Transcriptome Analysis Identifies Downstream Genes of CLAVATA3 in Tomato
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Identification of Transgenic Plants
2.3. Phenotypic Analysis of Flowers and Fruits
2.4. Transcriptome Sequencing and Data Analysis
2.5. Total RNA Extraction and Quantitative RT-PCR
3. Results
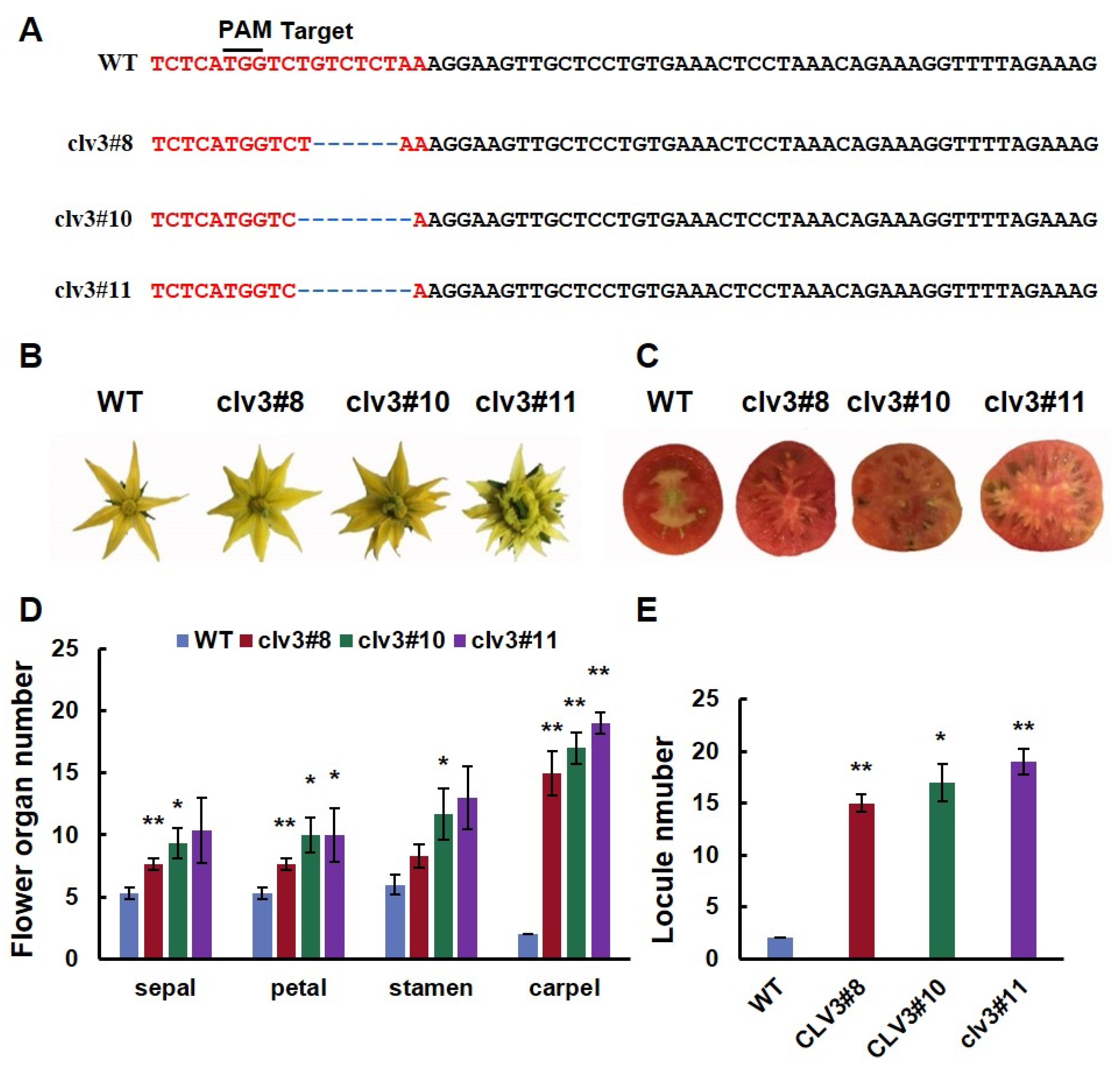
3.1. CRISPR/Cas9-Engineered Mutations in SlCLV3 Cause an Increase in the Number of Carpels and Fruit Locules
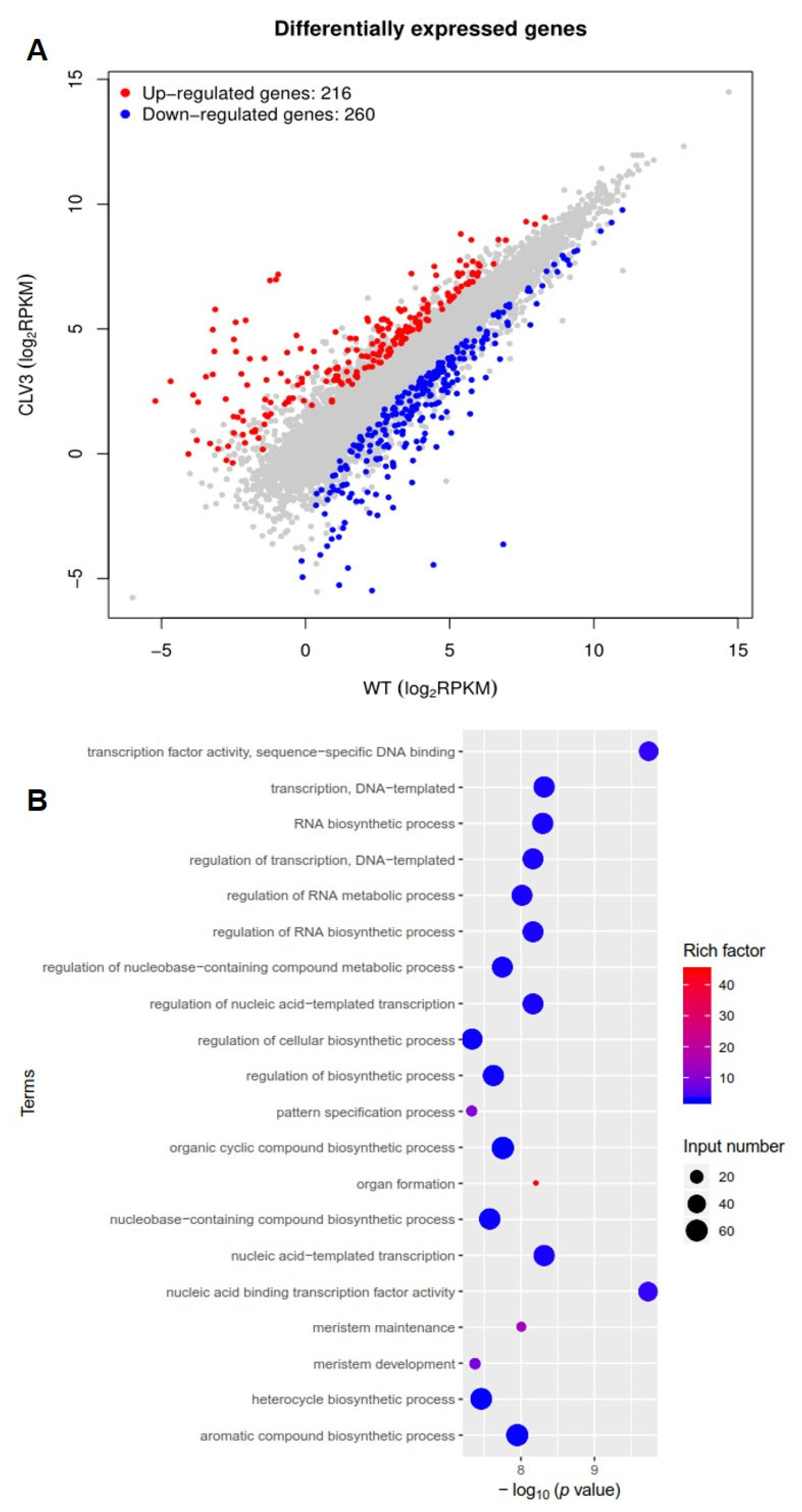
3.2. Transcriptome Analysis of WT and clv3 Mutants
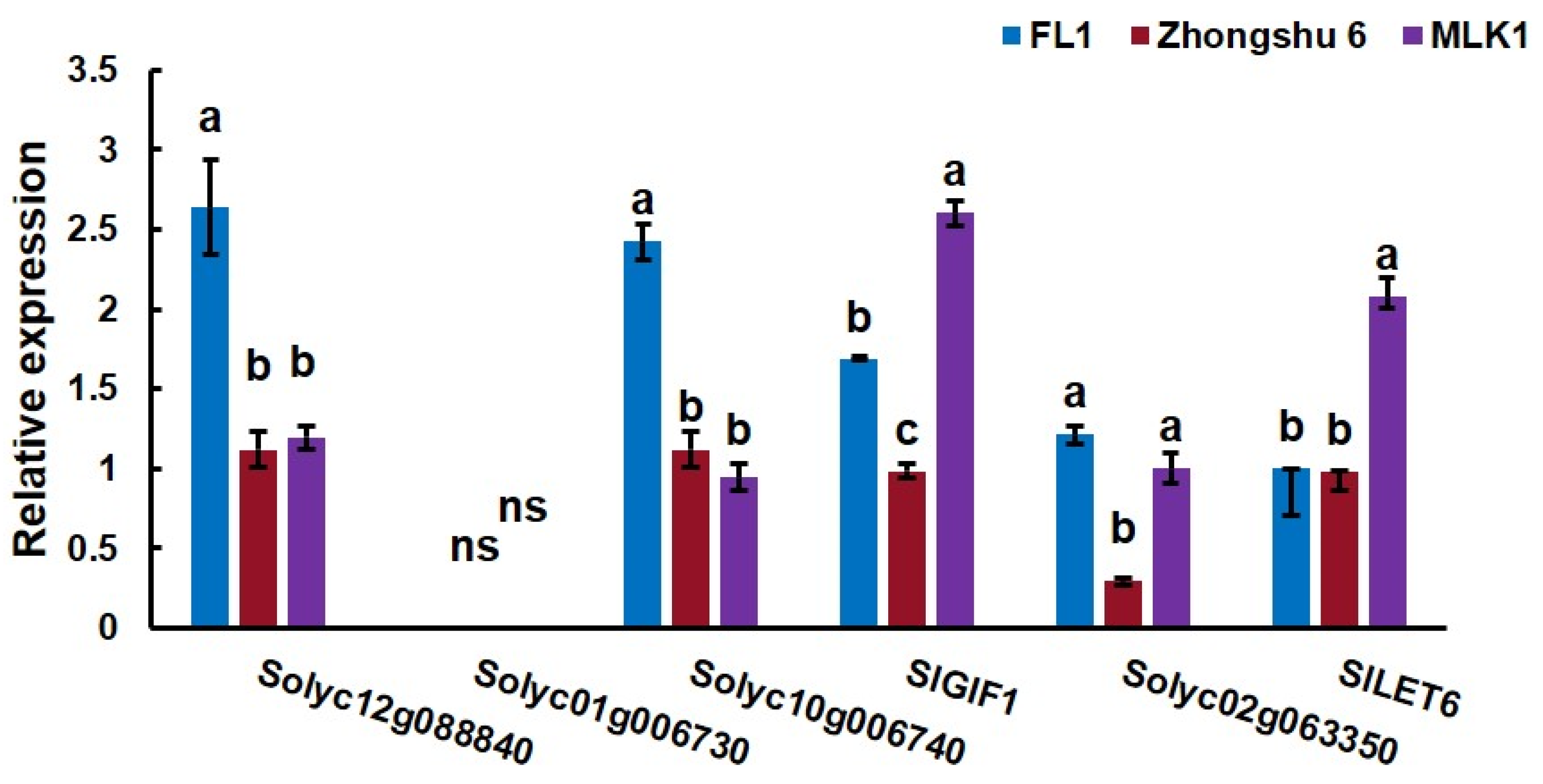
3.3. Higher Expression of SlLET6 and SlGIF1 Is Found on a Fas Mutation Background
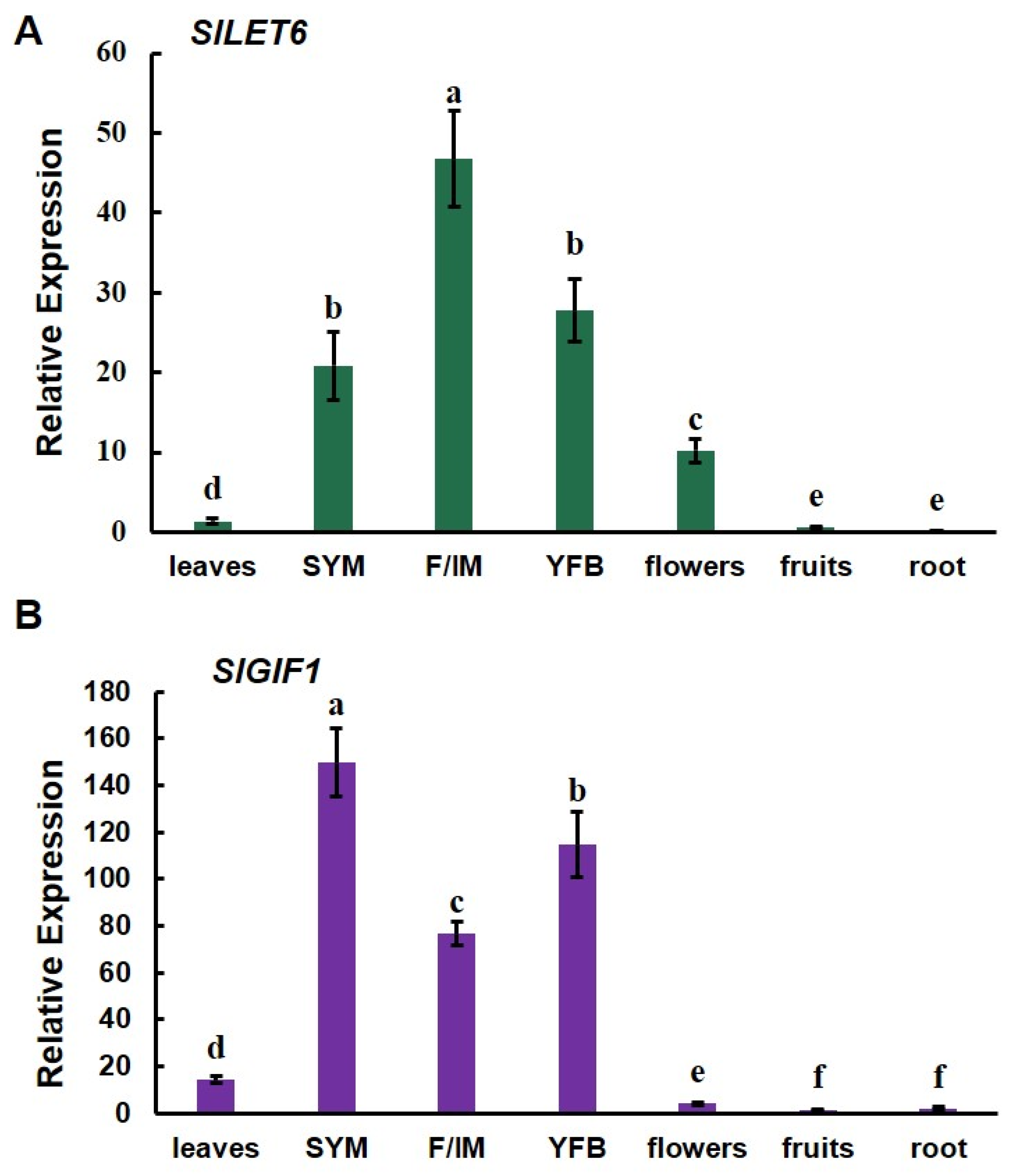
3.4. SlLET6 and SlGIF1 Were Highly Expressed during Flower Development
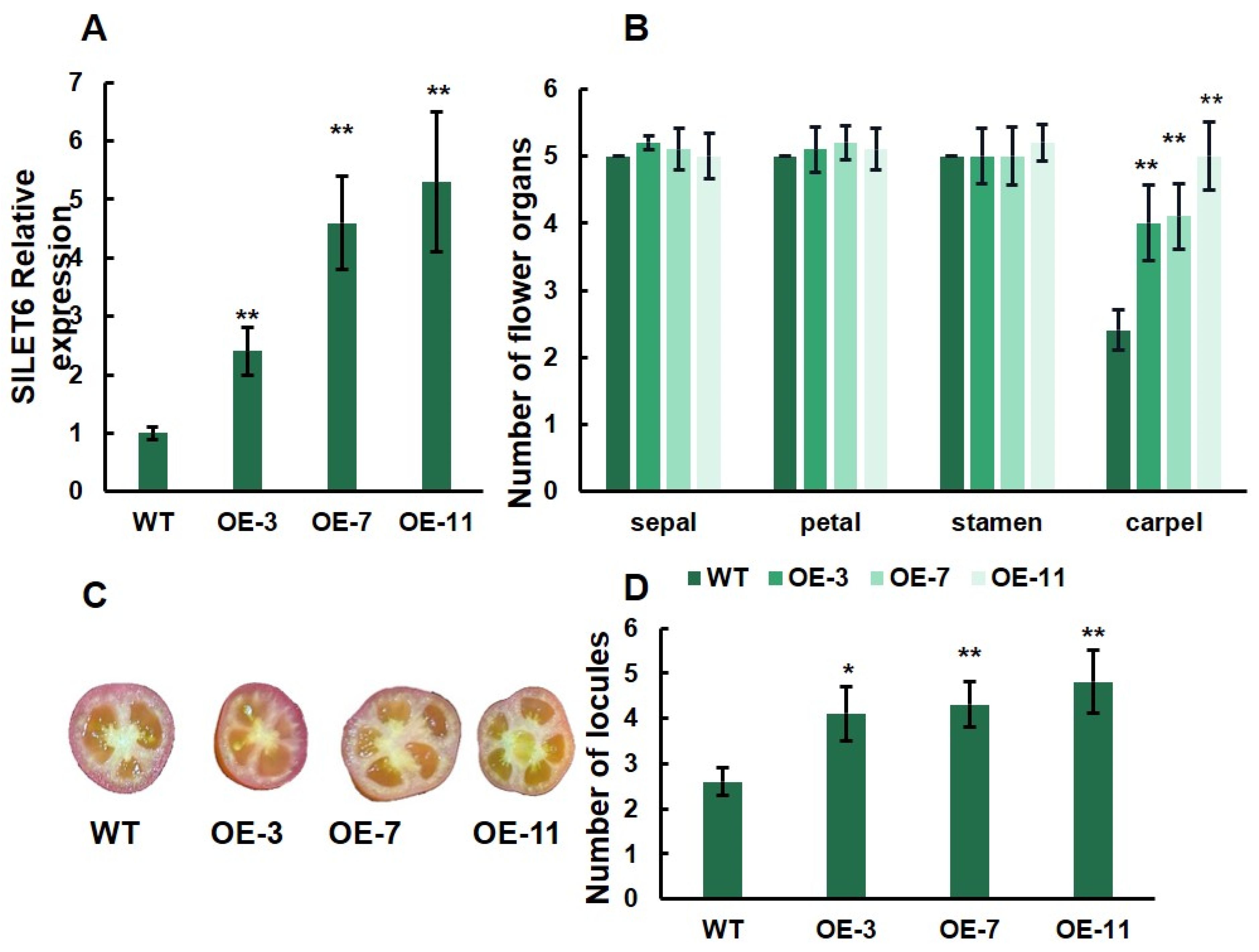
3.5. The Overexpression of SlLET6 Increased Tomato Fruit Locules
4. Discussion
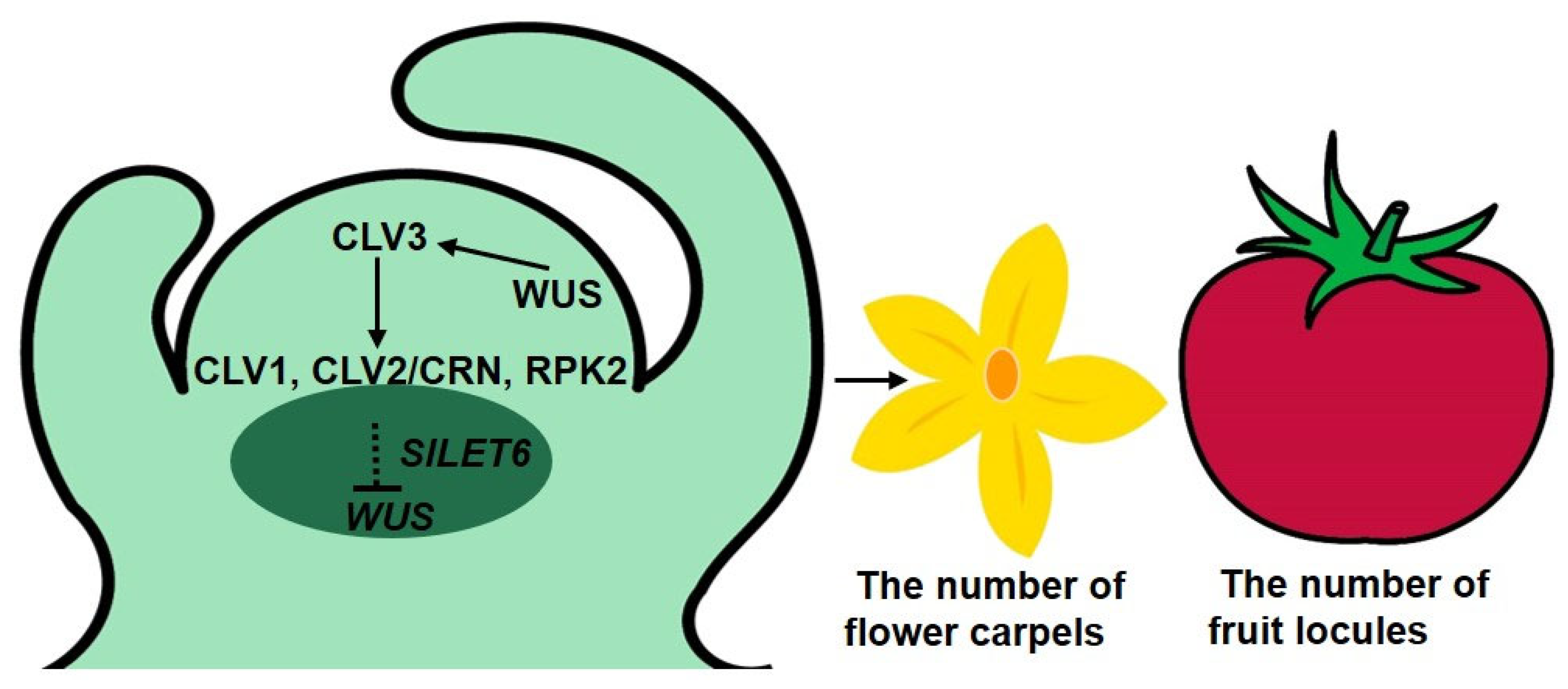
4.1. The Role of CLV3 in Regulating the Number of Flower Organs and Fruit Locules in Tomato
4.2. SlLET6 Plays an Important Role in Regulating the Flower Development of Tomato
4.3. SlLET6 May Play an Important Role Downstream of CLV3
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Tanksley, S.D. The genetic, developmental, and molecular bases of fruit size and shape variation in tomato. Plant Cell 2004, 16 (Suppl. 1), S181–S189. [Google Scholar] [CrossRef] [PubMed]
