Abstract
As the key transcription factors regulating auxin responsive genes expression, auxin response factors (ARFs) play critical roles in diverse aspects of plant growth and development. Betula pendula is a valuable ornamental tree, and the information on ARF gene family of B. pendula is needed for better understanding. The publication of the genome sequence of B. pendula enable to analyze the bioinformatics information and expression pattern of BpeARF gene family on the genome-wide basis. In this study, physical and chemical properties, chromosome location, phylogenetic relationship, gene structure, conserved domain, motif composition, and cis-acting element of BpeARF gene family were analyzed, and expression patterns of BpeARF genes were investigated using completely random design in different tissues and under exogenous NAA and drought treatments. A total of 17 BpeARF genes was identified from B. pendula genome, which were unevenly distributed on 13 chromosomes and encoded adequate proteins ranging from 613 to 1135 amino acids in length. Three BpeARF gene pairs were formed by segmental duplication, and the Ka/Ks values of these BpeARF gene pairs were less than 1. According to the phylogenetic relationship among B. pendula, Betula platyphylla, Populus trichocarpa, and Arabidopsis thaliana, the BpeARF genes were divided into four classes, and the intron/exon structure, conserved domain, and motif composition showed high similarity among the BpeARF genes within the same class. The cis-acting elements in the promoter regions of BpeARF genes were related to tissue development, hormone response, and stress resistance. Quantitative real-time PCR exhibited diverse expression patterns of BpeARF genes in different tissues and in response to exogenous auxin treatment and drought stress. The expressions of one, ten, seven, and three BpeARF genes were the high levels in buds, young leaves, stems, and roots, respectively. Under exogenous NAA treatment, six BpeARF genes in stems and roots were upregulated expression at all timepoints. Under drought stress, BpeARF7 and BpeARF15 were upregulated in stems and roots, and BpeARF5 and BpeARF6 were downregulated in leaves, stems, and roots. Our results provided valuable information for the classification and putative functions of BpeARF gene family, which may be helpful for selecting candidate genes and verifying gene function in the genetic engineering of birch trees in further research.
1. Introduction
Betula pendula, a cold-hardy deciduous arbor species belongs to the Betulaceae family, is mainly distributed in Europe, Caucasus, Siberia, and Northwest China. This species is recognized as a valuable ornamental tree due to white bark and pendulous branches, and is often planted decoratively in parks and gardens [1]. As the earliest discovered plant hormone, auxin plays an important role in diverse aspects of plant growth and development, such as embryogenesis, apical dominance, leaf expansion and aging, shoot elongation, root architecture, flower and fruit development, and abiotic stress responses, by regulating cell growth, division, and differentiation [2]. Recently, several genes involved in auxin transport and signal transduction were reported in B. pendula, such as PIN-formed (PIN) [3] and Gretchen Hagen 3 (GH3) [4], and a complex auxin signaling regulatory network was constructed at the molecular level in plants [5]. In the absence of auxin, auxin/indole-3-acetic acid (Aux/IAA) proteins formed heterodimers with auxin response factor (ARF) proteins to restrain ARF activity, which inhibited the transcription of auxin responsive genes, including AUX/IAA, GH3, and small auxin-up RNA (SAUR) [6]. When auxin level increases, AUX/IAA proteins bound to the SCFTIR1/AFB complex and were ubiquitinated and degraded by E3 ubiquitin ligase and 26S proteasome, respectively, which released ARF proteins from the heterodimers [7]. Being a vital component of auxin signaling pathway, ARF proteins could bind to auxin-responsive elements (AuxREs) in the promoter and regulated auxin responsive genes expression [8].
It is reported that ARF gene family was first discovered in Arabidopsis thaliana, and was widely present in plants as transcription factor genes [9]. A typical ARF protein contained three conserved domains, namely N-terminal B3-like DNA binding domain (DBD), Auxin Resp middle region (MR), and C-terminal Aux/IAA domain (CTD). The DBD domain was responsible for the recognition of AuxREs in the promoter of auxin responsive genes. The CTD domain, consisting of two highly conserved dimerization domains III and IV, facilitated the protein-protein interactions, such as homodimerization of ARFs or the heterodimerization of ARFs and Aux/IAAs. The MR domain, located between DBD and CTD, regulated the expression of auxin responsive genes as a transcriptional activator or repressor. The activator MR was enriched with glutamine, and the repressor MR was enriched with serine, leucine, proline, glycine, and tryptophan [10]. Nevertheless, several ARF proteins only had parts of the three conserved domains. For instance, HvARF4, -5, -14, -15, -16, -17, -18, -19, and -20 were only consists of DBD and MR in Hordeum vulgare [11], and OsARF2, -3, -13, -14, -15, and -20 were lack of CTD in Oryza sativa [12].
The biochemical and genetic evidences demonstrated that ARF genes played a crucial function in multiple auxin dependent processes. In A. thaliana, AtARF2, -4, and -5 regulated gametophyte development and seed weight [13,14]. AtARF6, -7, and -8 were activators of hypocotyl elongation to affect leaf and inflorescence growth [15,16]. In Gossypium hirsutum, GhARF2 and GhARF18 influenced fiber cell initiation by regulating several transcription factors [17]. In Solanum lycopersicon, SlARF7 controlled cell division at the early stage of fruit development [18], and SlARF4 and SlARF10 regulated sugar accumulation and chlorophyll biosynthesis in fruit [19,20]. Moreover, some ARF genes participated in the response to abiotic stresses. For example, BdARF4 and BdARF8 were upregulated under heavy metals and salicylic acid conditions in Brachypodium distachyon [21], and AhARF18 increased salt tolerance via posttranscriptional regulation of miR160 in Arachis hypogaea [22]. In a word, ARF gene family is largely involved in regulating plant growth and responding to environmental stress.
