Computational Investigation to Design Ofloxacin-Loaded Hybridized Nanocellulose/Lipid Nanogels for Accelerated Skin Repair
Abstract
1. Introduction
2. Results and Discussion
2.1. Characterization of the Ofloxacin-Loaded Hybridized NC/Lipid Nanogels
2.1.1. Particle Size Analysis
2.1.2. Encapsulation Efficiency (EE%)
2.1.3. In Vitro Release Study
2.2. Characterization of the Optimum Formula
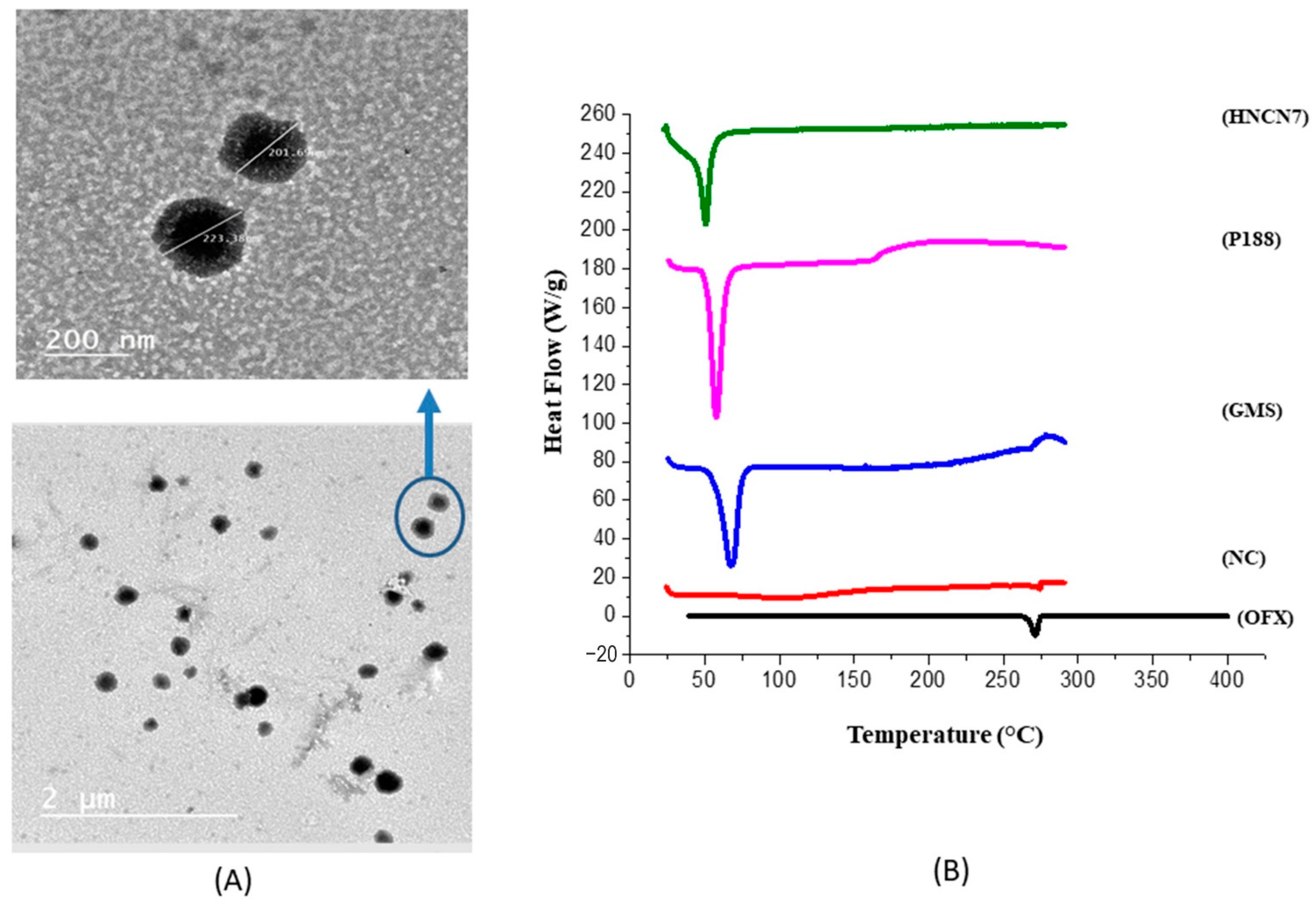
2.2.1. Morphological Examination
2.2.2. Differential Scanning Calorimetry
2.2.3. pH Measurement
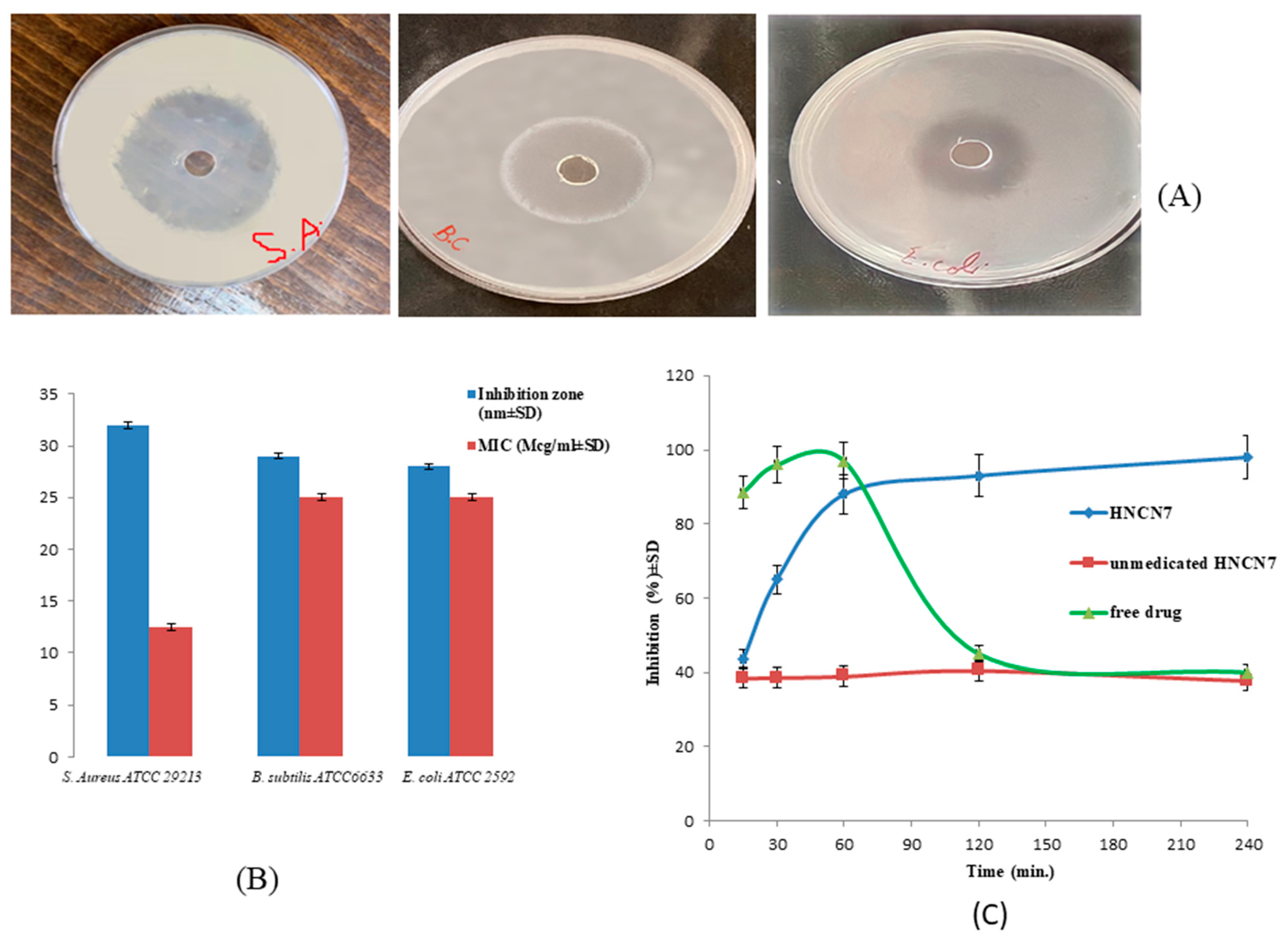
2.3. Microbiological Studies
2.3.1. Sensitivity Test and Measurement of the MIC
2.3.2. In-Vitro Time-Dependent Antibacterial Assay
2.4. Biological In Vitro Assay
2.4.1. Cytotoxicity Study
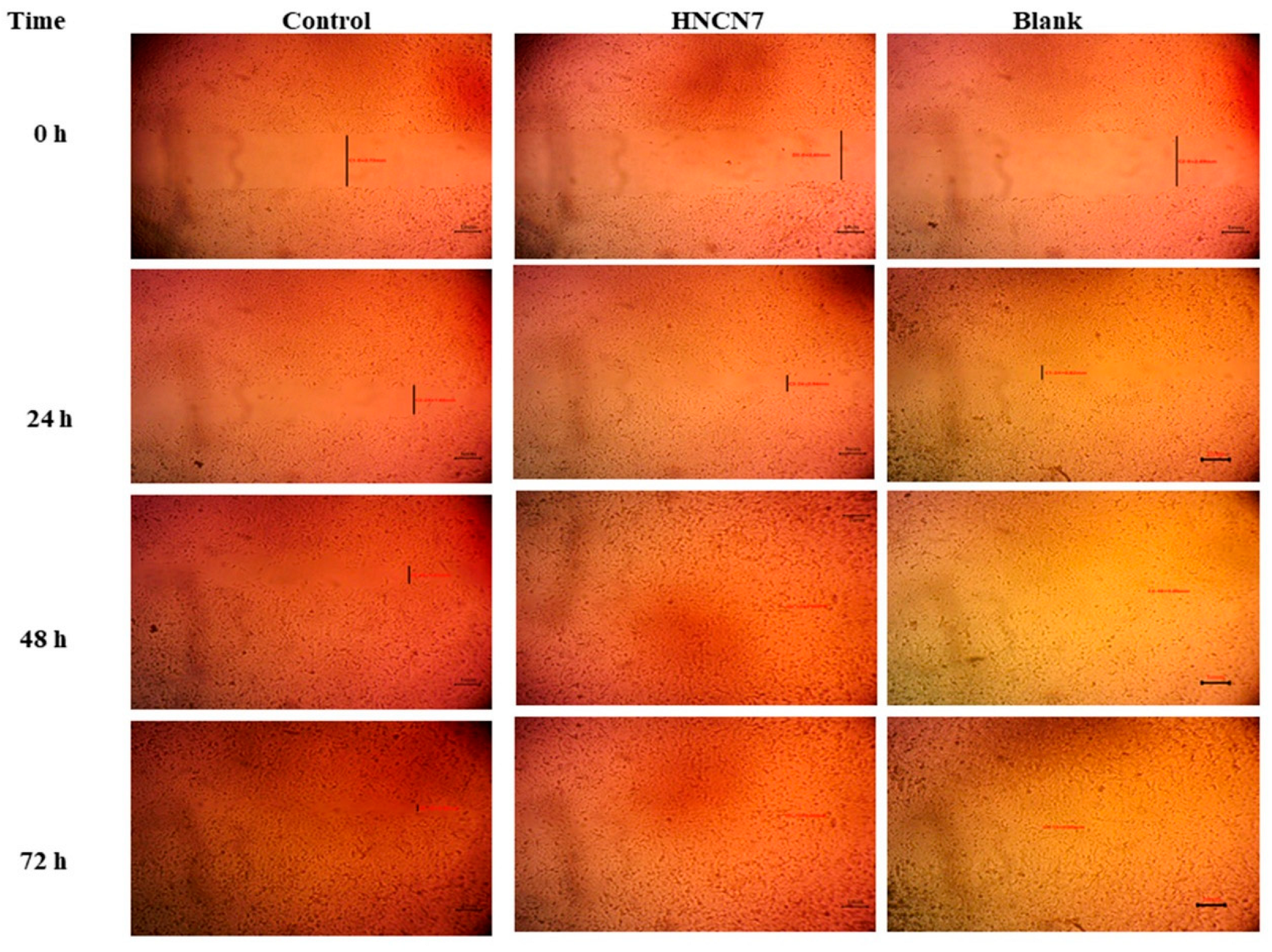
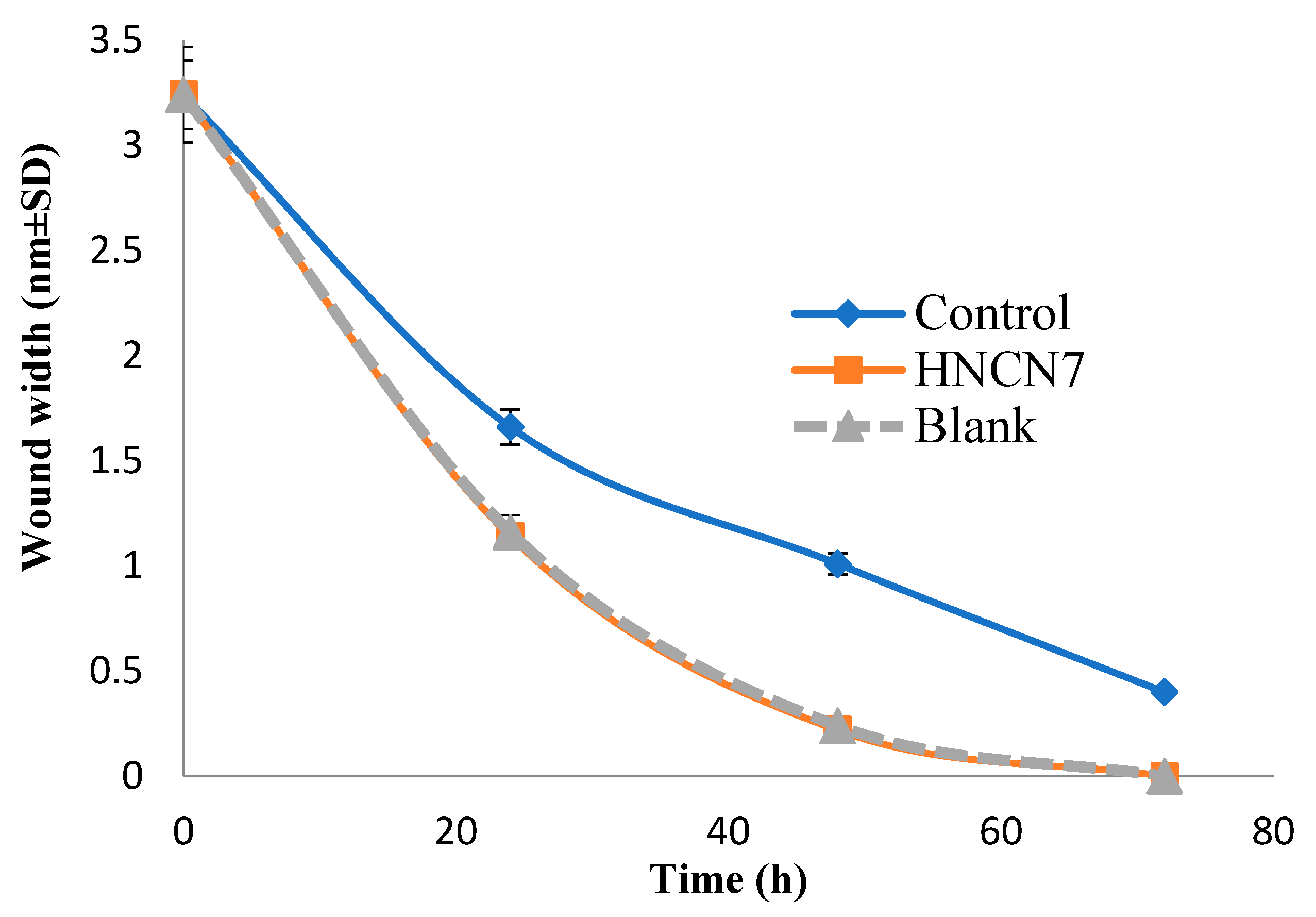
2.4.2. Experimental Scratch Assay
3. Conclusions
4. Materials and Methods
4.1. Materials
4.2. Methods
4.2.1. Design of Experiments (DOE) and Regression Analysis
4.2.2. Preparation of Ofloxacin-Loaded Hybridized NC/Lipid Nanogels
4.2.3. Evaluation of Ofloxacin-Loaded Hybridized NC/Lipid Nanogels
Encapsulation Efficiency (EE)
Estimation of Particle Size, Zeta Potential, and Polydispersity Index Analysis
In-Vitro Release Studies
4.2.4. Characterization of the Optimized HNCN
Differential Scanning Calorimetry (DSC)
pH Measurement
Morphological Examination
4.2.5. Microbiological Studies
Antimicrobial Sensitivity Test
Minimal Inhibitory Concentration (MIC) Measurement
In-Vitro Time-Dependent Antibacterial Assay
4.2.6. Biological In-Vitro Assay
Cytotoxicity Study
Experimental Scratch Assay
Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lei, H.; Zhu, C.; Fan, D. Optimization of human-like collagen composite polysaccharide hydrogel dressing preparation using response surface for burn repair. Carbohydr. Polym. 2020, 239, 116249. [Google Scholar] [CrossRef]
- Sheng, L.; Zhang, Z.; Zhang, Y.; Wang, E.; Ma, B.; Xu, Q.; Ma, L.; Zhang, M.; Pei, G.; Chang, J. A novel “hot spring”-mimetic hydrogel with excellent angiogenic properties for chronic wound healing. Biomaterials 2021, 264, 120414. [Google Scholar] [CrossRef]
- Leppänen, I.; Vikman, M.; Harlin, A.; Orelma, H. Enzymatic Degradation and Pilot-Scale Composting of Cellulose-Based Films with Different Chemical Structures. J. Polym. Environ. 2020, 28, 458–470. [Google Scholar] [CrossRef]
- Tottoli, E.M.; Dorati, R.; Genta, I.; Chiesa, E.; Pisani, S.; Conti, B. Skin wound healing process and new emerging technologies for skin wound care and regeneration. Pharmaceutics 2020, 12, 735. [Google Scholar] [CrossRef]