- Lippman, Z.; Tanksley, S.D. Dissecting the Genetic Pathway to Extreme Fruit Size in Tomato Using a Cross Between the Small-Fruited Wild Species Lycopersicon pimpinellifolium and L. esculentum var. Giant Heirloom. Genetics 2001, 158, 413–422. [Google Scholar] [CrossRef] [PubMed]
- Barrero, L.S.; Tanksley, S.D. Evaluating the genetic basis of multiple-locule fruit in a broad cross section of tomato cultivars. Theor. Appl. Genet. 2004, 109, 669–679. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, G.R.; Munos, S.; Anderson, C.; Sim, S.C.; Michel, A.; Causse, M.; Gardener, B.B.; Francis, D.; van der Knaap, E. Distribution of SUN, OVATE, LC, and FAS in the tomato germplasm and the relationship to fruit shape diversity. Plant Physiol. 2011, 156, 275–285. [Google Scholar] [CrossRef] [PubMed]
- Barrero, L.S.; Cong, B.; Wu, F.; Tanksley, S.D. Developmental characterization of the fasciated locus and mapping of Arabidopsis candidate genes involved in the control of floral meristem size and carpel number in tomato. Genome 2006, 49, 991–1006. [Google Scholar] [CrossRef] [PubMed]
- Munos, S.; Ranc, N.; Botton, E.; Berard, A.; Rolland, S.; Duffe, P.; Carretero, Y.; Le Paslier, M.C.; Delalande, C.; Bouzayen, M.; et al. Increase in tomato locule number is controlled by two single-nucleotide polymorphisms located near WUSCHEL. Plant Physiol. 2011, 156, 2244–2254. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Kim, Y.J.; Muller, R.; Yumul, R.E.; Liu, C.; Pan, Y.; Cao, X.; Goodrich, J.; Chen, X. AGAMOUS terminates floral stem cell maintenance in Arabidopsis by directly repressing WUSCHEL through recruitment of Polycomb Group proteins. Plant Cell 2011, 23, 3654–3670. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Leal, D.; Lemmon, Z.H.; Man, J.; Bartlett, M.E.; Lippman, Z.B. Engineering Quantitative Trait Variation for Crop Improvement by Genome Editing. Cell 2017, 171, 470–480. [Google Scholar] [CrossRef]
- Li, H.; Qi, M.; Sun, M.; Liu, Y.; Liu, Y.; Xu, T.; Li, Y.; Li, T. Tomato Transcription Factor SlWUS Plays an Important Role in Tomato Flower and Locule Development. Front. Plant Sci. 2017, 8, 457. [Google Scholar] [CrossRef]
- Cong, B.; Barrero, L.S.; Tanksley, S.D. Regulatory change in YABBY-like transcription factor led to evolution of extreme fruit size during tomato domestication. Nat. Genet. 2008, 40, 800–804. [Google Scholar] [CrossRef]
- Huang, Z.; Knaap, E.V.D. Tomato fruit weight 11.3 maps close to fasciated on the bottom of chromosome 11. Theor. Appl. Genet. 2011, 123, 465–474. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Liberatore, K.L.; MacAlister, C.A.; Huang, Z.; Chu, Y.H.; Jiang, K.; Brooks, C.; Ogawa-Ohnishi, M.; Xiong, G.; Pauly, M.; et al. A cascade of arabinosyltransferases controls shoot meristem size in tomato. Nat. Genet. 2015, 47, 784–792. [Google Scholar] [CrossRef] [PubMed]
