Modeling of 3D Blood Flows with Physics-Informed Neural Networks: Comparison of Network Architectures
Abstract
:1. Introduction
Physics-Informed Neural Networks
2. Methods
2.1. Fluid Model and Geometries
2.2. Network Architectures
2.2.1. Fully Connected Neural Network (FCNN)
2.2.2. Fully Connected Neural Network with Adaptive Activation (FCNNaa)
2.2.3. Fully Connected Neural Network with Skip Connections (FCNNskip)
2.2.4. Fourier Network (FN)
2.2.5. Modified Fourier Network (modFN)
2.2.6. Multiplicative Filter Network (MFN)
2.2.7. Deep Galerkin Method (DGM)
2.3. Network Details
2.4. Computational Fluid Dynamics (CFD)
2.5. Error Analysis
3. Results
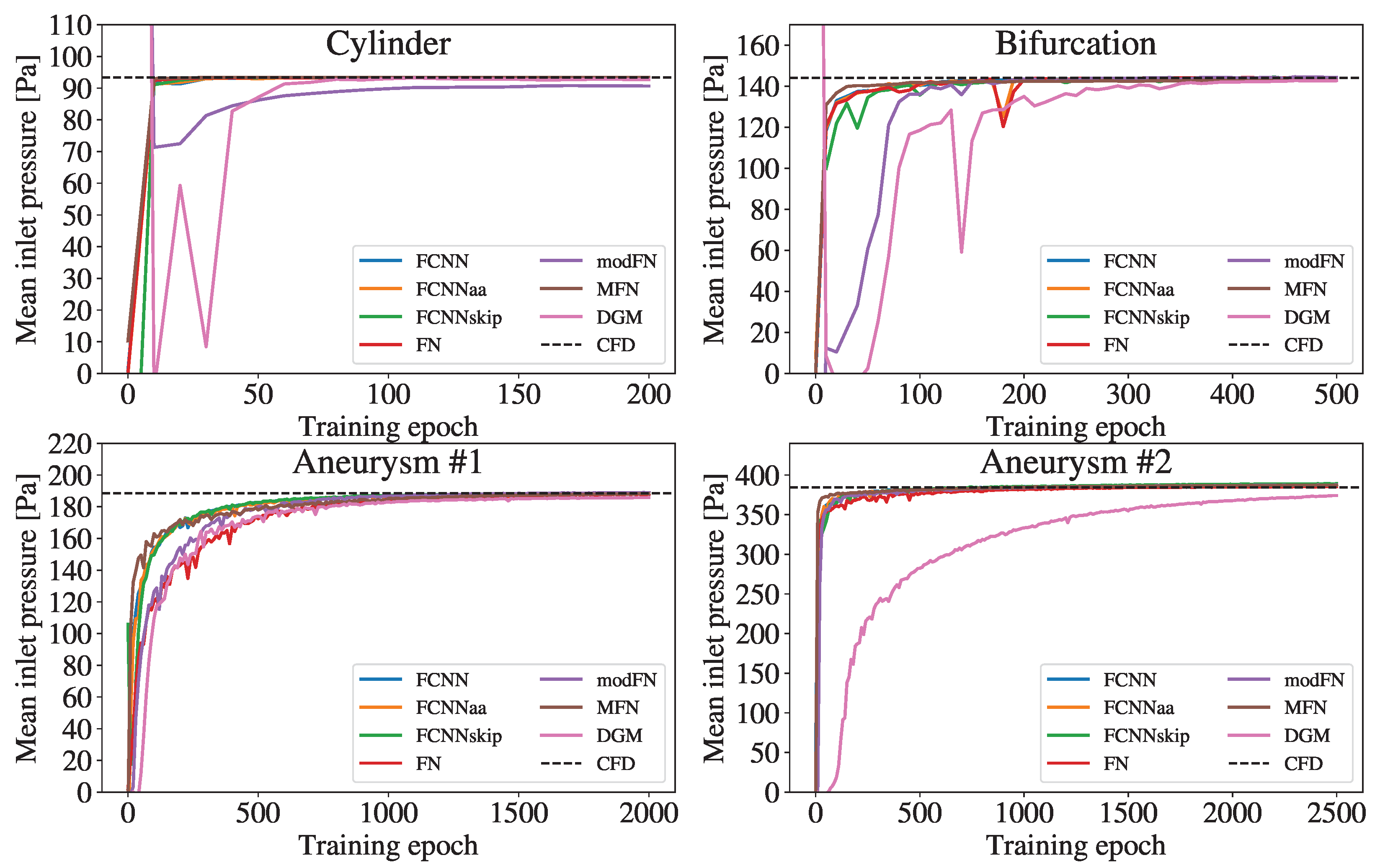
3.1. Comparison of Accuracy
3.2. Comparison of Training and Inference Runtimes
4. Discussion
Limitations
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| PINN | Physics-informed neural network |
| PDE | Partial differential equation |
| DL | Deep learning |
| FCNN | Fully connected neural network |
| FCNNaa | Fully connected neural network with adaptive activations |
| FCNNskip | Fully connected neural network with skip connections |
| FN | Fourier network |
| modFN | Modified Fourier network |
| MFN | Multiplicative filter network |
| DGM | Deep Galerkin Method |
| CNN | Convolutional neural network |
| CFD | Computational fluid dynamics |
| WSS | Wall shear stress |
References
- Brunton, S.L.; Noack, B.R.; Koumoutsakos, P. Machine Learning for Fluid Mechanics. Annu. Rev. Fluid Mech. 2020, 52, 477–508. [Google Scholar] [CrossRef] [Green Version]
- Han, J.; Jentzen, A.; E, W. Solving high-dimensional partial differential equations using deep learning. Proc. Natl. Acad. Sci. USA 2018, 115, 8505–8510. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, G.; Zhu, Y.; Yin, Y.; Su, M.; Li, M. Association of wall shear stress with intracranial aneurysm rupture: Systematic review and meta-analysis. Sci. Rep. 2017, 7, 5331. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cai, S.; Mao, Z.; Wang, Z.; Yin, M.; Karniadakis, G.E. Physics-informed neural networks (PINNs) for fluid mechanics: A review. Acta Mech. Sin. 2021, 37, 1727–1738. [Google Scholar] [CrossRef]