- Tamer, T.M.; Valachová, K.; Hassan, M.A.; Omer, A.M.; El-Shafeey, M.; Mohy Eldin, M.S.; Šoltés, L. Chitosan/hyaluronan/edaravone membranes for anti-inflammatory wound dressing: In vitro and in vivo evaluation studies. Mater. Sci. Eng. C 2018, 90, 227–235. [Google Scholar] [CrossRef]
- Zinge, C.; Kandasubramanian, B. Nanocellulose based Biodegradable Polymers. Eur. Polym. J. 2020, 133, 109758. [Google Scholar] [CrossRef]
- Chaudhari, A.A.; Vig, K.; Baganizi, D.R.; Sahu, R.; Dixit, S.; Dennis, V.; Singh, S.R.; Pillai, S.R.; Chaudhari, A.A.; Vig, K.; et al. Future prospects for scaffolding methods and biomaterials in skin tissue engineering: A review. Int. J. Mol. Sci. 2016, 17, 1974. [Google Scholar] [CrossRef]
- Brovold, M.; Almeida, J.I.; Pla-Palacín, I.; Sainz-Arnal, P.; Sánchez-Romero, N.; Rivas, J.J.; Almeida, H.; Dachary, P.R.; Serrano-Aulló, T.; Soker, S.; et al. Naturally-Derived Biomaterials for Tissue Engineering Applications. In Advances in Experimental Medicine and Biology; Springer: Singapore, 2018; Volume 1077, pp. 421–449. [Google Scholar]
- Lopez-Polo, J.; Silva-Weiss, A.; Zamorano, M.; Osorio, F.A. Humectability and physical properties of hydroxypropyl methylcellulose coatings with liposome-cellulose nanofibers: Food application. Carbohydr. Polym. 2020, 231, 115702. [Google Scholar] [CrossRef]
- Curvello, R.; Raghuwanshi, V.S.; Garnier, G. Engineering nanocellulose hydrogels for biomedical applications. Adv. Colloid Interface Sci. 2019, 267, 47–61. [Google Scholar] [CrossRef]
- Bai, L.; Huan, S.; Xiang, W.; Rojas, O.J. Pickering emulsions by combining cellulose nanofibrils and nanocrystals: Phase behavior and depletion stabilization. Green Chem. 2018, 20, 1571–1582. [Google Scholar] [CrossRef]
- Capron, I.; Rojas, O.J.; Bordes, R. Behavior of nanocelluloses at interfaces. Curr. Opin. Colloid Interface Sci. 2017, 29, 83–95. [Google Scholar] [CrossRef]
- Kamel, R.; El-Wakil, N.A.; Dufresne, A.; Elkasabgy, N.A. Nanocellulose: From an agricultural waste to a valuable pharmaceutical ingredient. Int. J. Biol. Macromol. 2020, 163, 1579–1590. [Google Scholar] [CrossRef] [PubMed]
- Letchford, K.; Jackson, J.K.; Wasserman, B.Z.; Ye, L.; Hamad, W.Y.; Burt, H.M. The use of nanocrystalline cellulose for the binding and controlled release of drugs. Int. J. Nanomed. 2011, 6, 321. [Google Scholar] [CrossRef] [PubMed]
- Kamel, R.; El-Wakil, N.A.; Abdelkhalek, A.A.; Elkasabgy, N.A. Nanofibrillated cellulose/cyclodextrin based 3D scaffolds loaded with raloxifene hydrochloride for bone regeneration. Int. J. Biol. Macromol. 2020, 156, 704–716. [Google Scholar] [CrossRef] [PubMed]