- Aguirre, L.; Hendelman, A.; Hutton, S.F.; McCandlish, D.M.; Lippman, Z.B. Idiosyncratic and dose-dependent epistasis drives variation in tomato fruit size. Science 2023, 328, 315–320. [Google Scholar] [CrossRef] [PubMed]
- Stahl, Y.; Simon, R. Plant primary meristems: Shared functions and regulatory mechanisms. Curr. Opin. Plant Biol. 2010, 13, 53–58. [Google Scholar] [CrossRef] [PubMed]
- Schoof, H.; Lenhard, M.; Haecker, A.; Mayer, K.F.; Jürgens, G.; Laux, T. The stem cell population of Arabidopsis shoot meristems in maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell 2000, 100, 635–644. [Google Scholar] [CrossRef] [PubMed]
- Gou, X.; He, K.; Yang, H.; Yuan, T.; Lin, H.; Clouse, S.D.; Li, J. Genome-wide cloning and sequence analysis of leucine-rich repeat receptor-like protein kinase genes in Arabidopsis thaliana. BMC Genom. 2010, 11, 19. [Google Scholar] [CrossRef] [PubMed]
- Muller, R.; Bleckmann, A.R. The receptor kinase CORYNE of Arabidopsis transmits the stem cell-limiting signal CLAVATA3 independently of CLAVATA1. Plant Cell 2008, 20, 934–946. [Google Scholar] [CrossRef] [PubMed]
- Atsuko, K.; Shigeyuki, B.; Yuriko, O.; Shinji, M.; Shingo, N.; Yvonne, S.; Rüdiger, S.; Kazuko, Y.S.; Hiroo, F.; Shinichiro, S. RPK2 is an essential receptor-like kinase that transmits the CLV3 signal in Arabidopsis. Development 2010, 137, 3911–3920. [Google Scholar]
- Williams, R.W.; Wilson, J.M.; Meyerowitz, E.M. A possible role for kinase-associated protein phosphatase in the Arabidopsis CLAVATA1 signaling pathway. Proc. Natl. Acad. Sci. USA 1997, 94, 10467–10472. [Google Scholar] [CrossRef]