With the development of high-throughput sequencing technology, a large number of plant genomes have been sequenced, which provides feasibility for ARF gene family identification on the genome-wide in plants [23]. Recently, researches on ARF gene family have been reported in several trees with the sequenced genomes, such as Populus trichocarpa [24], Betula platyphylla [25], Osmanthus fragrans [26], and Elaeis guineensis [27]. Considering the importance of ARF genes in plant growth and development, the information on ARF gene family of B. pendula is needed for better understanding. The genome of B. pendula with the size of 440 Mb has been constructed in 2017 [28]. In the present study, 17 BpeARF genes were identified based on B. pendula genome sequence, and physical and chemical property, gene structure, domain distribution, motif type, and cis-acting element composition of each BpeARF gene were determined. Moreover, the phylogenetic relationship of the ARF gene family in B. pendula, B. platyphylla, P. trichocarpa, and A. thaliana was also compared. To reveal the potential effect of BpeARF gene family on regulating birch growth and responding to abiotic stress, the expression patterns of BpeARF genes were analyzed by quantitative real-time PCR (qRT-PCR) in different tissues and under exogenous auxin and drought treatment. These results provided valuable information for the classification and putative functions of BpeARF gene family in birch growth, development, and stress response, which may be helpful for selecting candidate genes in the genetic engineering of birch trees.
2. Materials and Methods
2.1. Identification of ARF Genes in Betula pendula
The genome sequence of B. pendula was obtained from CoGe Database (https://genomevolution.org (accessed on 26 November 2023)). A systematic identification of the ARF family members in B. pendula (BpeARF) was conducted while using the full-length A. thaliana ARF (AtARF) proteins as initial Basic Local Alignment Search Tool (BLAST) queries out of the B. pendula genome with the e-value of 10−10 [29]. The Hidden Markov Model (HMM) (http://hmmer.org/download.html (accessed on 26 November 2023)) and Proterin Families Database (Pfam) (http://pfam.xfam.org/ (accessed on 26 November 2023)) were further used to search B. pendula genome sequences with DBD domain (Pfam 02362), MR domain (Pfam 06507), and CTD domain (Pfam 02309) [30,31]. Moreover, the NCBI Conserved Domain Database (https://www.ncbi.nlm.nih.gov/cdd/ (accessed on 26 November 2023)) was used to further confirm the position and integrity of the detected conserved domains in individual BpeARFs [32].
2.2. Amino Acid Sequence, Gene Structure and Subcellular Localization
The number of amino acids, molecular weight and theoretical isoelectric point of the BpeARF proteins were predicted with ProtParam tool (https://web.expasy.org/protparam/ (accessed on 26 November 2023)) [33]. The coding DNA sequence (CDS), intron, and exon structures of BpARF genes were analyzed by GSDS 2.0 (http://gsds.gao-lab.org/ (accessed on 26 November 2023)) [34]. The subcellular localization predictions of the BpeARFs were performed on the WoLF PSORT (https://wolfpsort.hgc.jp/ (accessed on 26 November 2023)) [35].
2.3. Chromosomal Localization, Gene Duplication and Phylogenetic Analysis
All the BpeARFs chromosomal locations were obtained from the annotation general feature format files of B. pendula genome sequence using MG2C (http://mg2c.iask.in/mg2c_v2.0 (accessed on 26 November 2023)) [36]. Gene duplication events were conducted by MCScanX software (http://chibba.pgml.uga.edu/mcscan2/ (accessed on 26 November 2023)) with the e-value of 10−5 [37]. The synteny relationship of ARF genes in B. pendula, B. platyphylla, P. trichocarpa, and A. thaliana genomes was constructed using Dual Synteny Plotter [38]. The substitution of nonsynonymous (Ka) and synonymous (Ks) for each repeated ARF gene were calculated using the KaKs_Calculator 3.0 [39]. To reveal the phylogenetic relationship of orthologous ARF genes in B. pendula, B. platyphylla, P. trichocarpa, and A. thaliana, the phylogenetic tree was constructed and drafted with MEGA X (https://www.megasoftware.net/ (accessed on 26 November 2023)), using the neighbor-joining method with a bootstrap of 1000 replicates [40]. Classified and annotated of ARF protein sequences by using iTOL (https://itol.embl.de/itol.cgi (accessed on 26 November 2023)) [41].
2.4. Conservative Motif and Promoter Cis-Acting Element
The conserved motifs of BpeARF proteins were predicted using MEME Suite (https://meme-suite.org/tools/meme (accessed on 26 November 2023)), and the search options were as follows: motif distribution among sequences was 0 to 1 occurrence, the motif width range was from 6 to 50 amino acids, and the maximum motifs per sequences was 20 [42]. A Perl script was used to retrieve 2000 bp sequences upstream of the transcription start sites of BpeARF genes. The cis-acting elements in promoters of the BpeARF genes were analyzed on PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/ (accessed on 26 November 2023)) [43]. The gene structure, conserved domains, conserved motifs, and cis-acting elements of each BpeARF gene were visualized by TBtools (https://github.com/CJ-Chen/TBtools (accessed on 26 November 2023)) [44].