- Raissi, M.; Perdikaris, P.; Karniadakis, G.E. Physics-informed neural networks: A deep learning framework for solving forward and inverse problems involving nonlinear partial differential equations. J. Comput. Phys. 2019, 378, 686–707. [Google Scholar] [CrossRef]
- Sirignano, J.; Spiliopoulos, K. DGM: A deep learning algorithm for solving partial differential equations. J. Comput. Phys. 2018, 375, 1339–1364. [Google Scholar] [CrossRef] [Green Version]
- Cuomo, S.; Di Cola, V.S.; Giampaolo, F.; Rozza, G.; Raissi, M.; Piccialli, F. Scientific Machine Learning Through Physics–Informed Neural Networks: Where we are and What’s Next. J. Sci. Comput. 2022, 92, 88. [Google Scholar] [CrossRef]
- Taebi, A. Deep Learning for Computational Hemodynamics: A Brief Review of Recent Advances. Fluids 2022, 7, 197. [Google Scholar] [CrossRef]
- Arzani, A.; Wang, J.X.; D’Souza, R.M. Uncovering near-wall blood flow from sparse data with physics-informed neural networks. Phys. Fluids 2021, 33, 071905. [Google Scholar] [CrossRef]
- Kissas, G.; Yang, Y.; Hwuang, E.; Witschey, W.R.; Detre, J.A.; Perdikaris, P. Machine learning in cardiovascular flows modeling: Predicting arterial blood pressure from non-invasive 4D flow MRI data using physics-informed neural networks. Comput. Methods Appl. Mech. Eng. 2020, 358, 112623. [Google Scholar] [CrossRef]
- Aliakbari, M.; Mahmoudi, M.; Vadasz, P.; Arzani, A. Predicting high-fidelity multiphysics data from low-fidelity fluid flow and transport solvers using physics-informed neural networks. Int. J. Heat Fluid Flow 2022, 96, 109002. [Google Scholar] [CrossRef]
- Fathi, M.F.; Perez-Raya, I.; Baghaie, A.; Berg, P.; Janiga, G.; Arzani, A.; D’Souza, R.M. Super-resolution and denoising of 4D-Flow MRI using physics-Informed deep neural nets. Comput. Methods Programs Biomed. 2020, 197, 105729. [Google Scholar] [CrossRef]
- Jin, X.; Cai, S.; Li, H.; Karniadakis, G.E. NSFnets (Navier-Stokes flow nets): Physics-informed neural networks for the incompressible Navier-Stokes equations. J. Comput. Phys. 2021, 426, 109951. [Google Scholar] [CrossRef]
- Eivazi, H.; Tahani, M.; Schlatter, P.; Vinuesa, R. Physics-informed neural networks for solving Reynolds-averaged Navier–Stokes equations. Phys. Fluids 2022, 34, 075117. [Google Scholar] [CrossRef]
- Oldenburg, J.; Borowski, F.; Öner, A.; Schmitz, K.P.; Stiehm, M. Geometry aware physics informed neural network surrogate for solving Navier–Stokes equation (GAPINN). Adv. Model. Simul. Eng. Sci. 2022, 9, 8. [Google Scholar] [CrossRef]
