Cancer Exosomes: An Overview and the Applications of Flow
Abstract
:1. Introduction
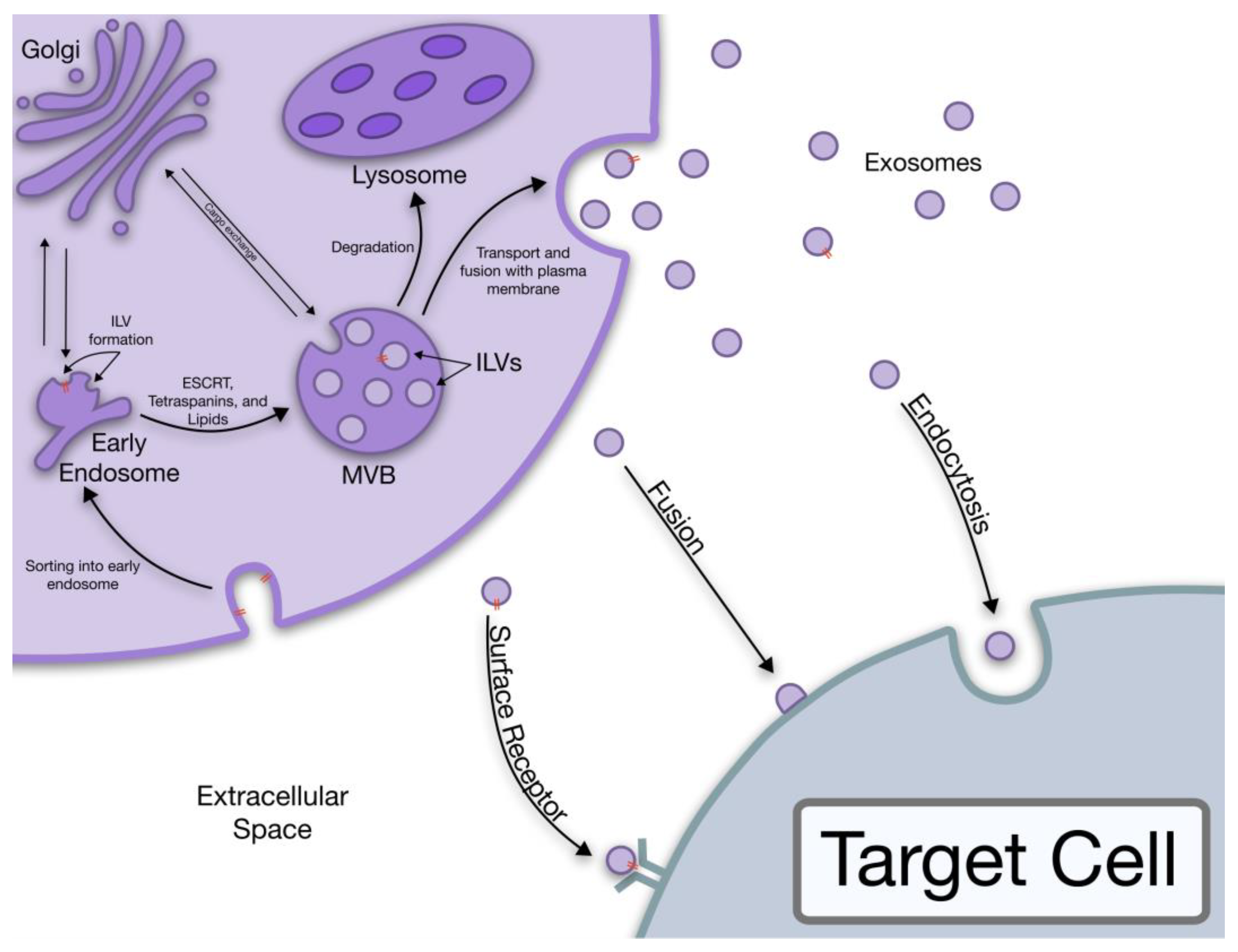
1.1. Exosome Biogenesis
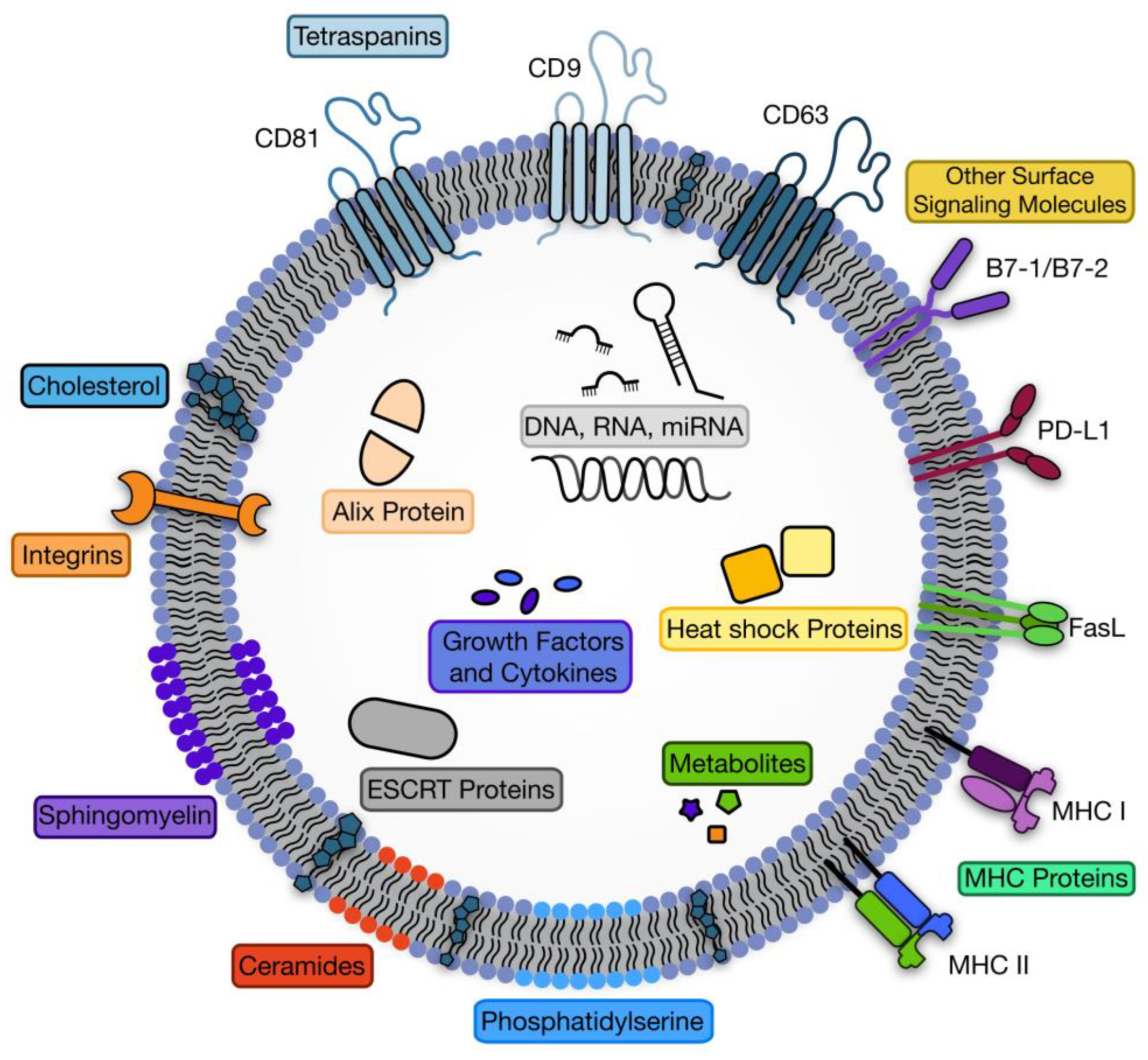
1.2. Exosome Structure
1.3. Exosome Cargo
2. Cancer Derived Exosome Cargo
3. Cancer Exosomes Role in Immune Suppression
3.1. Exosomal PD-L1 Protein
3.2. Exosomal B7 Proteins
3.3. Exosomal FasL Protein
4. Cancer Exosomes and Metastasis Progression
4.1. Tumor Microenvironment
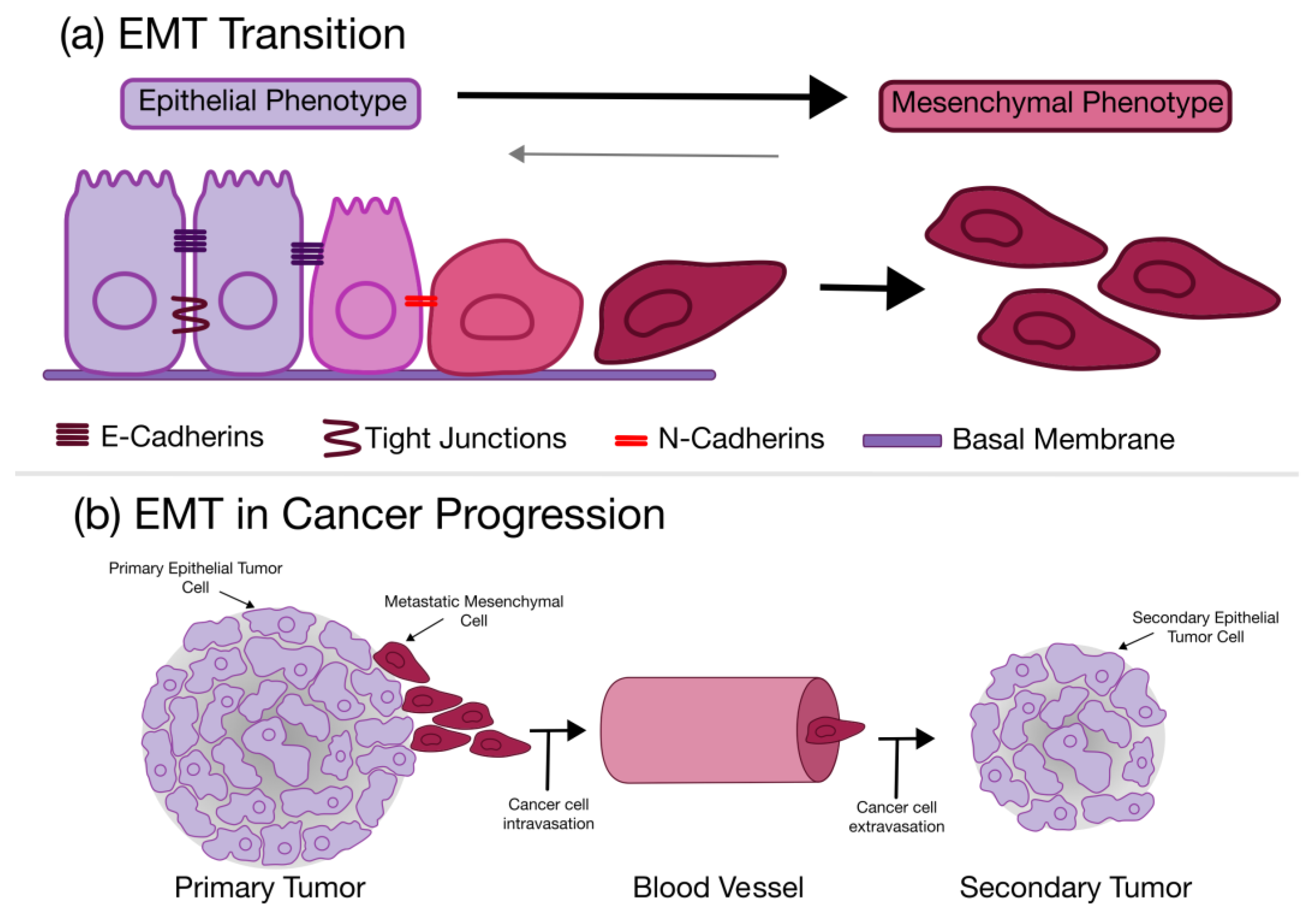
4.2. Epithelial to Mesenchymal Transition
4.3. Premetastatic Niche Formation
5. Exosomes and Flow
5.1. Microfluidics and Exosomes
5.2. Bioreactors and Exosomes
5.3. Flow Cytometry and Exosomes
6. Conclusions and Future Directions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer Statistics, 2019. CA A Cancer J. Clin. 2019, 69, 7–34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA A Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Rajagopal, C.; Harikumar, K.B. The Origin and Functions of Exosomes in Cancer. Front. Oncol. 2018, 8, 66. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gurung, S.; Perocheau, D.; Touramanidou, L.; Baruteau, J. The Exosome Journey: From Biogenesis to Uptake and Intracellular Signalling. Cell Commun. Signal. 2021, 19, 47. [Google Scholar] [CrossRef] [PubMed]
- Hessvik, N.P.; Llorente, A. Current Knowledge on Exosome Biogenesis and Release. Cell Mol. Life Sci. 2018, 75, 193–208. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Liu, Y.; Liu, H.; Tang, W.H. Exosomes: Biogenesis, Biologic Function and Clinical Potential. Cell Biosci. 2019, 9, 19. [Google Scholar] [CrossRef]