2.5. Plant Materials and Treatments
The saplings of B. pendula clone were planted at Xinshan Nursery Stock Cooperative, located in Jiangmifeng Town, Longtan District, Jilin City, China. In mid-June 2021, the healthy and semi-lignified branches with a bud and a leaf were collected as softwood cuttings. The basal regions of the cuttings were submerged in the rooting solution (a mixture containing 0.30 mmol/L 3-indolebutyric acid (IBA) and 0.21 mmol/L 1-naphthlcetic acid (NAA)) for 2 h, and then were cultivated in a seedbed filled with perlite as growth substrate. In cultivation, substrate water content (10 cm in depth) was maintained at 80% by auto-spraying device during the experimental period. After 45 days, the cuttings were transplanted into plastic bowls filled with humus soil and sandy soil (The ratio of humus soil to sandy soil was 3:1) in the greenhouse using completely random design, and the surviving cuttings were used for exogenous auxin and drought stress treatments. The leaves of cuttings were sprayed with 0.1 mmol/L NAA as exogenous auxin treatment, and the adventitious roots of cuttings were irrigated with 30 mmol/L polyethylene glycol 6000 (PEG6000) as drought stress treatment. Each treatment was applied for 0 h, 6 h, 12 h, 24 h, and 48 h, respectively. The leaves, stems, and roots of cuttings were harvested at the corresponding timepoints, and immediately frozen in liquid nitrogen and stored at −80 °C for qRT-PCR. In early-July 2022, the buds, young leaves, mature leaves, stems, and roots were collected from cuttings without NAA and PEG6000 treatments, respectively, and immediately frozen in liquid nitrogen and stored at −80 °C for qRT-PCR.
2.6. RNA Extraction and qRT-PCR Analysis