- Song, S.K.; Clark, S.E. POL and related phosphatases are dosage-sensitive regulators of meristem and organ development in Arabidopsis. Dev. Biol. 2005, 285, 272–284. [Google Scholar] [CrossRef]
- Takashi, I.; Ryo, T.; Masashi, Y.; Mitsuhiro, A.; Kanako, M.; Masayuki, F.; Katsushi, Y.; Shuji, S.; Masayuki, H.; Hiroyuki, T. Heterotrimeric G proteins control stem cell proliferation through CLAVATA signaling in Arabidopsis. EMBO Rep. 2015, 15, 1202–1209. [Google Scholar]
- Shigeyuki, B.; Fuminori, T.; Atsuko, K.; Hiroki, M.; Kazuo, S.; Hiroo, F.; Shinichiro, S. Mitogen-activated protein kinase regulated by the CLAVATA receptors contributes to shoot apical meristem homeostasis. Plant Cell Physiol. 2011, 52, 14–29. [Google Scholar]
- Chou, H.; Zhu, Y.; Ma, Y.; Berkowitz, G.A. The CLAVATA signaling pathway mediating stem cell fate in shoot meristems requires Ca2+ as a secondary cytosolic messenger. Plant J. 2016, 85, 494–506. [Google Scholar] [CrossRef]
- Clark, S.E.; Jacobsen, S.E.; Levin, J.Z.; Meyerowitz, E.M. The CLAVATA and SHOOT MERISTEMLESS loci competitively regulate meristem activity in Arabidopsis. Development 1996, 122, 1567–1575. [Google Scholar] [CrossRef]
- Zhang, Y.; Yang, S.; Song, Y.; Wang, J. Genome-wide characterization, expression and functional analysis of CLV3/ESR gene family in tomato. BMC Genom. 2014, 15, 827. [Google Scholar] [CrossRef]