- Filipazzi, P.; Bürdek, M.; Villa, A.; Rivoltini, L.; Huber, V. Recent Advances on the Role of Tumor Exosomes in Immunosuppression and Disease Progression. Semin. Cancer Biol. 2012, 22, 342–349. [Google Scholar] [CrossRef] [Green Version]
- Johnstone, R.M.; Adam, M.; Hammond, J.R.; Orr, L.; Turbide, C. Vesicle Formation during Reticulocyte Maturation. Association of Plasma Membrane Activities with Released Vesicles (Exosomes). J. Biol. Chem. 1987, 262, 9412–9420. [Google Scholar] [CrossRef]
- Johnstone, R.M. Revisiting the Road to the Discovery of Exosomes. Blood Cells Mol. Dis. 2005, 34, 214–219. [Google Scholar] [CrossRef]
- Colombo, M.; Raposo, G.; Théry, C. Biogenesis, Secretion, and Intercellular Interactions of Exosomes and Other Extracellular Vesicles. Annu. Rev. Cell Dev. Biol. 2014, 30, 255–289. [Google Scholar] [CrossRef]
- Frydrychowicz, M.; Kolecka-Bednarczyk, A.; Madejczyk, M.; Yasar, S.; Dworacki, G. Exosomes—Structure, Biogenesis and Biological Role in Non-Small-Cell Lung Cancer. Scand. J. Immunol. 2015, 81, 2–10. [Google Scholar] [CrossRef]
- Mashouri, L.; Yousefi, H.; Aref, A.R.; Ahadi, A.M.; Molaei, F.; Alahari, S.K. Exosomes: Composition, Biogenesis, and Mechanisms in Cancer Metastasis and Drug Resistance. Mol. Cancer 2019, 18, 75. [Google Scholar] [CrossRef] [Green Version]
- The ESCRT Pathway—ScienceDirect. Available online: https://www.sciencedirect.com/science/article/pii/S1534580711002073?via%3Dihub (accessed on 19 October 2022).