- Wen, C.; Yuan, Q.; Liang, H.; Vriesekoop, F. Preparation and stabilization of d-limonene Pickering emulsions by cellulose nanocrystals. Carbohydr. Polym. 2014, 112, 695–700. [Google Scholar] [CrossRef] [PubMed]
- Kamel, R.; El-Wakil, N.A.; Abdelkhalek, A.A.; Elkasabgy, N.A. Topical cellulose nanocrystals-stabilized nanoemulgel loaded with ciprofloxacin HCl with enhanced antibacterial activity and tissue regenerative properties. J. Drug Deliv. Sci. Technol. 2021, 64, 102553. [Google Scholar] [CrossRef]
- Jebali, A.; Hekmatimoghaddam, S.; Behzadi, A.; Rezapor, I.; Mohammadi, B.H.; Jasemizad, T.; Yasini, S.A.; Javadzadeh, M.; Amiri, A.; Soltani, M.; et al. Antimicrobial activity of nanocellulose conjugated with allicin and lysozyme. Cellulose 2013, 20, 2897–2907. [Google Scholar] [CrossRef]
- Yuan, H.; Chen, L.; Hong, F.F. Evaluation of wet nanocellulose membranes produced by different bacterial strains for healing full-thickness skin defects. Carbohydr. Polym. 2022, 285, 119218. [Google Scholar] [CrossRef]
- Pittella, C.Q.; Vitoria, E.M.; Mendes, M.F.; Coli, T.C.; Delage, M.D.; Junior, G.F.; Santos, K.B.; Nascimento, T.C. The Use of Bacterial Nanocellulose as Wound Healing Dressing: A Scoping Review. Res. Sq. 2022. [Google Scholar] [CrossRef]
- Patel, D.K.; Ganguly, K.; Hexiu, J.; Dutta, S.D.; Patil, T.V.; Lim, K.-T. Functionalized chitosan/spherical nanocellulose-based hydrogel with superior antibacterial efficiency for wound healing. Carbohydr. Polym. 2022, 284, 119202. [Google Scholar] [CrossRef]
- Abbas, H.; Kamel, R.; El-Sayed, N. Dermal anti-oxidant, anti-inflammatory and anti-aging effects of Compritol ATO-based Resveratrol colloidal carriers prepared using mixed surfactants. Int. J. Pharm. 2018, 541, 37–47. [Google Scholar] [CrossRef] [PubMed]
- Kamel, R.; Abbas, H.; Fayez, A. Diosmin/essential oil combination for dermal photo-protection using a lipoid colloidal carrier. J. Photochem. Photobiol. B 2017, 170, 49–57. [Google Scholar] [CrossRef] [PubMed]
- Fadeel, D.A.A.; Kamel, R.; Fadel, M. PEGylated lipid nanocarrier for enhancing photodynamic therapy of skin carcinoma using curcumin: In-vitro/in-vivo studies and histopathological examination. Sci. Rep. 2020, 10, 10435. [Google Scholar] [CrossRef]
- Kamel, R.; Mostafa, D.M. Rutin nanostructured lipid cosmeceutical preparation with sun protective potential. J. Photochem. Photobiol. B Biol. 2015, 153, 59–66. [Google Scholar] [CrossRef] [PubMed]
- Jindal, S.; Kumar, A.; Goyal, K.; Awasthi, R.; Kulkarni, G.T. Lipid nanocarriers for dermal delivery of lutein. In Nanomedicine for Bioactives; Springer: Singapore, 2020; pp. 341–366. [Google Scholar]