To explore the expression patterns of BpeARF genes in tissues and treatments, qRT-PCR was performed and primers for these genes were designed manually, and 18S rRNA and α-Tublin were used as internal reference genes (Table 1). The total RNA from each sample was extracted using an improved hexadecyl trimethyl ammonium bromide (CTAB) method and treated with DNase I to remove contaminating DNA [45]. The first-strand cDNA was synthesized using a PrimeScript™ RT reagent Kit (Takara, Dalian, China). The reactions were performed in a real-time PCR system (ABI QuantStudio5, Applied Biosystems, Carlsbad, CA, USA) and the relative expression level was calculated by 2−ΔΔCt method [46]. The expressions of buds and control samples (0 h) were set to 1 for normalization, respectively, and all experiments were conducted with three biological replicates for each sample.

Table 1.
Primers used for quantitative real-time PCR.
2.7. Statistical Analysis
The relative expression data was analyzed by one-way analysis of variance (ANOVA) and Duncan test using Statistics Analysis System (SAS, v9.21, SAS Institute Inc., Raleigh, NC, USA), and the data underwent logarithmic conversion with a base of 2.
3. Results
3.1. Identification of BpeARF Gene Family
To identify BpeARF gene family, BLASTP using AtARF protein sequences as queries, and HMM searching with DBD, MR, and CTD against the B. pendula genome. A total 17 BpeARF genes were identified in B. pendula, including ten complete genes and seven truncated genes, and these gene were provisionally named BpeARF1 to BpeARF17. The CDS lengths of the BpeARFs ranged from 1842 bp to 3408 bp, which produced adequate proteins ranging from 613 to 1135 amino acids in length with molecular weight of 65.209–127.009 kDa. The theoretical isoelectric point of the BpeARFs ranged from 4.71 to 7.96, including two basic proteins and fifteen acidic proteins. All the BpeARF proteins was predicted to be located in the nucleus (Table 2).

Table 2.
Position and properties of BpeARF gene family in Betula pendula.
3.2. Chromosome Distribution and Synteny Relationship of BpeARF Genes
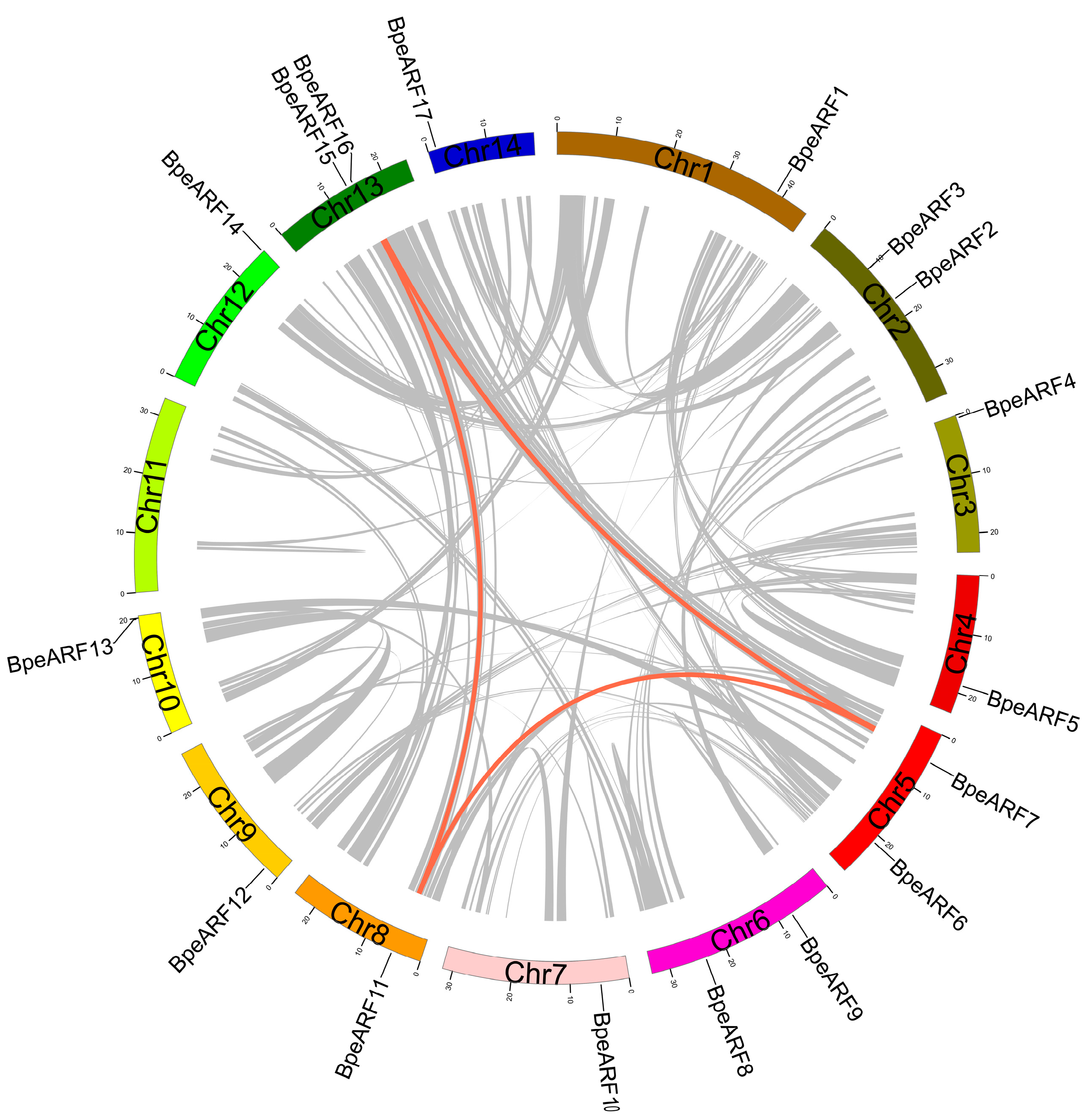