- Villarroya-Beltri, C.; Baixauli, F.; Gutiérrez-Vázquez, C.; Sánchez-Madrid, F.; Mittelbrunn, M. Sorting It out: Regulation of Exosome Loading. Semin. Cancer Biol. 2014, 28, 3–13. [Google Scholar] [CrossRef] [Green Version]
- Yue, B.; Yang, H.; Wang, J.; Ru, W.; Wu, J.; Huang, Y.; Lan, X.; Lei, C.; Chen, H. Exosome Biogenesis, Secretion and Function of Exosomal MiRNAs in Skeletal Muscle Myogenesis. Cell Prolif. 2020, 53, e12857. [Google Scholar] [CrossRef]
- Babst, M. MVB Vesicle Formation: ESCRT-Dependent, ESCRT-Independent and Everything in Between. Curr. Opin. Cell Biol. 2011, 23, 452–457. [Google Scholar] [CrossRef] [Green Version]
- Trajkovic, K.; Hsu, C.; Chiantia, S.; Rajendran, L.; Wenzel, D.; Wieland, F.; Schwille, P.; Brügger, B.; Simons, M. Ceramide Triggers Budding of Exosome Vesicles into Multivesicular Endosomes. Science 2008, 319, 1244–1247. [Google Scholar] [CrossRef]
- Mathivanan, S.; Fahner, C.J.; Reid, G.E.; Simpson, R.J. ExoCarta 2012: Database of Exosomal Proteins, RNA and Lipids. Nucleic Acids Res. 2012, 40, D1241–D1244. [Google Scholar] [CrossRef] [Green Version]
- Hannafon, B.N.; Ding, W.-Q. Intercellular Communication by Exosome-Derived MicroRNAs in Cancer. Int. J. Mol. Sci. 2013, 14, 14240–14269. [Google Scholar] [CrossRef] [Green Version]
- Bobrie, A.; Colombo, M.; Raposo, G.; Théry, C. Exosome Secretion: Molecular Mechanisms and Roles in Immune Responses. Traffic 2011, 12, 1659–1668. [Google Scholar] [CrossRef]
- Chen, G.; Huang, A.C.; Zhang, W.; Zhang, G.; Wu, M.; Xu, W.; Yu, Z.; Yang, J.; Wang, B.; Sun, H.; et al. Exosomal PD-L1 Contributes to Immunosuppression and Is Associated with Anti-PD-1 Response. Nature 2018, 560, 382–386. [Google Scholar] [CrossRef]
- Zhang, H.; Dai, Z.; Wu, W.; Wang, Z.; Zhang, N.; Zhang, L.; Zeng, W.-J.; Liu, Z.; Cheng, Q. Regulatory Mechanisms of Immune Checkpoints PD-L1 and CTLA-4 in Cancer. J. Exp. Clin. Cancer Res. 2021, 40, 184. [Google Scholar] [CrossRef] [PubMed]
- Fang, Z.; Ding, Y.; Xue, Z.; Li, P.; Li, J.; Li, F. Roles of Exosomes as Drug Delivery Systems in Cancer Immunotherapy: A Mini-Review. Discov. Oncol. 2022, 13, 74. [Google Scholar] [CrossRef] [PubMed]
- MacFarlane, L.-A.; Murphy, P.R. MicroRNA: Biogenesis, Function and Role in Cancer. Curr. Genom. 2010, 11, 537–561. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gebert, L.F.R.; MacRae, I.J. Regulation of MicroRNA Function in Animals. Nat. Rev. Mol. Cell Biol 2019, 20, 21–37. [Google Scholar] [CrossRef] [PubMed]
- Valinezhad Orang, A.; Safaralizadeh, R.; Kazemzadeh-Bavili, M. Mechanisms of MiRNA-Mediated Gene Regulation from Common Downregulation to MRNA-Specific Upregulation. Int. J. Genom. 2014, 2014, 970607. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Q.; Li, Y.; Zhao, M.; Lin, H.; Wang, W.; Li, D.; Cui, W.; Zhou, C.; Zhong, J.; Huang, C. MiR-494 Acts as a Tumor Promoter by Targeting CASP2 in Non-Small Cell Lung Cancer. Sci. Rep. 2019, 9, 3008. [Google Scholar] [CrossRef]
- Kalluri, R.; LeBleu, V.S. Discovery of Double-Stranded Genomic DNA in Circulating Exosomes. Cold Spring Harb. Symp. Quant. Biol. 2016, 81, 275–280. [Google Scholar] [CrossRef] [Green Version]
- Thakur, B.K.; Zhang, H.; Becker, A.; Matei, I.; Huang, Y.; Costa-Silva, B.; Zheng, Y.; Hoshino, A.; Brazier, H.; Xiang, J.; et al. Double-Stranded DNA in Exosomes: A Novel Biomarker in Cancer Detection. Cell Res. 2014, 24, 766–769. [Google Scholar] [CrossRef] [Green Version]
- Ghanam, J.; Chetty, V.K.; Barthel, L.; Reinhardt, D.; Hoyer, P.-F.; Thakur, B.K. DNA in Extracellular Vesicles: From Evolution to Its Current Application in Health and Disease. Cell Biosci. 2022, 12, 37. [Google Scholar] [CrossRef]
- Tang, Y.; Zong, S.; Zeng, H.; Ruan, X.; Yao, L.; Han, S.; Hou, F. MicroRNAs and Angiogenesis: A New Era for the Management of Colorectal Cancer. Cancer Cell Int. 2021, 21, 221. [Google Scholar] [CrossRef]
- Hu, H.-Y.; Yu, C.-H.; Zhang, H.-H.; Zhang, S.-Z.; Yu, W.-Y.; Yang, Y.; Chen, Q. Exosomal MiR-1229 Derived from Colorectal Cancer Cells Promotes Angiogenesis by Targeting HIPK2. Int. J. Biol. Macromol. 2019, 132, 470–477. [Google Scholar] [CrossRef]
- Yamada, N.; Tsujimura, N.; Kumazaki, M.; Shinohara, H.; Taniguchi, K.; Nakagawa, Y.; Naoe, T.; Akao, Y. Colorectal Cancer Cell-Derived Microvesicles Containing MicroRNA-1246 Promote Angiogenesis by Activating Smad 1/5/8 Signaling Elicited by PML down-Regulation in Endothelial Cells. Biochim. Biophys. Acta (BBA)-Gene Regul. Mech. 2014, 1839, 1256–1272. [Google Scholar] [CrossRef]
- Sun, X.; Lin, F.; Sun, W.; Zhu, W.; Fang, D.; Luo, L.; Li, S.; Zhang, W.; Jiang, L. Exosome-Transmitted MiRNA-335-5p Promotes Colorectal Cancer Invasion and Metastasis by Facilitating EMT via Targeting RASA1. Mol. Ther.-Nucleic Acids 2021, 24, 164–174. [Google Scholar] [CrossRef]
- Dai, W.; Zhou, J.; Wang, H.; Zhang, M.; Yang, X.; Song, W. MiR-424-5p Promotes the Proliferation and Metastasis of Colorectal Cancer by Directly Targeting SCN4B. Pathol.-Res. Pract. 2020, 216, 152731. [Google Scholar] [CrossRef]