- Su, D.; Xiang, W.; Wen, L.; Lu, W.; Shi, Y.; Liu, Y.; Li, Z. Genome-wide identification, characterization and expression analysis of BES1 gene family in tomato. BMC Plant Biol. 2021, 21, 161. [Google Scholar] [CrossRef]
- Su, D.; Wen, L.; Xiang, W.; Shi, Y.; Lu, W.; Liu, Y.; Xian, Z.; Li, Z. Tomato transcriptional repressor SlBES1.8 influences shoot apical meristem development by inhibiting the DNA binding ability of SlWUS. Plant. J. 2022, 110, 482–498. [Google Scholar] [CrossRef]
- Roth, O.; Alvarez, J.P.; Levy, M.; Bowman, J.L.; Ori, N.; Shani, E. The KNOXI Transcription Factor SHOOT MERISTEMLESS Regulates Floral Fate in Arabidopsis. Plant Cell 2018, 30, 1309–1321. [Google Scholar] [CrossRef]
- Scofield, S.; Murison, A.; Jones, A.; Fozard, J.; Aida, M.; Band, L.R.; Bennett, M.; Murray, J.A.H. Coordination of meristem and boundary functions by transcription factors in the SHOOT MERISTEMLESS regulatory network. Development 2018, 145, 157081. [Google Scholar] [CrossRef]
- Janssen, B.J.; Williams, A.; Chen, J.J.; Mathern, J.; Hake, S.; Sinha, N. Isolation and characterization of two knotted-like homeobox genes from tomato. Plant Mol. Biol. 1998, 36, 417–425. [Google Scholar] [CrossRef]
- Janssen, B.J.; Lund, L.; Sinha, N. Overexpression of a homeobox gene, LeT6, reveals indeterminate features in the tomato compound leaf. Plant Physiol. 1998, 117, 771–786. [Google Scholar] [CrossRef]