- Montenegro, L. Lipid-based nanoparticles as carriers for dermal delivery of antioxidants. Curr. Drug Metab. 2017, 18, 469–480. [Google Scholar] [CrossRef]
- Rodenak-Kladniew, B.; Montoto, S.S.; Sbaraglini, M.; Di Ianni, M.; Ruiz, M.; Talevi, A.; Alvarez, V.; Durán, N.; Castro, G.; Islan, G. Hybrid Ofloxacin/eugenol co-loaded solid lipid nanoparticles with enhanced and targetable antimicrobial properties. Int. J. Pharm. 2019, 569, 118575. [Google Scholar] [CrossRef]
- Agubata, C.O.; Okereke, C.; Nzekwe, I.T.; Onoja, R.I.; Obitte, N.C. Development and evaluation of wound healing hydrogels based on a quinolone, hydroxypropyl methylcellulose and biodegradable microfibres. Eur. J. Pharm. Sci. 2016, 89, 1–10. [Google Scholar] [CrossRef]
- Mahmood, H.; Khan, I.U.; Asif, M.; Khan, R.U.; Asghar, S.; Khalid, I.; Khalid, S.H.; Irfan, M.; Rehman, F.; Shahzad, Y.; et al. In vitro and in vivo evaluation of gellan gum hydrogel films: Assessing the co impact of therapeutic oils and ofloxacin on wound healing. Int. J. Biol. Macromol. 2021, 166, 483–495. [Google Scholar] [CrossRef]
- Das, S.; Dey, T.K.; De, A.; Banerjee, A.; Chakraborty, S.; Das, B.; Mukhopadhyay, A.K.; Mukherjee, B.; Samanta, A. Antimicrobial loaded gum odina—gelatin based biomimetic spongy scaffold for accelerated wound healing with complete cutaneous texture. Int. J. Pharm. 2021, 606, 120892. [Google Scholar] [CrossRef]
- Kota, S.; Jahangir, M.A.; Ahmed, M.; Kazmi, I.; Bhavani, P.; Muheem, A.; Saleem, M. Development and evaluation of ofloxacin topical gel containing wound healing modifiers from natural sources. Der Pharm. Lett. 2015, 7, 226–233. [Google Scholar]
- Cevc, G.; Vierl, U. Nanotechnology and the transdermal route: A state of the art review and critical appraisal. J. Control. Release 2010, 141, 277–299. [Google Scholar] [CrossRef] [PubMed]
- Müller, R.H.; Jacobs, C.; Kayser, O. Nanosuspensions as particulate drug formulations in therapy: Rationale for development and what we can expect for the future. Adv. Drug Deliv. Rev. 2001, 47, 3–19. [Google Scholar] [CrossRef]
- Heurtault, B.; Saulnier, P.; Pech, B.; Proust, J.-E.; Benoit, J.-P. Physico-chemical stability of colloidal lipid particles. Biomaterials 2003, 24, 4283–4300. [Google Scholar] [CrossRef]
- Xu, H.-N.; Tang, Y.-Y.; Ouyang, X.-K. Shear-induced breakup of cellulose nanocrystal aggregates. Langmuir 2017, 33, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Danaei, M.; Dehghankhold, M.; Ataei, S.; Hasanzadeh Davarani, F.; Javanmard, R.; Dokhani, A.; Khorasani, S.; Mozafari, M.R. Impact of Particle Size and Polydispersity Index on the Clinical Applications of Lipidic Nanocarrier Systems. Pharmaceutics 2018, 10, 57. [Google Scholar] [CrossRef]