- Zeng, Z.; Li, Y.; Pan, Y.; Lan, X.; Song, F.; Sun, J.; Zhou, K.; Liu, X.; Ren, X.; Wang, F.; et al. Cancer-Derived Exosomal MiR-25-3p Promotes Pre-Metastatic Niche Formation by Inducing Vascular Permeability and Angiogenesis. Nat. Commun. 2018, 9, 5395. [Google Scholar] [CrossRef] [Green Version]
- Li, B.; Cao, Y.; Sun, M.; Feng, H. Expression, Regulation, and Function of Exosome-Derived MiRNAs in Cancer Progression and Therapy. FASEB J. 2021, 35, e21916. [Google Scholar] [CrossRef]
- Paskeh, M.D.A.; Entezari, M.; Mirzaei, S.; Zabolian, A.; Saleki, H.; Naghdi, M.J.; Sabet, S.; Khoshbakht, M.A.; Hashemi, M.; Hushmandi, K.; et al. Emerging Role of Exosomes in Cancer Progression and Tumor Microenvironment Remodeling. J. Hematol. Oncol. 2022, 15, 83. [Google Scholar] [CrossRef]
- Lin, Q.; Zhou, C.-R.; Bai, M.-J.; Zhu, D.; Chen, J.-W.; Wang, H.-F.; Li, M.-A.; Wu, C.; Li, Z.-R.; Huang, M.-S. Exosome-Mediated MiRNA Delivery Promotes Liver Cancer EMT and Metastasis. Am. J. Transl. Res. 2020, 12, 1080–1095. [Google Scholar]
- Mao, G.; Liu, Y.; Fang, X.; Liu, Y.; Fang, L.; Lin, L.; Liu, X.; Wang, N. Tumor-Derived MicroRNA-494 Promotes Angiogenesis in Non-Small Cell Lung Cancer. Angiogenesis 2015, 18, 373–382. [Google Scholar] [CrossRef]
- Berchem, G.; Noman, M.Z.; Bosseler, M.; Paggetti, J.; Baconnais, S.; Le Cam, E.; Nanbakhsh, A.; Moussay, E.; Mami-Chouaib, F.; Janji, B.; et al. Hypoxic Tumor-Derived Microvesicles Negatively Regulate NK Cell Function by a Mechanism Involving TGF-β and MiR23a Transfer. OncoImmunology 2016, 5, e1062968. [Google Scholar] [CrossRef] [Green Version]
- Sruthi, T.V.; Edatt, L.; Raji, G.R.; Kunhiraman, H.; Shankar, S.S.; Shankar, V.; Ramachandran, V.; Poyyakkara, A.; Kumar, S.V.B. Horizontal Transfer of MiR-23a from Hypoxic Tumor Cell Colonies Can Induce Angiogenesis. J. Cell. Physiol. 2018, 233, 3498–3514. [Google Scholar] [CrossRef] [PubMed]
- Shang, D.; Xie, C.; Hu, J.; Tan, J.; Yuan, Y.; Liu, Z.; Yang, Z. Pancreatic Cancer Cell–Derived Exosomal MicroRNA-27a Promotes Angiogenesis of Human Microvascular Endothelial Cells in Pancreatic Cancer via BTG2. J. Cell. Mol. Med. 2020, 24, 588–604. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, L.; Li, C.; Wang, S.; Wang, Z.; Jiang, J.; Wang, W.; Li, X.; Chen, J.; Liu, K.; Li, C.; et al. Exosomes Derived from Hypoxic Oral Squamous Cell Carcinoma Cells Deliver MiR-21 to Normoxic Cells to Elicit a Prometastatic Phenotype. Cancer Res. 2016, 76, 1770–1780. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Tang, K.; Chen, L.; Du, M.; Qu, Z. Exosomal CircGDI2 Suppresses Oral Squamous Cell Carcinoma Progression Through the Regulation of MiR-424-5p/SCAI Axis. Cancer Manag. Res. 2020, 12, 7501–7514. [Google Scholar] [CrossRef] [PubMed]
- Lucero, R.; Zappulli, V.; Sammarco, A.; Murillo, O.D.; Cheah, P.S.; Srinivasan, S.; Tai, E.; Ting, D.T.; Wei, Z.; Roth, M.E.; et al. Glioma-Derived MiRNA-Containing Extracellular Vesicles Induce Angiogenesis by Reprogramming Brain Endothelial Cells. Cell Rep. 2020, 30, 2065–2074.e4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scarth, J.A.; Patterson, M.R.; Morgan, E.L.; Macdonald, A. The Human Papillomavirus Oncoproteins: A Review of the Host Pathways Targeted on the Road to Transformation. J. Gen. Virol. 2021, 102, 001540. [Google Scholar] [CrossRef]
- Chiantore, M.V.; Mangino, G.; Iuliano, M.; Zangrillo, M.S.; De Lillis, I.; Vaccari, G.; Accardi, R.; Tommasino, M.; Columba Cabezas, S.; Federico, M.; et al. Human Papillomavirus E6 and E7 Oncoproteins Affect the Expression of Cancer-Related MicroRNAs: Additional Evidence in HPV-Induced Tumorigenesis. J. Cancer Res. Clin. Oncol. 2016, 142, 1751–1763. [Google Scholar] [CrossRef]
- Konstantinell, A.; Bruun, J.-A.; Olsen, R.; Aspar, A.; Škalko-Basnet, N.; Sveinbjørnsson, B.; Moens, U. Secretomic Analysis of Extracellular Vesicles Originating from Polyomavirus-Negative and Polyomavirus-Positive Merkel Cell Carcinoma Cell Lines. Proteomics 2016, 16, 2587–2591. [Google Scholar] [CrossRef]
- Luga, V.; Zhang, L.; Viloria-Petit, A.M.; Ogunjimi, A.A.; Inanlou, M.R.; Chiu, E.; Buchanan, M.; Hosein, A.N.; Basik, M.; Wrana, J.L. Exosomes Mediate Stromal Mobilization of Autocrine Wnt-PCP Signaling in Breast Cancer Cell Migration. Cell 2012, 151, 1542–1556. [Google Scholar] [CrossRef] [Green Version]
- Hofmann, L.; Ludwig, S.; Vahl, J.M.; Brunner, C.; Hoffmann, T.K.; Theodoraki, M.-N. The Emerging Role of Exosomes in Diagnosis, Prognosis, and Therapy in Head and Neck Cancer. Int. J. Mol. Sci. 2020, 21, 4072. [Google Scholar] [CrossRef]
- Han, Y.; Liu, D.; Li, L. PD-1/PD-L1 Pathway: Current Researches in Cancer. Am. J. Cancer Res. 2020, 10, 727–742. [Google Scholar]
- Iwai, Y.; Hamanishi, J.; Chamoto, K.; Honjo, T. Cancer Immunotherapies Targeting the PD-1 Signaling Pathway. J. Biomed. Sci. 2017, 24, 26. [Google Scholar] [CrossRef]
- Sholl, L.M. Biomarkers of Response to Checkpoint Inhibitors beyond PD-L1 in Lung Cancer. Mod. Pathol. 2022, 35, 66–74. [Google Scholar] [CrossRef]