The 17 members of the BpeARF gene family were unevenly distributed in 13 chromosomes of B. pendula, with no BpeARF gene distribution in chromosome 11 (Table 2). Most BpeARF genes were on the beginning and end regions of the chromosomes. There were two BpeARF genes each on Chromosome 2, -5, -6, and -13, and one BpeARF gene on Chromosome 1, -3, -4, -7, -8, -9, -10, -12, and -14. Chromosome 2, -7, and -11, as the similar chromosome size, contained different number of BpeARF genes, indicating that the number of BpeARF genes on each chromosome was irrelevant to the chromosome size. In addition, collinearity analysis of the BpeARF genes showed three pairs (BpARF7/11, BpARF7/15, and BpARF11/15, respectively) of homologous BpeARF genes, which illustrated the three BpeARF gene pairs were formed by segmental duplication (Figure 1). The Ka/Ks of BpeARF7/11, BpeARF7/15, and BpeARF11/15 were 0.145, 0.196, and 0.157, respectively, suggesting their function conservation.
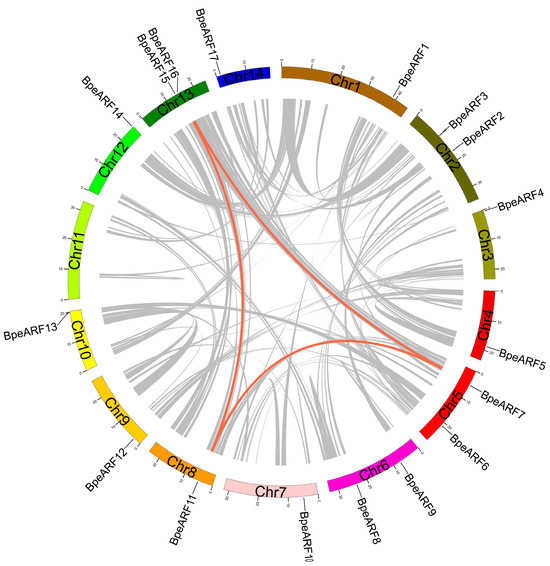
Figure 1.
Chromosome distribution and gene duplication of BpeARF gene family. Each gene was mapped to the chromosome based on its physical location. The red lines emphasized collinear pairs of BpeARF genes, while the gray lines indicated collinear blocks of B. pendula genes.
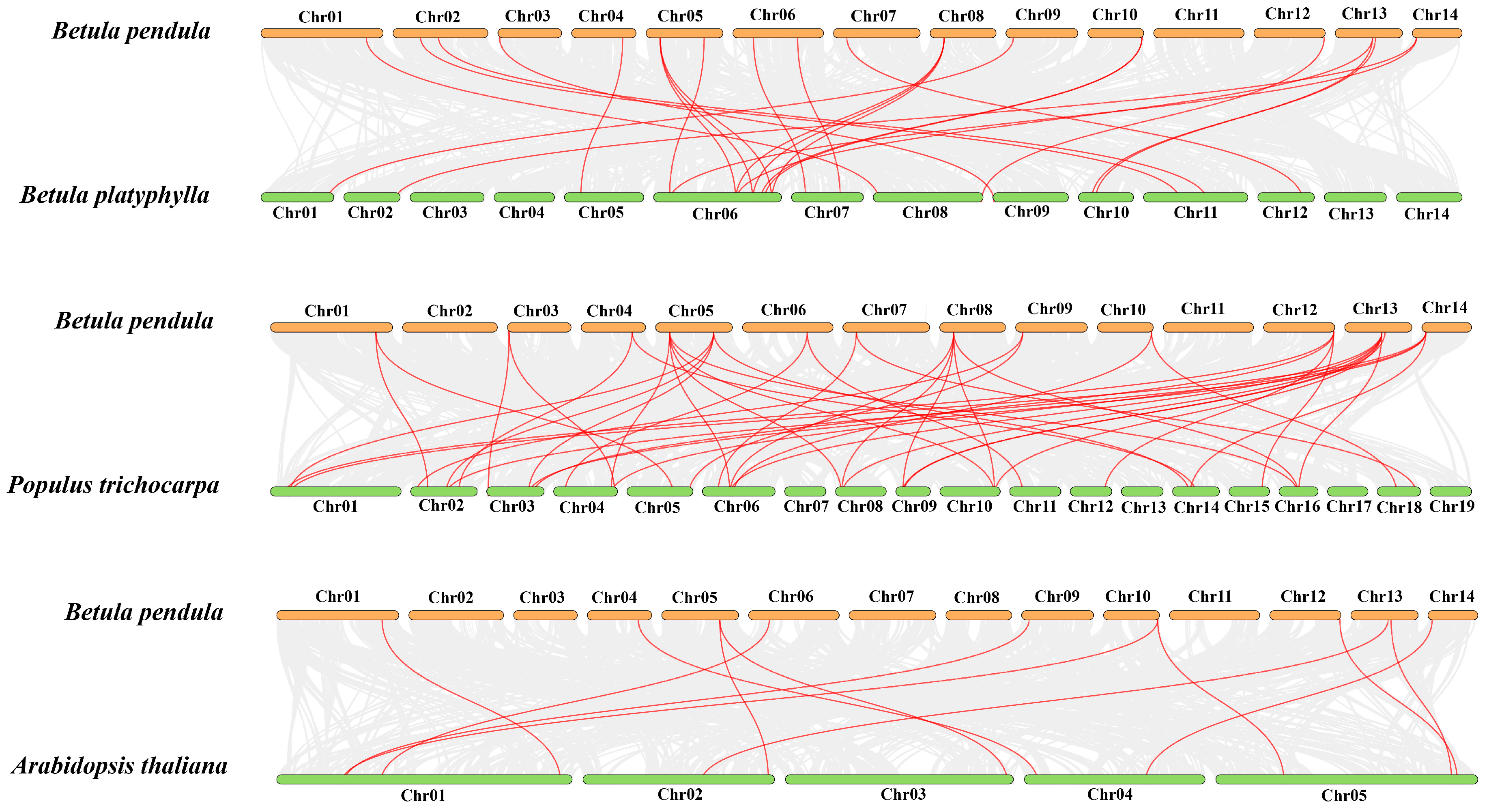
To further examine the orthologous relationships of ARF genes, the synteny analysis was conducted among three trees (B. pendula, B. platyphylla, and P. trichocarpa) and one herb (A. thaliana) (Figure 2). The results suggested that there were 24 pairs, 43 pairs, and 12 pairs orthologous genes between B. pendula and B. platyphylla, B. pendula and P. trichocarpa, and B. pendula and A. thaliana, respectively. Moreover, nine BpeARF genes (BpeARF1, -5, -6, -12, -13, -14, -15, -16, and -17) were associated with gene pairs among B. pendula, B. platyphylla, P. trichocarpa, and A. thaliana, suggesting that these genes may play an important role in the evolution of the ARF gene family. The Ka/Ks values of most orthologous ARF gene pairs were less than 1, demonstrating their function relative conservation in B. pendula, B. platyphylla, P. trichocarpa, and A. thaliana.