- Mudalige, T.; Qu, H.; Van Haute, D.; Ansar, S.M.; Paredes, A.; Ingle, T. Characterization of Nanomaterials: Tools and Challenges. Nanomater. In Nanomaterials for Food Applications; Elsevier: Amsterdam, The Netherlands, 2019; pp. 313–353. [Google Scholar]
- Cherhal, F.; Cousin, F.; Capron, I. Structural description of the interface of Pickering emulsions stabilized by cellulose nanocrystals. Biomacromolecules 2016, 17, 496–502. [Google Scholar] [CrossRef]
- Li, P.; Jin, F.; Sun, Q.; Ma, J. The application of second-order approximation of Taylor series in thickness shear vibration analysis of quartz crystal microbalances. Ultrasonics 2015, 58, 96–103. [Google Scholar] [CrossRef]
- Kulkarni, A.R.; Soppimath, K.S.; Aminabhavi, T.M.; Rudzinski, W.E. In-vitro release kinetics of cefadroxil-loaded sodium alginate interpenetrating network beads. Eur. J. Pharm. Biopharm. 2001, 51, 127–133. [Google Scholar] [CrossRef]
- Raina, H.; Kaur, S.; Jindal, A.B. Development of efavirenz loaded solid lipid nanoparticles: Risk assessment, quality-by-design (QbD) based optimisation and physicochemical characterisation. J. Drug Deliv. Sci. Technol. 2017, 39, 180–191. [Google Scholar] [CrossRef]
- Moon, R.J.; Martini, A.; Nairn, J.; Simonsen, J.; Youngblood, J. Cellulose nanomaterials review: Structure, properties and nanocomposites. Chem. Soc. Rev. 2011, 40, 3941–3994. [Google Scholar] [CrossRef]
- Plasencia, I.; Norlen, L.; Bagatolli, L.A. Direct visualization of lipid domains in human skin stratum corneum’s lipid membranes: Effect of pH and temperature. Biophys. J. 2007, 93, 3142–3155. [Google Scholar] [CrossRef]
- Alavarse, A.C.; de Oliveira Silva, F.W.; Colque, J.T.; da Silva, V.M.; Prieto, T.; Venancio, E.C.; Bonvent, J.-J. Tetracycline hydrochloride-loaded electrospun nanofibers mats based on PVA and chitosan for wound dressing. Mater Sci. Eng. C Mater Biol. Appl. 2017, 77, 271–281. [Google Scholar] [CrossRef]
- Moes, J.; Koolen, S.; Huitema, A.; Schellens, J.; Beijnen, J.; Nuijen, B. Pharmaceutical development and preliminary clinical testing of an oral solid dispersion formulation of docetaxel (ModraDoc001). Int. J. Pharm. 2011, 420, 244–250. [Google Scholar] [CrossRef]
- Kamel, R.; Basha, M.; El Awdan, S. Development and evaluation of long-acting epidural “smart” thermoreversible injection loaded with spray-dried polymeric nanospheres using experimental design. J. Drug Target. 2013, 21, 277–290. [Google Scholar] [CrossRef]