- Song, S.K.; Lee, M.M.; Clark, S.E. POL and PLL1 phosphatases are CLAVATA1 signaling intermediates required for Arabidopsis shoot and floral stem cells. Development 2006, 133, 4691–4698. [Google Scholar] [CrossRef]
- Lenhard, M.; Jürgens, G.; Laux, T. The WUSCHEL and SHOOTMERISTEMLESS genes fulfil complementary roles in Arabidopsis shoot meristem regulation. Development 2002, 129, 3195. [Google Scholar] [CrossRef]




| Locus Name | Primers Sequences (5′–3′) | |
|---|---|---|
| SlGIF1 | Sense: | ATGCAGCAGCAGCACCTGATG |
| Antisense: | TTAATTCCCATCTTCAGAAGA | |
| SlLET6 | Sense: | ATGGAGGGTGGTTCTAGTGGA |
| Antisense: | TCAGAGGAGAGAGGGTGTCATATC |
| Locus Name | Primers Sequences (5′–3′) | |
|---|---|---|
| SlGIF1 | Sense: | TGTGCAGAGTCCCAAGCTAA |
| Antisense: | GCTGCTCATTCCAAGTTGCT | |
| SlLET6 | Sense: | TCGAAAAACTGCTGTCTGAGGA |
| Antisense: | AGCAAAGTCAACAAGCACACC | |
| Solyc12g088840 | Sense: | TCGAAAAACTGCTGTCTGAGGA- |
| Antisense: | AGCAAAGTCAACAAGCACACC | |
| Solyc01g006730 | Sense: | TCATCGGTTGGTTCGTCGTT |
| Antisense: | CCCAATGCTCGATACTCGCT | |
| Solyc10g006740 | Sense: | TGCATGGACGCGGTTGATTA- |
| Antisense: | TGAGAGCTCCATCTCCATCCA | |
| Solyc02g063350 | Sense: | ACGGCGACGGAAAAATCTCT |
| Antisense: | AACTCCGACGCCGAAATCTT- | |
| Locus Name | Description | Log FC | Up/Down Regulated | p-Value |
|---|---|---|---|---|
| Solyc02g083950 | WUSCHEL protein | 1.21 | up | 0.004 |
| Solyc11g071810 | YABBY2b protein | 1.52 | down | 0.001 |
| AG1 | AGAMOUS protein | 1.81 | up | 0.003 |
| Solyc12g088840 | calcium ion binding protein | −2.2250 | down | 0.00121 |