- Kythreotou, A.; Siddique, A.; Mauri, F.A.; Bower, M.; Pinato, D.J. Pd-L1. J. Clin. Pathol. 2018, 71, 189–194. [Google Scholar] [CrossRef]
- Dou, D.; Ren, X.; Han, M.; Xu, X.; Ge, X.; Gu, Y.; Wang, X. Cancer-Associated Fibroblasts-Derived Exosomes Suppress Immune Cell Function in Breast Cancer via the MiR-92/PD-L1 Pathway. Front. Immunol. 2020, 11, 2026. [Google Scholar] [CrossRef]
- Theodoraki, M.-N.; Yerneni, S.S.; Hoffmann, T.K.; Gooding, W.E.; Whiteside, T.L. Clinical Significance of PD-L1+ Exosomes in Plasma of Head and Neck Cancer Patients. Clin. Cancer Res. 2018, 24, 896–905. [Google Scholar] [CrossRef] [Green Version]
- Sharma, P.; Diergaarde, B.; Ferrone, S.; Kirkwood, J.M.; Whiteside, T.L. Melanoma Cell-Derived Exosomes in Plasma of Melanoma Patients Suppress Functions of Immune Effector Cells. Sci. Rep. 2020, 10, 92. [Google Scholar] [CrossRef] [Green Version]
- Araki, Y.; Mimura, T. The Mechanisms Underlying Chronic Inflammation in Rheumatoid Arthritis from the Perspective of the Epigenetic Landscape. J. Immunol. Res. 2016, 2016, e6290682. [Google Scholar] [CrossRef] [Green Version]
- Poggio, M.; Hu, T.; Pai, C.-C.; Chu, B.; Belair, C.D.; Chang, A.; Montabana, E.; Lang, U.E.; Fu, Q.; Fong, L.; et al. Suppression of Exosomal PD-L1 Induces Systemic Anti-Tumor Immunity and Memory. Cell 2019, 177, 414–427.e13. [Google Scholar] [CrossRef] [Green Version]
- Chan, D.V.; Gibson, H.M.; Aufiero, B.M.; Wilson, A.J.; Hafner, M.S.; Mi, Q.-S.; Wong, H.K. Differential CTLA-4 Expression in Human CD4+ versus CD8+ T Cells Is Associated with Increased NFAT1 and Inhibition of CD4+ Proliferation. Genes Immun. 2014, 15, 25–32. [Google Scholar] [CrossRef] [Green Version]
- Xing, C.; Li, H.; Li, R.-J.; Yin, L.; Zhang, H.-F.; Huang, Z.-N.; Cheng, Z.; Li, J.; Wang, Z.-H.; Peng, H.-L. The Roles of Exosomal Immune Checkpoint Proteins in Tumors. Mil. Med. Res. 2021, 8, 56. [Google Scholar] [CrossRef] [PubMed]
- Vackova, J.; Polakova, I.; Johari, S.D.; Smahel, M. CD80 Expression on Tumor Cells Alters Tumor Microenvironment and Efficacy of Cancer Immunotherapy by CTLA-4 Blockade. Cancers 2021, 13, 1935. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Ye, Y.; Yu, H.; Lin, S.-H.; Tu, H.; Liang, D.; Chang, D.W.; Huang, M.; Wu, X. Immune Checkpoint-Related Serum Proteins and Genetic Variants Predict Outcomes of Localized Prostate Cancer, a Cohort Study. Cancer Immunol. Immunother. 2021, 70, 701–712. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Bai, W.; Zhang, L. The Overexpression of CD80 and ISG15 Are Associated with the Progression and Metastasis of Breast Cancer by a Meta-Analysis Integrating Three Microarray Datasets. Pathol. Oncol. Res. 2020, 26, 443–452. [Google Scholar] [CrossRef] [PubMed]
- Payandeh, Z.; Pirpour Tazehkand, A.; Mansoori, B.; Khaze, V.; Asadi, M.; Baradaran, B.; Samadi, N. The Impact of Nrf2 Silencing on Nrf2-PD-L1 Axis to Overcome Oxaliplatin Resistance as Well as Migration in Colon Cancer. Avicenna J. Med. Biotechnol. 2021, 13, 116–122. [Google Scholar] [CrossRef]
- Azambuja, J.H.; Ludwig, N.; Yerneni, S.; Rao, A.; Braganhol, E.; Whiteside, T.L. Molecular Profiles and Immunomodulatory Activities of Glioblastoma-Derived Exosomes. Neurooncol. Adv. 2020, 2, vdaa056. [Google Scholar] [CrossRef]
- Sansom, D.M. CD28, CTLA-4 and Their Ligands: Who Does What and to Whom? Immunology 2000, 101, 169–177. [Google Scholar] [CrossRef]
- Facciabene, A.; Motz, G.T.; Coukos, G. T-Regulatory Cells: Key Players in Tumor Immune Escape and Angiogenesis. Cancer Res. 2012, 72, 2162–2171. [Google Scholar] [CrossRef] [Green Version]
- Selby, M.J.; Engelhardt, J.J.; Quigley, M.; Henning, K.A.; Chen, T.; Srinivasan, M.; Korman, A.J. Anti-CTLA-4 Antibodies of IgG2a Isotype Enhance Antitumor Activity through Reduction of Intratumoral Regulatory T Cells. Cancer Immunol. Res. 2013, 1, 32–42. [Google Scholar] [CrossRef] [Green Version]
- Ribas, A.; Kefford, R.; Marshall, M.A.; Punt, C.J.A.; Haanen, J.B.; Marmol, M.; Garbe, C.; Gogas, H.; Schachter, J.; Linette, G.; et al. Phase III Randomized Clinical Trial Comparing Tremelimumab With Standard-of-Care Chemotherapy in Patients With Advanced Melanoma. JCO 2013, 31, 616–622. [Google Scholar] [CrossRef] [Green Version]
- Johnson, M.L.; Cho, B.C.; Luft, A.; Alatorre-Alexander, J.; Geater, S.L.; Laktionov, K.; Kim, S.-W.; Ursol, G.; Hussein, M.; Lim, F.L.; et al. Durvalumab With or Without Tremelimumab in Combination With Chemotherapy as First-Line Therapy for Metastatic Non–Small-Cell Lung Cancer: The Phase III POSEIDON Study. JCO 2022, JCO.22.00975. [Google Scholar] [CrossRef]
- Kugeratski, F.G.; Kalluri, R. Exosomes as Mediators of Immune Regulation and Immunotherapy in Cancer. FEBS J. 2021, 288, 10–35. [Google Scholar] [CrossRef]
- Huber, V.; Fais, S.; Iero, M.; Lugini, L.; Canese, P.; Squarcina, P.; Zaccheddu, A.; Colone, M.; Arancia, G.; Gentile, M.; et al. Human Colorectal Cancer Cells Induce T-Cell Death Through Release of Proapoptotic Microvesicles: Role in Immune Escape. Gastroenterology 2005, 128, 1796–1804. [Google Scholar] [CrossRef]