- Kamel, R. Study of the influence of selected variables on the preparation of prolonged release bioadhesive vaginal carbohydrate microspheres using experimental design. J. Drug Deliv. Sci. Technol. 2013, 23, 247–254. [Google Scholar] [CrossRef]
- Kamel, R.; Salama, A.; Shaffie, N.M.; Salah, N.M. Cerebral effect of optimized Allium sativum oil-loaded chitosan nanorods: GC-MS analysis and in vitro/in vivo evaluation. Food Funct. 2020, 1, 5357–5376. [Google Scholar] [CrossRef]
- Mehnert, W.; Mader, K. Solid lipid nanoparticles: Production, characterization and applications. Adv. Drug Deliv. Rev. 2001, 47, 165–196. [Google Scholar] [CrossRef]
- Sandri, G.; Bonferoni, M.C.; Gökçe, E.H.; Ferrari, F.; Rossi, S.; Patrini, M.; Caramella, C. Chitosan-associated SLN: In vitro and ex vivo characterization of cyclosporine A loaded ophthalmic systems. J. Microencapsul. 2010, 27, 735–746. [Google Scholar] [CrossRef]
- Salama, A.H.; AbouSamra, M.M.; Awad, G.E.A.; Mansy, S.S. Promising bioadhesive ofloxacin-loaded polymeric nanoparticles for the treatment of ocular inflammation: Formulation and in vivo evaluation. Drug Deliv. Transl. Res. 2020, 11, 1943–1957. [Google Scholar] [CrossRef]
- Souto, E.B.; Mehnert, W.; Muller, R.H. Polymorphic behaviour of Compritol888 ATO as bulk lipid and as SLN and NLC. J. Microencapsul. 2006, 23, 417–433. [Google Scholar] [CrossRef]
- D’Souza, S. A Review of In Vitro Drug Release Test Methods for Nano-Sized Dosage Forms. Adv. Pharm. 2014, 2014, 304757. [Google Scholar] [CrossRef]
- Mittal, N.; Kaur, G. Leucaena leucocephala (Lam.) galactomannan nanoparticles: Optimization and characterization for ocular delivery in glaucoma treatment. Int. J. Biol. Macromol. 2019, 139, 1252–1262. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Pizarro, R.; Silva-Abreu, M.; Calpena, A.C.; Egea, M.A.; Espina, M.; García, M.L. Development of fluorometholone-loaded PLGA nanoparticles for treatment of inflammatory disorders of anterior and posterior segments of the eye. Int. J. Pharm. 2018, 547, 338–346. [Google Scholar] [CrossRef] [PubMed]
- Varshosaz, J.; Tavakoli, N.; Kheirolahi, F. Use of hydrophilic natural gums in formulation of sustained-release matrix tablets of tramadol hydrochloride. AAPS PharmSciTech 2006, 7, E168–E174. [Google Scholar] [CrossRef] [PubMed]
- Amin, K.M.; El-masry, A.H.; Mohamed, N.A.; Awad, G.E.; Habib, B.S. Synthesis, characterization and antimicrobial activity of some novel isoindole-1,3-dione derivatives. Der Pharm. Chem. 2013, 5, 97–108. [Google Scholar]