- Amalinadhi, C.; Palar, P.S.; Stevenson, R.; Zuhal, L. On Physics-Informed Deep Learning for Solving Navier-Stokes Equations. In Proceedings of the AIAA SCITECH 2022 Forum, San Diego, CA, USA, 3–7 January 2022. [Google Scholar] [CrossRef]
- Raissi, M.; Yazdani, A.; Karniadakis, G.E. Hidden fluid mechanics: Learning velocity and pressure fields from flow visualizations. Science 2020, 367, 1026–1030. [Google Scholar] [CrossRef]
- Ma, H.; Zhang, Y.; Thuerey, N.; Hu, X.Y.; Haidn, O.J. Physics-Driven Learning of the Steady Navier-Stokes Equations using Deep Convolutional Neural Networks. Commun. Comput. Phys. 2022, 32, 715–736. [Google Scholar] [CrossRef]
- Eichinger, M.; Heinlein, A.; Klawonn, A. Stationary Flow Predictions Using Convolutional Neural Networks; Springer: Berlin/Heidelberg, Germany, 2021; pp. 541–549. [Google Scholar] [CrossRef]
- Guo, X.; Li, W.; Iorio, F. Convolutional Neural Networks for Steady Flow Approximation. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, KDD ’16, San Francisco, CA, USA, 13–17 August 2016; Association for Computing Machinery: New York, NY, USA, 2016; pp. 481–490. [Google Scholar] [CrossRef]
- Gao, H.; Sun, L.; Wang, J.X. PhyGeoNet: Physics-informed geometry-adaptive convolutional neural networks for solving parameterized steady-state PDEs on irregular domain. J. Comput. Phys. 2021, 428, 110079. [Google Scholar] [CrossRef]
- Choudhary, K.; DeCost, B.; Chen, C.; Jain, A.; Tavazza, F.; Cohn, R.; Park, C.W.; Choudhary, A.; Agrawal, A.; Billinge, S.J.L.; et al. Recent advances and applications of deep learning methods in materials science. npj Comput. Mater. 2022, 8, 1–26. [Google Scholar] [CrossRef]
- Markidis, S. The Old and the New: Can Physics-Informed Deep-Learning Replace Traditional Linear Solvers? Front. Big Data 2021, 4. [Google Scholar] [CrossRef]
- Karniadakis, G.E.; Kevrekidis, I.G.; Lu, L.; Perdikaris, P.; Wang, S.; Yang, L. Physics-informed machine learning. Nat. Rev. Phys. 2021, 3, 422–440. [Google Scholar] [CrossRef]
- Raissi, M.; Karniadakis, G.E. Hidden physics models: Machine learning of nonlinear partial differential equations. J. Comput. Phys. 2018, 357, 125–141. [Google Scholar] [CrossRef] [Green Version]