- Andreola, G.; Rivoltini, L.; Castelli, C.; Huber, V.; Perego, P.; Deho, P.; Squarcina, P.; Accornero, P.; Lozupone, F.; Lugini, L.; et al. Induction of Lymphocyte Apoptosis by Tumor Cell Secretion of FasL-Bearing Microvesicles. J. Exp. Med. 2002, 195, 1303–1316. [Google Scholar] [CrossRef] [Green Version]
- Gao, Y.; Ma, H.; Lv, C.; Lan, F.; Wang, Y.; Deng, Y. Exosomes and Exosomal MicroRNA in Non-Targeted Radiation Bystander and Abscopal Effects in the Central Nervous System. Cancer Lett. 2021, 499, 73–84. [Google Scholar] [CrossRef]
- Boisvert, F.-M.; Hendzel, M.J.; Bazett-Jones, D.P. Promyelocytic Leukemia (Pml) Nuclear Bodies Are Protein Structures That Do Not Accumulate RNA. J. Cell Biol. 2000, 148, 283–292. [Google Scholar] [CrossRef] [Green Version]
- Rankin, E.B.; Giaccia, A.J. Hypoxic Control of Metastasis. Science 2016, 352, 175–180. [Google Scholar] [CrossRef] [Green Version]
- Ramteke, A.; Ting, H.; Agarwal, C.; Mateen, S.; Somasagara, R.; Hussain, A.; Graner, M.; Frederick, B.; Agarwal, R.; Deep, G. Exosomes Secreted under Hypoxia Enhance Invasiveness and Stemness of Prostate Cancer Cells by Targeting Adherens Junction Molecules. Mol. Carcinog. 2015, 54, 554–565. [Google Scholar] [CrossRef] [Green Version]
- Kalluri, R.; Weinberg, R.A. The Basics of Epithelial-Mesenchymal Transition. J. Clin. Investig. 2009, 119, 1420–1428. [Google Scholar] [CrossRef] [Green Version]
- Ribatti, D.; Tamma, R.; Annese, T. Epithelial-Mesenchymal Transition in Cancer: A Historical Overview. Transl. Oncol. 2020, 13, 100773. [Google Scholar] [CrossRef]
- Kaufhold, S.; Bonavida, B. Central Role of Snail1 in the Regulation of EMT and Resistance in Cancer: A Target for Therapeutic Intervention. J. Exp. Clin. Cancer Res. 2014, 33, 62. [Google Scholar] [CrossRef] [PubMed]
- Noble, A.R.; Hogg, K.; Suman, R.; Berney, D.M.; Bourgoin, S.; Maitland, N.J.; Rumsby, M.G. Phospholipase D2 in Prostate Cancer: Protein Expression Changes with Gleason Score. Br. J. Cancer 2019, 121, 1016–1026. [Google Scholar] [CrossRef] [PubMed]
- Ganesan, R.; Mallets, E.; Gomez-Cambronero, J. The Transcription Factors Slug (SNAI2) and Snail (SNAI1) Regulate Phospholipase D (PLD) Promoter in Opposite Ways towards Cancer Cell Invasion. Mol. Oncol. 2016, 10, 663–676. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mani, S.A.; Guo, W.; Liao, M.-J.; Eaton, E.N.; Ayyanan, A.; Zhou, A.Y.; Brooks, M.; Reinhard, F.; Zhang, C.C.; Shipitsin, M.; et al. The Epithelial-Mesenchymal Transition Generates Cells with Properties of Stem Cells. Cell 2008, 133, 704–715. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- May, C.D.; Sphyris, N.; Evans, K.W.; Werden, S.J.; Guo, W.; Mani, S.A. Epithelial-Mesenchymal Transition and Cancer Stem Cells: A Dangerously Dynamic Duo in Breast Cancer Progression. Breast Cancer Res. 2011, 13, 202. [Google Scholar] [CrossRef] [Green Version]
- Lobb, R.J.; Lima, L.G.; Möller, A. Exosomes: Key Mediators of Metastasis and Pre-Metastatic Niche Formation. Semin. Cell Dev. Biol. 2017, 67, 3–10. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Xiang, X.; Zhuang, X.; Zhang, S.; Liu, C.; Cheng, Z.; Michalek, S.; Grizzle, W.; Zhang, H.-G. Contribution of MyD88 to the Tumor Exosome-Mediated Induction of Myeloid Derived Suppressor Cells. Am. J. Pathol. 2010, 176, 2490–2499. [Google Scholar] [CrossRef]
- Wang, J.; De Veirman, K.; Faict, S.; Frassanito, M.A.; Ribatti, D.; Vacca, A.; Menu, E. Multiple Myeloma Exosomes Establish a Favourable Bone Marrow Microenvironment with Enhanced Angiogenesis and Immunosuppression. J. Pathol. 2016, 239, 162–173. [Google Scholar] [CrossRef]
- Liu, Y.; Gu, Y.; Han, Y.; Zhang, Q.; Jiang, Z.; Zhang, X.; Huang, B.; Xu, X.; Zheng, J.; Cao, X. Tumor Exosomal RNAs Promote Lung Pre-Metastatic Niche Formation by Activating Alveolar Epithelial TLR3 to Recruit Neutrophils. Cancer Cell 2016, 30, 243–256. [Google Scholar] [CrossRef] [Green Version]
- Olejarz, W.; Kubiak-Tomaszewska, G.; Chrzanowska, A.; Lorenc, T. Exosomes in Angiogenesis and Anti-Angiogenic Therapy in Cancers. Int. J. Mol. Sci. 2020, 21, 5840. [Google Scholar] [CrossRef]
- Zhang, W.; Xing, J.; Liu, T.; Zhang, J.; Dai, Z.; Zhang, H.; Wang, D.; Tang, D. Small Extracellular Vesicles: From Mediating Cancer Cell Metastasis to Therapeutic Value in Pancreatic Cancer. Cell Commun. Signal. 2022, 20, 1. [Google Scholar] [CrossRef]
- Itoh, Y. Membrane-Type Matrix Metalloproteinases: Their Functions and Regulations. Matrix Biol. 2015, 44–46, 207–223. [Google Scholar] [CrossRef]
- Barberis, E.; Vanella, V.V.; Falasca, M.; Caneapero, V.; Cappellano, G.; Raineri, D.; Ghirimoldi, M.; De Giorgis, V.; Puricelli, C.; Vaschetto, R.; et al. Circulating Exosomes Are Strongly Involved in SARS-CoV-2 Infection. Front. Mol. Biosci. 2021, 8, 632290. [Google Scholar] [CrossRef]
- Livshits, M.A.; Khomyakova, E.; Evtushenko, E.G.; Lazarev, V.N.; Kulemin, N.A.; Semina, S.E.; Generozov, E.V.; Govorun, V.M. Isolation of Exosomes by Differential Centrifugation: Theoretical Analysis of a Commonly Used Protocol. Sci. Rep. 2015, 5, 17319. [Google Scholar] [CrossRef] [Green Version]