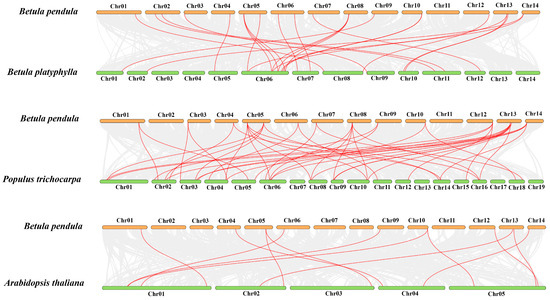
Figure 2.
Synteny relationships of the ARF genes between B. pendula and three other plant species. The red lines emphasized the syntenic ARF gene pairs, and the gray lines indicated the collinear blocks within B. pendula and other plant genomes.
3.3. Phylogeny and Classification of BpeARF Genes
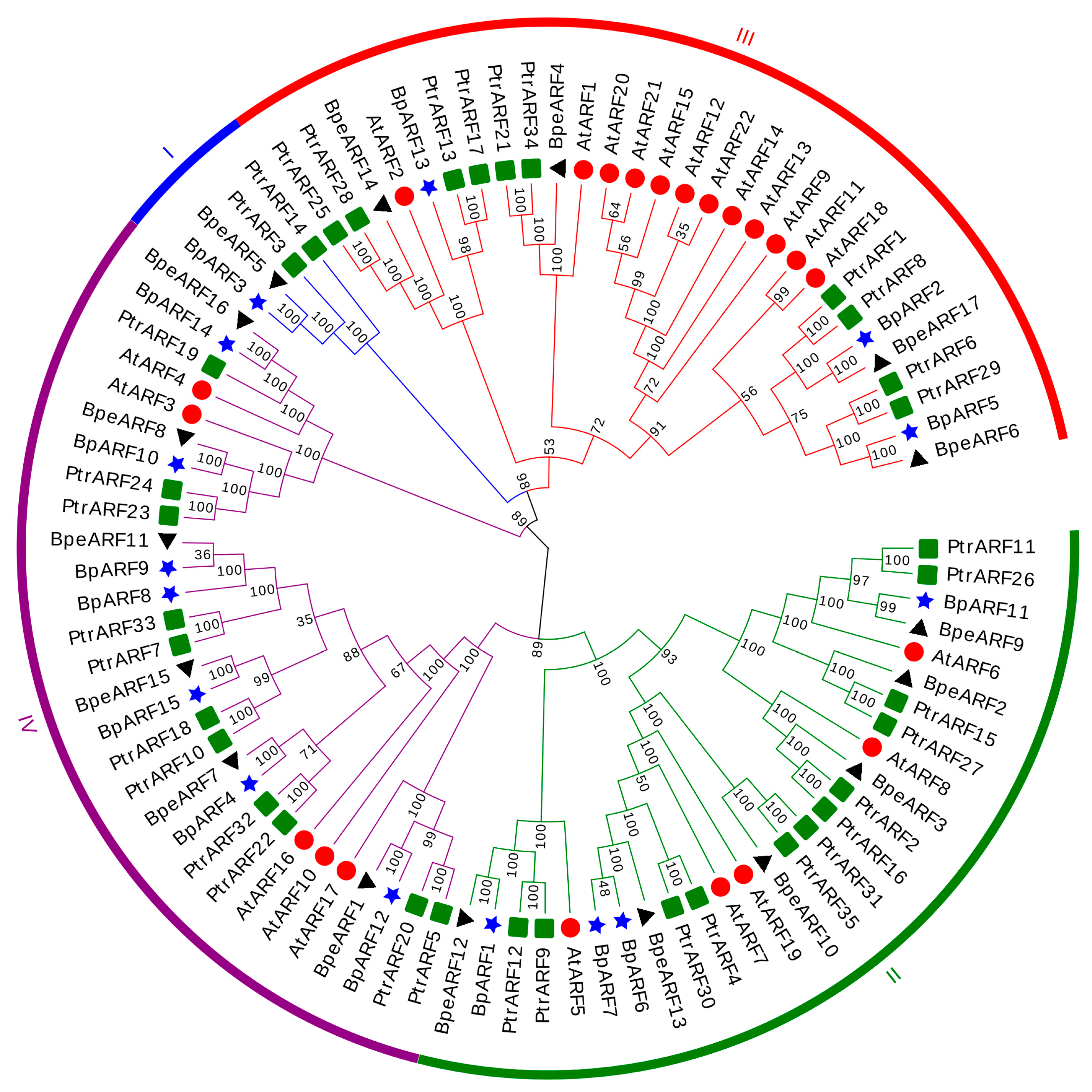
To reveal the phylogenetic relationship and grouping pattern of BpeARFs, a phylogenetic tree was constructed based on 17 BpeARF, 15 BpARF, 35 PtrARF, and 22 AtARF members using the neighbor-joining method. All the ARFs was divided into four classes, named Class I, Class II, Class III, and Class IV (Figure 3). Class III (four BpeARFs, three BpARFs, ten PtrARFs, and twelve AtARFs) and Class IV (six BpeARFs, seven BpARFs, eleven PtrARFs, and five AtARFs) contained 29 ARFs, respectively, followed by Class II (27 ARFs, including six BpeARFs, four BpARFs, twelve PtrARFs, and five AtARFs). Class I was an isolated branch which only contained BpeARF5, BpARF3, PtrARF3 and PtrARF14, and lacked of AtARF members. Furthermore, most BpeARF genes were first grouped with BpARF genes, which suggested that ARF genes might have a closer evolutionary relationship in birch. Nevertheless, BpeARF14 and AtARF2, BpeARF4 and AtARF1, and BpeARF3 and AtARF8 were clustered together, respectively, indicating that the evolutionary relationship of some ARF genes between B. pendula and A. thaliana were more conserved than that between trees and herbs.
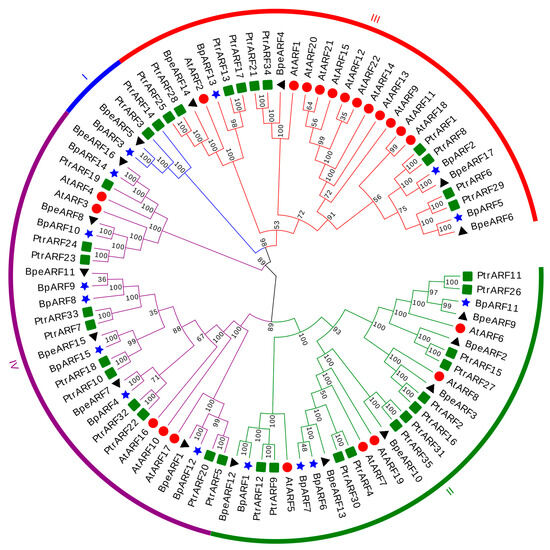
Figure 3.
Phylogenetic relationship and classification ARF genes from B. pendula, B. platyphylla, P. trichocarpa, and A. thaliana. All ARF genes were classified into four classes with different colors. Four plant species were indicated by distinct symbols, respectively. Triangle, B. pendula; star, B. platyphylla; square, P. trichocarpa; round, A. thaliana. Bootstrap values were indicated by different numbers.