| Solyc01g006730 | calcium-dependent protein kinase 20-like | 4.2036 | up | 0.00026 |
| Solyc10g006740 | calcium-binding protein PBP1-like | 1.9243 | up | 8 × 10−5 |
| Solyc11g006230 | GRF1-interacting factor 1 (SlGIF1) | 1.9224 | up | 1.67 × 10−5 |
| Solyc02g063350 | probable calcium-binding protein CML18 | −1.7595 | down | 0.00025 |
| Solyc02g081120 | class 1 knotted-like homeodomain protein (SlLET6) | 1.8474 | up | 0.00016 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Sun, H.; Tian, L.; Miao, Y.; Hou, L.; Sun, M.; Qi, M.; Li, T. Transcriptome Analysis Identifies Downstream Genes of CLAVATA3 in Tomato. Horticulturae 2024, 10, 11. https://doi.org/10.3390/horticulturae10010011
Zhang Y, Sun H, Tian L, Miao Y, Hou L, Sun M, Qi M, Li T. Transcriptome Analysis Identifies Downstream Genes of CLAVATA3 in Tomato. Horticulturae. 2024; 10(1):11. https://doi.org/10.3390/horticulturae10010011
Chicago/Turabian StyleZhang, Yaofeng, Huixian Sun, Linlin Tian, Yanxiu Miao, Leiping Hou, Meihua Sun, Mingfang Qi, and Tianlai Li. 2024. "Transcriptome Analysis Identifies Downstream Genes of CLAVATA3 in Tomato" Horticulturae 10, no. 1: 11. https://doi.org/10.3390/horticulturae10010011
APA StyleZhang, Y., Sun, H., Tian, L., Miao, Y., Hou, L., Sun, M., Qi, M., & Li, T. (2024). Transcriptome Analysis Identifies Downstream Genes of CLAVATA3 in Tomato. Horticulturae, 10(1), 11. https://doi.org/10.3390/horticulturae10010011