- Scott, J.A.G.; Hall, A.J.; Hannington, A.; Edwards, R.; Mwarumba, S.; Lowe, B.; Griffiths, D.; Crook, D.; Marsh, K. Serotype distribution and prevalence of resistance to benzylpenicillin in three representative populations of Streptococcus pneumoniae isolates from the coast of Kenya. Clin. Infect. Dis. 1998, 27, 1442–1450. [Google Scholar] [CrossRef]
- Kamel, R.; Abbas, H. Rational for the use of coconut oil-based anti-mycotic pessaries to combat recurrent vaginal infection: In-vitro/in-vivo evaluation and preliminary prospective clinical investigation. Int. J. Appl. Pharm. 2018, 10, 159–166. [Google Scholar] [CrossRef]
- Kamel, R.; Abbas, H. A multi-microcarrier of Metronidazole-biopolymers complexes as a potential vaginal delivery system. Int. J. Polym. Mater. Polym. Biomater. 2018, 67, 236–245. [Google Scholar] [CrossRef]
- Skehan, P.; Storeng, R.; Scudiero, D.; Monks, A.; McMahon, J.; Vistica, D.; Warren, J.T.; Bokesch, H.; Kenney, S.; Boyd, M.R. New colorimetric cytotoxicity assay for anticancer-drug screening. J. Natl. Cancer Inst. 1990, 82, 1107–1112. [Google Scholar] [CrossRef]
- Martinotti, S.; Ranzato, E. Scratch Wound Healing Assay. In Methods in Molecular Biology; Humana: New York, NY, USA, 2020; Volume 2109, pp. 225–229. [Google Scholar]
- Jonkman, J.; Cathcart, J.A.; Xu, F.; Bartolini, M.E.; Amon, J.E.; Stevens, K.M.; Colarusso, P. An introduction to the wound healing assay using live-cell microscopy. Cell Adh. Migr. 2014, 8, 440–451. [Google Scholar] [CrossRef]
- Rodriguez, L.G.; Wu, X.; Guan, J.L. Wound-healing assay. In Methods in Molecular Biology; Humana: New York, NY, USA, 2005; Volume 294, pp. 23–29. [Google Scholar]



| Formulae | PS ± SD (nm) | PDI ± SD | ZP ± SD | EE (%) ± SD | MRT ± SD (h) |
|---|---|---|---|---|---|
| HNCN1 | 244.9 ± 6.45 | 0.6 ± 0.03 | −26.6 ± 1.22 | 95.80 ± 2.01 | 0.89 ± 0.02 |
| HNCN2 | 265.1 ± 10.01 | 0.4 ± 0.05 | −26.1 ± 0.03 | 97.13 ± 1.23 | 0.54 ± 0.03 |
| HNCN3 | 254.2 ± 8.22 | 0.4 ± 0.01 | −27.3 ± 1.25 | 97.73 ± 1.36 | 0.94 ± 0.10 |
| HNCN4 | 268.3 ± 11.32 | 0.6 ± 0.02 | −22.3 ± 1.01 | 96.56 ± 2.30 | 0.74 ± 0.06 |
| HNCN5 | 173.5 ± 11.01 | 0.4 ± 0.02 | −27.9 ± 0.50 | 97.05 ± 2.01 | 0.83 ± 0.07 |
| HNCN6 | 190.1 ± 5.50 | 0.5 ± 0.01 | −29.6 ± 0.84 | 96.56 ± 1.22 | 0.46 ± 0.03 |
| HNCN7 | 200.2 ± 6.74 | 0.5 ± 0.01 | −26.4 ± 0.50 | 97.53 ± 1.56 | 0.93 ± 0.11 |
| HNCN8 | 214.5 ± 5.90 | 0.6 ± 0.03 | −26.5 ± 0.77 | 97.86 ± 2.02 | 0.73 ± 0.07 |
| Best fitting equation | 226.25 + 8.15X1 + 7.88X2 − 31.73X3 − 1.03X1X2 − 0.38X1X3 + 4.85X2X3 | 0.53 + 0.028X1 + 0.022X2 + 0.007X3 + 0.065X1X2 + 0.025X1X3 − 0.045X2X3 | −26.59 + 0.46X1 + 0.96X2 − 1.01X3 + 0.76X1X2 − 0.91X1X3 + 0.19X2X3 | 97.02 + 0.0001X1 + 0.39X2 + 0.22X3 − 0.21X1X2 − 0.04X1X3 + 0.05X2X3 | 0.76 − 0.14X1 + 0.08X2 − 0.02X3 + 0.04X1X2 − 0.001X1X3 + 0.01X2X3 |