- Fallahi, H.; Zhang, J.; Phan, H.-P.; Nguyen, N.-T. Flexible Microfluidics: Fundamentals, Recent Developments, and Applications. Micromachines 2019, 10, 830. [Google Scholar] [CrossRef] [Green Version]
- Tai, Y.; Chen, K.; Hsieh, J.; Shen, T. Exosomes in Cancer Development and Clinical Applications. Cancer Sci. 2018, 109, 2364–2374. [Google Scholar] [CrossRef] [Green Version]
- Oliveira-Rodríguez, M.; López-Cobo, S.; Reyburn, H.T.; Costa-García, A.; López-Martín, S.; Yáñez-Mó, M.; Cernuda-Morollón, E.; Paschen, A.; Valés-Gómez, M.; Blanco-López, M.C. Development of a Rapid Lateral Flow Immunoassay Test for Detection of Exosomes Previously Enriched from Cell Culture Medium and Body Fluids. J. Extracell. Vesicles 2016, 5, 31803. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.; Skog, J.; Hsu, C.-H.; Lessard, R.T.; Balaj, L.; Wurdinger, T.; Carter, B.S.; Breakefield, X.O.; Toner, M.; Irimia, D. Microfluidic Isolation and Transcriptome Analysis of Serum Microvesicles. Lab Chip 2010, 10, 505–511. [Google Scholar] [CrossRef] [Green Version]
- Li, Q.; Wang, Y.; Xue, Y.; Qiao, L.; Yu, G.; Liu, Y.; Yu, S. Ultrasensitive Analysis of Exosomes Using a 3D Self-Assembled Nanostructured SiO2 Microfluidic Chip. ACS Appl. Mater. Interfaces 2022, 14, 14693–14702. [Google Scholar] [CrossRef]
- Kanwar, S.S.; Dunlay, C.J.; Simeone, D.M.; Nagrath, S. Microfluidic Device (ExoChip) for on-Chip Isolation, Quantification and Characterization of Circulating Exosomes. Lab Chip 2014, 14, 1891–1900. [Google Scholar] [CrossRef]
- Zhang, J.Z.; Nagrath, S. Microfluidics and Cancer: Are We There Yet? Biomed. Microdevices 2013, 15, 595–609. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Z.; Yang, Y.; Zeng, Y.; He, M. A Microfluidic ExoSearch Chip for Multiplexed Exosome Detection towards Blood-Based Ovarian Cancer Diagnosis. Lab Chip 2016, 16, 489–496. [Google Scholar] [CrossRef]
- Davies, R.T.; Kim, J.; Jang, S.C.; Choi, E.-J.; Gho, Y.S.; Park, J. Microfluidic Filtration System to Isolate Extracellular Vesicles from Blood. Lab Chip 2012, 12, 5202–5210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, Y.; Wang, Y.; Lu, Y.; Luo, X.; Huang, Y.; Xie, T.; Pilarsky, C.; Dang, Y.; Zhang, J. Microfluidic Technology for the Isolation and Analysis of Exosomes. Micromachines 2022, 13, 1571. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zhu, Q.; Cheng, L.; Wang, Y.; Li, M.; Yang, Q.; Hu, L.; Lou, D.; Li, J.; Dong, X.; et al. Exosome Detection via the Ultrafast-Isolation System: EXODUS. Nat. Methods 2021, 18, 212–218. [Google Scholar] [CrossRef]
- Liu, F.; Vermesh, O.; Mani, V.; Ge, T.J.; Madsen, S.J.; Sabour, A.; Hsu, E.-C.; Gowrishankar, G.; Kanada, M.; Jokerst, J.V.; et al. The Exosome Total Isolation Chip. ACS Nano 2017, 11, 10712–10723. [Google Scholar] [CrossRef]
- Cho, S.; Jo, W.; Heo, Y.; Kang, J.Y.; Kwak, R.; Park, J. Isolation of Extracellular Vesicle from Blood Plasma Using Electrophoretic Migration through Porous Membrane. Sens. Actuators B Chem. 2016, 233, 289–297. [Google Scholar] [CrossRef]
- Wang, Z.; Wu, H.; Fine, D.; Schmulen, J.; Hu, Y.; Godin, B.; Zhang, J.X.J.; Liu, X. Ciliated Micropillars for the Microfluidic-Based Isolation of Nanoscale Lipid Vesicles. Lab Chip 2013, 13, 2879–2882. [Google Scholar] [CrossRef] [Green Version]
- Abreu, C.M.; Caballero, D.; Kundu, S.C.; Reis, R.L. From Exosomes to Circulating Tumor Cells: Using Microfluidics to Detect High Predictive Cancer Biomarkers. In Microfluidics and Biosensors in Cancer Research: Applications in Cancer Modeling and Theranostics; Caballero, D., Kundu, S.C., Reis, R.L., Eds.; Advances in Experimental Medicine and Biology; Springer: Cham, Switzerland, 2022; pp. 369–387. ISBN 978-3-031-04039-9. [Google Scholar]
- Wunsch, B.H.; Smith, J.T.; Gifford, S.M.; Wang, C.; Brink, M.; Bruce, R.L.; Austin, R.H.; Stolovitzky, G.; Astier, Y. Nanoscale Lateral Displacement Arrays for the Separation of Exosomes and Colloids down to 20 Nm. Nat. Nanotech 2016, 11, 936–940. [Google Scholar] [CrossRef]
- Wu, M.; Ouyang, Y.; Wang, Z.; Zhang, R.; Huang, P.-H.; Chen, C.; Li, H.; Li, P.; Quinn, D.; Dao, M.; et al. Isolation of Exosomes from Whole Blood by Integrating Acoustics and Microfluidics. Proc. Natl. Acad. Sci. USA 2017, 114, 10584–10589. [Google Scholar] [CrossRef] [Green Version]
- Lin, S.; Yu, Z.; Chen, D.; Wang, Z.; Miao, J.; Li, Q.; Zhang, D.; Song, J.; Cui, D. Progress in Microfluidics-Based Exosome Separation and Detection Technologies for Diagnostic Applications. Small 2020, 16, 1903916. [Google Scholar] [CrossRef]
- Jeong, K.; Jun Yu, Y.; Young You, J.; Jong Rhee, W.; Ah Kim, J. Exosome-Mediated MicroRNA-497 Delivery for Anti-Cancer Therapy in a Microfluidic 3D Lung Cancer Model. Lab Chip 2020, 20, 548–557. [Google Scholar] [CrossRef]
- Kimiz-Gebologlu, I.; Oncel, S.S. Exosomes: Large-Scale Production, Isolation, Drug Loading Efficiency, and Biodistribution and Uptake. J. Control. Release 2022, 347, 533–543. [Google Scholar] [CrossRef]