- Kingma, D.P.; Ba, J. Adam: A Method for Stochastic Optimization. In Proceedings of the International Conference on Learning Representations, San Diego, CA, USA, 7–9 May 2015. [Google Scholar] [CrossRef]
- Fenz, W.; Dirnberger, J.; Georgiev, I. Blood Flow Simulations with Application to Cerebral Aneurysms. In Proceedings of the Modeling and Simulation in Medicine Symposium, Pasadena, CA, USA, 3–6 April 2016; Society for Computer Simulation International: San Diego, CA, USA, 2016; p. 3. [Google Scholar] [CrossRef]
- Hennigh, O.; Narasimhan, S.; Nabian, M.A.; Subramaniam, A.; Tangsali, K.; Fang, Z.; Rietmann, M.; Byeon, W.; Choudhry, S. NVIDIA SimNet™: An AI-Accelerated Multi-Physics Simulation Framework. In International Conference on Computational Science, Proceedings of the Computational Science—ICCS 2021: 21st International Conference, Krakow, Poland, 16–18 June 2021; Springer: Berlin/Heidelberg, Germany, 2021; Part V; pp. 447–461. [Google Scholar] [CrossRef]
- Yu, C.C.; Tang, Y.C.; Liu, B.D. An adaptive activation function for multilayer feedforward neural networks. In Proceedings of the 2002 IEEE Region 10 Conference on Computers, Communications, Control and Power Engineering, TENCOM ’02. Beijing, China, 28–31 October 2002; Volume 1, pp. 645–650. [Google Scholar] [CrossRef]
- Qian, S.; Liu, H.; Liu, C.; Wu, S.; Wong, H.S. Adaptive Activation Functions in Convolutional Neural Networks. Neurocomput 2018, 272, 204–212. [Google Scholar] [CrossRef]
- Jagtap, A.D.; Kawaguchi, K.; Karniadakis, G.E. Adaptive activation functions accelerate convergence in deep and physics-informed neural networks. J. Comput. Phys. 2020, 404, 109136. [Google Scholar] [CrossRef] [Green Version]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar] [CrossRef] [Green Version]
- Rahaman, N.; Baratin, A.; Arpit, D.; Draxler, F.; Lin, M.; Hamprecht, F.A.; Bengio, Y.; Courville, A. On the Spectral Bias of Neural Networks. In Proceedings of the 36th International Conference on Machine Learning, Long Beach, CA, USA, 9–15 June 2019; Volume 97, pp. 5301–5310. [Google Scholar] [CrossRef]