3.4. Gene Structure, Conserved Domain and Motif Composition of BpeARF Genes
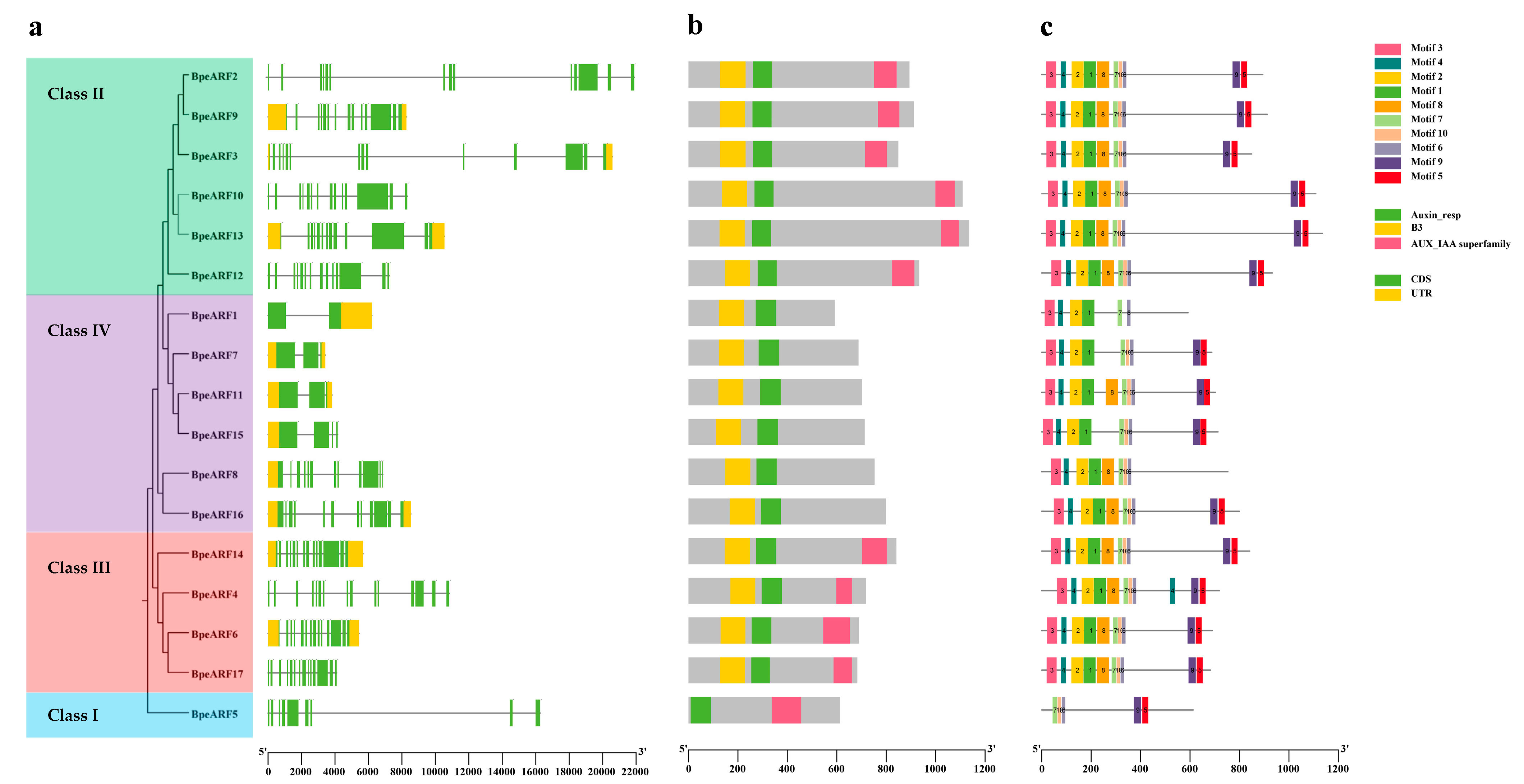
To understand the structural components of the BpeARF genes, the exons and introns of the BpeARF genes were obtained by comparison with the corresponding genomic DNA sequences (Figure 4a). Gene structural analysis provided further evidence to support the phylogeny and classification of BpeARF gene family. A similar ARF classification pattern to the phylogenetic tree was observed, with BpeARFs clustering into four classes according to their gene structure. The results suggested that the numbers of introns and exons of BpeARF genes ranged from 1 to 14 and 2 to 15 across the four classes, respectively. BpeARF5 in Class I had nine exons and eight introns, Class II contained 13–14 exons and 12–13 introns, Class III had 14–15 exons and 13–14 introns, and Class IV contained 2–12 exons and 1–11 introns, indicating that the gene structure was somewhat differentiated in different classes, whereas they were conserved among the BpeARF genes within a class.
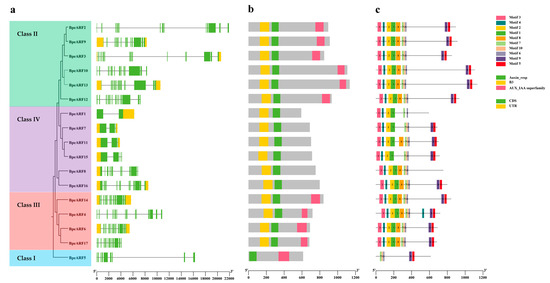
Figure 4.
Gene structure, conserved domain and motif composition of BpeARF gene family. (a) Gene structure. Green boxes, yellow boxes, and black lines represented CDS regions, UTR regions, and intron regions, respectively; (b) Conserved domain. DBD, MR, and CTD were marked in yellow, green, and pink, respectively; (c) Motif composition. Different motifs were indicated by different numbers in various color boxes.
It was reported that a typical ARF protein contained DBD, MR, and CTD domains. The results showed that 10 of 17 BpeARF proteins had these three typical domains (Figure 4b). According to the phylogenetic tree of BpeARFs, BpeARF5 in Class I contained a repressor MR and a CTD, but no DBD. BpeARF2, -3, -9, -10, -12, and -13 in Class II each had a DBD, an activator MR, and a CTD. BpeARF4, -6, -14, and -17 in Class III each had a DBD, a repressor MR, and a CTD. BpeARF1, -7, -8, -11, -15, and -16 in Class IV each contained a DBD, a repressor MR, but no CTD. It was suggested the BpeARF activator and repressor MRs unevenly distributed in different classes, with Class II containing activators, and Class I, III, and IV comprising repressors.
To explore the functional diversity of BpeARF proteins, motif locations in these proteins was performed using MEME program (Figure 4c). In the present work, a total of ten motifs was identified, and Motif 3, -7, and -8 were found in all the 17 BpeARF proteins. By comparing with the conserved domains, the results showed that Motif 1, -2, and -5 belong to DBD, and Motif 3, -7, and -8 correspond to MR. Furthermore, the BpeARF5 in Class I lacked Motif 1, -2, and -5 which was conserved in DBD, and all members in Class IV lacked Motif 9 except BpeARF8, while the members in Class II and III possessed all the motifs. The similar motif composition in a class from the phylogenetic tree, supported the reliability of BpeARF proteins classification.