| Significance | Significant | Not Significant | Not Significant | Not Significant | Significant |
| R2 | 0.9998 | - | - | - | 0.9998 |
| Adjusted R2 | 0.9986 | - | - | - | 0.9984 |
| Predicted R2 | 0.9876 | - | - | - | 0.9856 |
| Adeq Precision | 76.190 | - | - | - | 71.813 |
| Reduced model | 226.25 + 8.15X1 + 7.88X2 − 31.73X3 + 4.85X2X3 | - | - | - | 0.76 − 0.14X1 + 0.08X2 + 0.01X1X2 |
| Terms | Y1 (PS) | Y5 (MRT) | ||
|---|---|---|---|---|
| Effect | p Value for Coefficients of Factors | Effect | p Value for Coefficients of Factors | |
| b0 | 226.25 | 0.001 | 0.759 | 0.005 |
| b1 | 8.15 | 0.039 | −0.144 | 0.027 |
| b2 | 7.875 | 0.040 | 0.081 | 0.048 |
| b3 | −31.725 | 0.010 | −0.024 | 0.161 |
| b1b2 | −1.025 | 0.288 | 0.039 | 0.100 |
| b1b3 | −0.375 | 0.590 | −0.001 | 0.874 |
| b2b3 | 4.85 | 0.065 | 0.014 | 0.271 |
| Formulae | Variable Levels in Coded Form | ||
|---|---|---|---|
| X1 | X2 | X3 | |
| HNCN1 | −1 | −1 | −1 |
| HNCN2 | 1 | −1 | −1 |
| HNCN3 | −1 | 1 | −1 |
| HNCN4 | 1 | 1 | −1 |
| HNCN5 | −1 | −1 | 1 |
| HNCN6 | 1 | −1 | 1 |
| HNCN7 | −1 | 1 | 1 |
| HNCN8 | 1 | 1 | 1 |
| Translation of Coded Levels in Actual Units | |||
| Variable Levels | −1 (low) | 1 (high) | |
| X1: Lipid concentration (w/v) | 2% | 4% | |
| X2: Surfactant concentration (w/v) | 5% | 10% | |
| X3: NC concentration (w/v) | 1% | 2% | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
AbouSamra, M.M.; El Hoffy, N.M.; El-Wakil, N.A.; Awad, G.E.A.; Kamel, R. Computational Investigation to Design Ofloxacin-Loaded Hybridized Nanocellulose/Lipid Nanogels for Accelerated Skin Repair. Gels 2022, 8, 593. https://doi.org/10.3390/gels8090593
AbouSamra MM, El Hoffy NM, El-Wakil NA, Awad GEA, Kamel R. Computational Investigation to Design Ofloxacin-Loaded Hybridized Nanocellulose/Lipid Nanogels for Accelerated Skin Repair. Gels. 2022; 8(9):593. https://doi.org/10.3390/gels8090593
Chicago/Turabian StyleAbouSamra, Mona M., Nada M. El Hoffy, Nahla A. El-Wakil, Ghada E. A. Awad, and Rabab Kamel. 2022. "Computational Investigation to Design Ofloxacin-Loaded Hybridized Nanocellulose/Lipid Nanogels for Accelerated Skin Repair" Gels 8, no. 9: 593. https://doi.org/10.3390/gels8090593
APA StyleAbouSamra, M. M., El Hoffy, N. M., El-Wakil, N. A., Awad, G. E. A., & Kamel, R. (2022). Computational Investigation to Design Ofloxacin-Loaded Hybridized Nanocellulose/Lipid Nanogels for Accelerated Skin Repair. Gels, 8(9), 593. https://doi.org/10.3390/gels8090593