- Tancik, M.; Srinivasan, P.; Mildenhall, B.; Fridovich-Keil, S.; Raghavan, N.; Singhal, U.; Ramamoorthi, R.; Barron, J.; Ng, R. Fourier Features Let Networks Learn High Frequency Functions in Low Dimensional Domains. In Proceedings of the Advances in Neural Information Processing Systems, Virtual Event. 6–12 December 2020; Curran Associates, Inc.: Red Hook, NY, USA, 2020; Volume 33, pp. 7537–7547. [Google Scholar] [CrossRef]
- Mildenhall, B.; Srinivasan, P.P.; Tancik, M.; Barron, J.T.; Ramamoorthi, R.; Ng, R. NeRF: Representing Scenes as Neural Radiance Fields for View Synthesis. In Proceedings of the Computer Vision—ECCV 2020, Glasgow, UK, 23–28 August 2020; Lecture Notes in Computer Science; Springer International Publishing: Berlin/Heidelberg, Germany, 2020; pp. 405–421. [Google Scholar] [CrossRef]
- Wang, S.; Teng, Y.; Perdikaris, P. Understanding and Mitigating Gradient Flow Pathologies in Physics-Informed Neural Networks. SIAM J. Sci. Comput. 2021, 43, A3055–A3081. [Google Scholar] [CrossRef]
- Fathony, R.; Sahu, A.K.; Willmott, D.; Kolter, J.Z. Multiplicative Filter Networks. In Proceedings of the International Conference on Learning Representations, Virtual Event. 3–7 May 2021. [Google Scholar]
- Al-Aradi, A.; Correia, A.; Naiff, D.; Jardim, G.; Saporito, Y. Solving Nonlinear and High-Dimensional Partial Differential Equations via Deep Learning. arXiv 2018, arXiv:1811.08782. [Google Scholar]
- Ramachandran, P.; Zoph, B.; Le, Q.V. Searching for Activation Functions. arXiv 2017, arXiv:1710.05941. [Google Scholar]
- Fenz, W.; Dirnberger, J.; Watzl, C.; Krieger, M. Parallel simulation and visualization of blood flow in intracranial aneurysms. In Proceedings of the 11th IEEE/ACM International Conference on Grid Computing, Brussels, Belgium, 25–28 October 2010; pp. 153–160. [Google Scholar] [CrossRef]
- Gmeiner, M.; Dirnberger, J.; Fenz, W.; Gollwitzer, M.; Wurm, G.; Trenkler, J.; Gruber, A. Virtual Cerebral Aneurysm Clipping with Real-Time Haptic Force Feedback in Neurosurgical Education. World Neurosurg. 2018, 112, e313–e323. [Google Scholar] [CrossRef]
- Zienkiewicz, O.; Nithiarasu, P.; Codina, R.; Vázquez, M.; Ortiz, P. The characteristic-based-split procedure: An efficient and accurate algorithm for fluid problems. Int. J. Numer. Methods Fluids 1999, 31, 359–392. [Google Scholar] [CrossRef]
- Schöberl, J. NETGEN An advancing front 2D/3D-mesh generator based on abstract rules. Comput. Vis. Sci. 1997, 1, 41–52. [Google Scholar] [CrossRef] [Green Version]
