https://doi.org/10.3390/gels8040206
APA StyleKarakaya, E., Bider, F., Frank, A., Teßmar, J., Schöbel, L., Forster, L., Schrüfer, S., Schmidt, H.-W., Schubert, D. W., Blaeser, A., Boccaccini, A. R., & Detsch, R. (2022). Targeted Printing of Cells: Evaluation of ADA-PEG Bioinks for Drop on Demand Approaches. Gels, 8(4), 206. https://doi.org/10.3390/gels8040206