- Chen, Y.-S.; Lin, E.-Y.; Chiou, T.-W.; Harn, H.-J. Exosomes in Clinical Trial and Their Production in Compliance with Good Manufacturing Practice. Tzu. Chi Med. J. 2019, 32, 113–120. [Google Scholar] [CrossRef]
- Kim, H.; Kim, E.H.; Kwak, G.; Chi, S.-G.; Kim, S.H.; Yang, Y. Exosomes: Cell-Derived Nanoplatforms for the Delivery of Cancer Therapeutics. Int. J. Mol. Sci. 2021, 22, 14. [Google Scholar] [CrossRef]
- Watson, D.C.; Bayik, D.; Srivatsan, A.; Bergamaschi, C.; Valentin, A.; Niu, G.; Bear, J.; Monninger, M.; Sun, M.; Morales-Kastresana, A.; et al. Efficient Production and Enhanced Tumor Delivery of Engineered Extracellular Vesicles. Biomaterials 2016, 105, 195–205. [Google Scholar] [CrossRef] [Green Version]
- Wu, J.; Wu, D.; Wu, G.; Bei, H.-P.; Li, Z.; Xu, H.; Wang, Y.; Wu, D.; Liu, H.; Shi, S.; et al. Scale-out Production of Extracellular Vesicles Derived from Natural Killer Cells via Mechanical Stimulation in a Seesaw-Motion Bioreactor for Cancer Therapy. Biofabrication 2022, 14, 045004. [Google Scholar] [CrossRef]
- Jeske, R.; Liu, C.; Duke, L.; Canonicco Castro, M.L.; Muok, L.; Arthur, P.; Singh, M.; Jung, S.; Sun, L.; Li, Y. Upscaling Human Mesenchymal Stromal Cell Production in a Novel Vertical-Wheel Bioreactor Enhances Extracellular Vesicle Secretion and Cargo Profile. Bioact. Mater. 2022. [Google Scholar] [CrossRef]
- Hisey, C.L.; Artuyants, A.; Guo, G.; Chang, V.; Reshef, G.; Middleditch, M.; Jacob, B.; Chamley, L.W.; Blenkiron, C. Investigating the Consistency of Extracellular Vesicle Production from Breast Cancer Subtypes Using CELLine Adherent Bioreactors. bioRxiv 2022. [Google Scholar] [CrossRef]
- Karami, D.; Srivastava, A.; Ramesh, R.; Sikavitsas, V.I. Investigating Cancerous Exosomes’ Effects on CD8+ T-Cell IL-2 Production in a 3D Unidirectional Flow Bioreactor Using 3D Printed, RGD-Functionalized PLLA Scaffolds. J. Funct. Biomater. 2022, 13, 30. [Google Scholar] [CrossRef]
- Liu, X.; Zong, Z.; Xing, M.; Liu, X.; Li, J.; Liu, D. PH-Mediated Clustering of Exosomes: Breaking Through the Size Limit of Exosome Analysis in Conventional Flow Cytometry. Nano Lett. 2021, 21, 8817–8823. [Google Scholar] [CrossRef] [PubMed]
- Theodoraki, M.-N.; Hong, C.-S.; Donnenberg, V.S.; Donnenberg, A.D.; Whiteside, T.L. Evaluation of Exosome Proteins by On-Bead Flow Cytometry. Cytom. Part A 2021, 99, 372–381. [Google Scholar] [CrossRef] [PubMed]
- Moura, S.L.; Martín, C.G.; Martí, M.; Pividori, M.I. Multiplex Detection and Characterization of Breast Cancer Exosomes by Magneto-Actuated Immunoassay. Talanta 2020, 211, 120657. [Google Scholar] [CrossRef] [PubMed]
- Mastoridis, S.; Bertolino, G.M.; Whitehouse, G.; Dazzi, F.; Sanchez-Fueyo, A.; Martinez-Llordella, M. Multiparametric Analysis of Circulating Exosomes and Other Small Extracellular Vesicles by Advanced Imaging Flow Cytometry. Front. Immunol. 2018, 9, 1583. [Google Scholar] [CrossRef] [Green Version]
- Erdbrügger, U.; Rudy, C.K.; Etter, M.E.; Dryden, K.A.; Yeager, M.; Klibanov, A.L.; Lannigan, J. Imaging Flow Cytometry Elucidates Limitations of Microparticle Analysis by Conventional Flow Cytometry. Cytom. Part A 2014, 85, 756–770. [Google Scholar] [CrossRef]
- Nolan, J.; Sarimollaoglu, M.; Nedosekin, D.A.; Jamshidi-Parsian, A.; Galanzha, E.I.; Kore, R.A.; Griffin, R.J.; Zharov, V.P. In Vivo Flow Cytometry of Circulating Tumor-Associated Exosomes. Anal. Cell. Pathol. 2016, 2016, e1628057. [Google Scholar] [CrossRef]
| Type of Cancer | Name of miRNA | Main Function | Reference |
|---|---|---|---|
| Colorectal cancer | miR-1229 | Angiogenesis | [31,32] |
| miR-1246 | Angiogenesis | [33] | |
| miR-335-5p | EMT promotion | [34] | |
| miR-424-5p | Angiogenesis, metastasis, proliferation | [35] | |
| miR-25-3p | Angiogenesis, metastasis, vascular permeability | [36] | |
| Hepatocellular cancer | miR-210 | Angiogenesis | [37,38] |
| miR-374a-5p | EMT promotion | [39] | |
| Lung cancer | miR-494 | Angiogenesis, apoptosis prevention | [20,37,40] |
| miR-23a | Angiogenesis, metastasis | [41,42] | |
| Pancreatic cancer | miR-27a | Angiogenesis | [43] |
| Skin cancer | miR-21 | Angiogenesis, metastasis | [44] |
| miR-424-5p | Angiogenesis, metastasis, proliferation | [45] | |
| Glioma | miR-9-5p | Angiogenesis | [46] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bryant, P.; Sikavitsas, V.I. Cancer Exosomes: An Overview and the Applications of Flow. Fluids 2023, 8, 7. https://doi.org/10.3390/fluids8010007
Bryant P, Sikavitsas VI. Cancer Exosomes: An Overview and the Applications of Flow. Fluids. 2023; 8(1):7. https://doi.org/10.3390/fluids8010007
Chicago/Turabian StyleBryant, Parker, and Vassilios I. Sikavitsas. 2023. "Cancer Exosomes: An Overview and the Applications of Flow" Fluids 8, no. 1: 7. https://doi.org/10.3390/fluids8010007
APA StyleBryant, P., & Sikavitsas, V. I. (2023). Cancer Exosomes: An Overview and the Applications of Flow. Fluids, 8(1), 7. https://doi.org/10.3390/fluids8010007