- Blanchard, M.; Bennouna, M.A. The Representation Power of Neural Networks: Breaking the Curse of Dimensionality. arXiv 2021, arXiv:2012.05451. [Google Scholar]
- Srivastava, R.K.; Greff, K.; Schmidhuber, J. Highway Networks. arXiv 2015, arXiv:1505.00387. [Google Scholar]
- Cheng, C.; Zhang, G.T. Deep Learning Method Based on Physics Informed Neural Network with Resnet Block for Solving Fluid Flow Problems. Water 2021, 13, 423. [Google Scholar] [CrossRef]
- Rafiq, M.; Rafiq, G.; Choi, G.S. DSFA-PINN: Deep Spectral Feature Aggregation Physics Informed Neural Network. IEEE Access 2022, 10, 22247–22259. [Google Scholar] [CrossRef]
- Wong, J.C.; Ooi, C.; Gupta, A.; Ong, Y.S. Learning in Sinusoidal Spaces with Physics-Informed Neural Networks. arXiv 2022, arXiv:2109.09338. [Google Scholar]
- Li, J.; Yue, J.; Zhang, W.; Duan, W. The Deep Learning Galerkin Method for the General Stokes Equations. J. Sci. Comput. 2022, 93, 5. [Google Scholar] [CrossRef]
- Matsumoto, M. Application of Deep Galerkin Method to Solve Compressible Navier-Stokes Equations. Trans. Jpn. Soc. Aeronaut. Space Sci. 2021, 64, 348–357. [Google Scholar] [CrossRef]
- Bronstein, M.M.; Bruna, J.; LeCun, Y.; Szlam, A.; Vandergheynst, P. Geometric Deep Learning: Going beyond Euclidean data. IEEE Signal Process. Mag. 2017, 34, 18–42. [Google Scholar] [CrossRef] [Green Version]
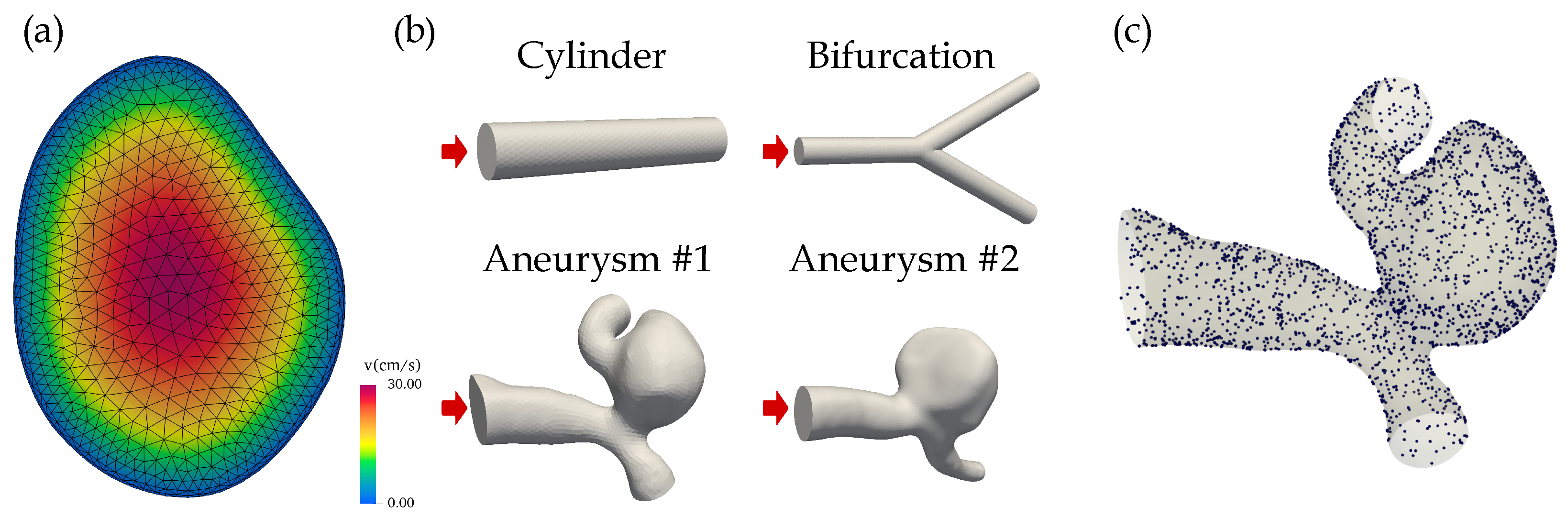


| Boundary Condition | #Points | |
|---|---|---|
| Inlet | Parabolic inlet velocity | 2000 |
| Outlet | Zero-pressure: | 1000 |
| Lateral surface | No-slip: | 2000 |
| Interior | Navier-Stokes residual | 3000 |
| Continuity plane | Mass flow continuity | 8000 |
| Cylinder | Bifurcation | |||||
|---|---|---|---|---|---|---|
| p [Pa] | abs vel [cm/s] | WSS [Pa] | p [Pa] | abs vel [cm/s] | WSS [Pa] | |
| FCNN | 0.640 ± 0.422 | 0.153 ± 0.121 | 0.085 ± 0.050 | 0.813 ± 0.338 | 0.210 ± 0.094 | 0.169 ± 0.215 |
| FCNNaa | 0.642 ± 0.428 | 0.153 ± 0.122 | 0.086 ± 0.050 | 0.826 ± 0.350 | 0.208 ± 0.091 | 0.167 ± 0.213 |
| FCNNskip | 0.642 ± 0.419 | 0.153 ± 0.121 | 0.085 ± 0.050 | 0.785 ± 0.329 | 0.203 ± 0.091 | 0.167 ± 0.218 |
| FN | 0.634 ± 0.425 | 0.154 ± 0.121 | 0.086 ± 0.050 | 0.945 ± 0.415 | 0.214 ± 0.094 | 0.171 ± 0.212 |
| modFN | 0.718 ± 0.425 | 0.116 ± 0.109 | 0.071 ± 0.061 | 0.898 ± 0.395 | 0.213 ± 0.091 | 0.173 ± 0.207 |
| MFN | 0.663 ± 0.421 | 0.154 ± 0.122 | 0.084 ± 0.053 | 0.628 ± 0.375 | 0.198 ± 0.138 | 0.157 ± 0.110 |
| DGM | 0.290 ± 0.257 | 0.107 ± 0.105 | 0.065 ± 0.054 | 0.136 ± 0.164 | 0.118 ± 0.072 | 0.145 ± 0.220 |
| Aneurysm | Aneurysm | |||||
| p [Pa] | abs vel [cm/s] | WSS [Pa] | p [Pa] | abs vel [cm/s] | WSS [Pa] | |
| FCNN | 0.924 ± 0.622 | 0.384 ± 0.323 | 0.607 ± 1.416 | 2.055 ± 0.672 | 1.125 ± 1.203 | 0.419 ± 0.458 |
| FCNNaa | 0.968 ± 0.625 | 0.386 ± 0.323 | 0.610 ± 1.426 | 2.056 ± 0.673 | 1.114 ± 1.201 | 0.417 ± 0.461 |
| FCNNskip | 0.868 ± 0.606 | 0.364 ± 0.311 | 0.611 ± 1.428 | 1.996 ± 0.659 | 1.123 ± 1.136 | 0.412 ± 0.433 |
| FN | 1.051 ± 0.731 | 0.512 ± 0.451 | 0.614 ± 1.412 | 2.348 ± 0.733 | 1.214 ± 1.617 | 0.491 ± 0.655 |
| modFN | 0.931 ± 0.647 | 0.408 ± 0.345 | 0.611 ± 1.424 | 2.182 ± 0.685 | 1.079 ± 1.263 | 0.441 ± 0.520 |
| MFN | 0.732 ± 0.729 | 0.369 ± 0.340 | 0.600 ± 1.370 | 2.203 ± 0.703 | 1.140 ± 1.391 | 0.446 ± 0.549 |
| DGM | 1.752 ± 1.229 | 0.363 ± 0.319 | 0.583 ± 1.380 | 5.489 ± 1.081 | 0.557 ± 0.768 | 0.205 ± 0.338 |
| FCNN | FCNNaa | FCNNskip | FN | modFN | MFN | DGM | |
|---|---|---|---|---|---|---|---|
| Runtime/FCNN | 100% | 115% | 102% | 101% | 169% | 114% | 363% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moser, P.; Fenz, W.; Thumfart, S.; Ganitzer, I.; Giretzlehner, M. Modeling of 3D Blood Flows with Physics-Informed Neural Networks: Comparison of Network Architectures. Fluids 2023, 8, 46. https://doi.org/10.3390/fluids8020046
Moser P, Fenz W, Thumfart S, Ganitzer I, Giretzlehner M. Modeling of 3D Blood Flows with Physics-Informed Neural Networks: Comparison of Network Architectures. Fluids. 2023; 8(2):46. https://doi.org/10.3390/fluids8020046
Chicago/Turabian StyleMoser, Philipp, Wolfgang Fenz, Stefan Thumfart, Isabell Ganitzer, and Michael Giretzlehner. 2023. "Modeling of 3D Blood Flows with Physics-Informed Neural Networks: Comparison of Network Architectures" Fluids 8, no. 2: 46. https://doi.org/10.3390/fluids8020046
APA StyleMoser, P., Fenz, W., Thumfart, S., Ganitzer, I., & Giretzlehner, M. (2023). Modeling of 3D Blood Flows with Physics-Informed Neural Networks: Comparison of Network Architectures. Fluids, 8(2), 46. https://doi.org/10.3390/fluids8020046