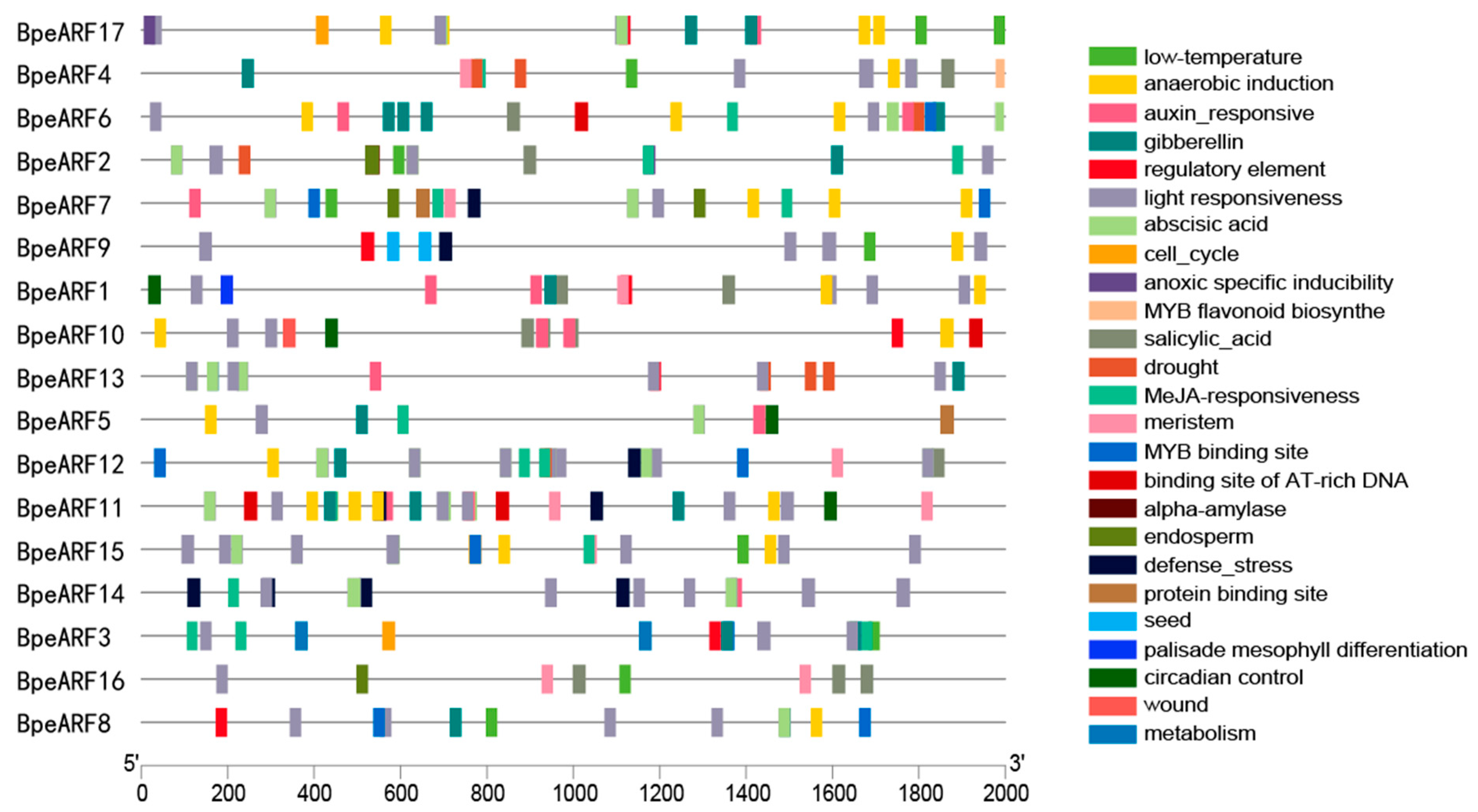
3.5. Cis-Acting Element in Promoter Region of BpeARF Genes
The gene expression is regulated by transcription factors through binding to the specific cis-acting elements. To identify the cis-acting elements of BpeARFs, the 2000 bp upstream sequence of each BpeARF gene was analyzed via the PlantCARE database (Figure 5). The results indicated that a total of 320 cis-acting elements was contained in 17 BpeARF genes, and most cis-acting elements were related to environmental factors and plant hormones. As the most abundant element, light responsiveness was identified in all BpeARF genes. Moreover, the elements involved in low temperature, drought, and defense stress also existed in promoter regions, and only BpeARF10 contained wound-responsive elements. As elements related to plant hormones, auxin-, gibberellin-, abscisic acid-, salicylic acid-, and methyl jasmonate (MeJA)-responsive elements were common in promoters, and all the BpeARF genes contained these elements except BpeARF9. A total of 17 elements corresponding to development was found in BpeARF promoters, which was involved in meristem, endosperm, seed, and palisade mesophyll differentiation. Additionally, the elements responding to binding site of AT-rich DNA, protein, and MYB transcription factor were also found in BpeARF3, -5, -6, -7, -8, -9, -10, -11, -12, and -15 promoter regions. In a word, the diversity of cis-acting elements suggested that BpeARF genes might participate in some biological processes, such as tissue development, hormone response, and defense stress.
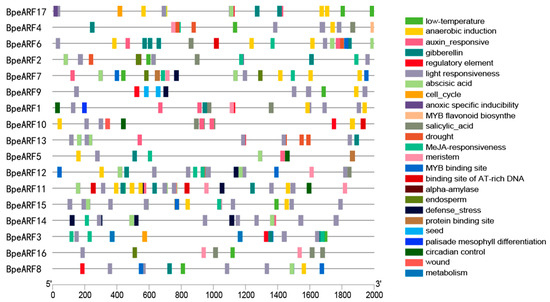
Figure 5.
The cis-acting elements in the promoter regions of BpeARF genes in B. pendula. Different cis-acting elements were indicated by various diagrams.
3.6. Expression Patterns of BpeARF Genes in Plant Tissues
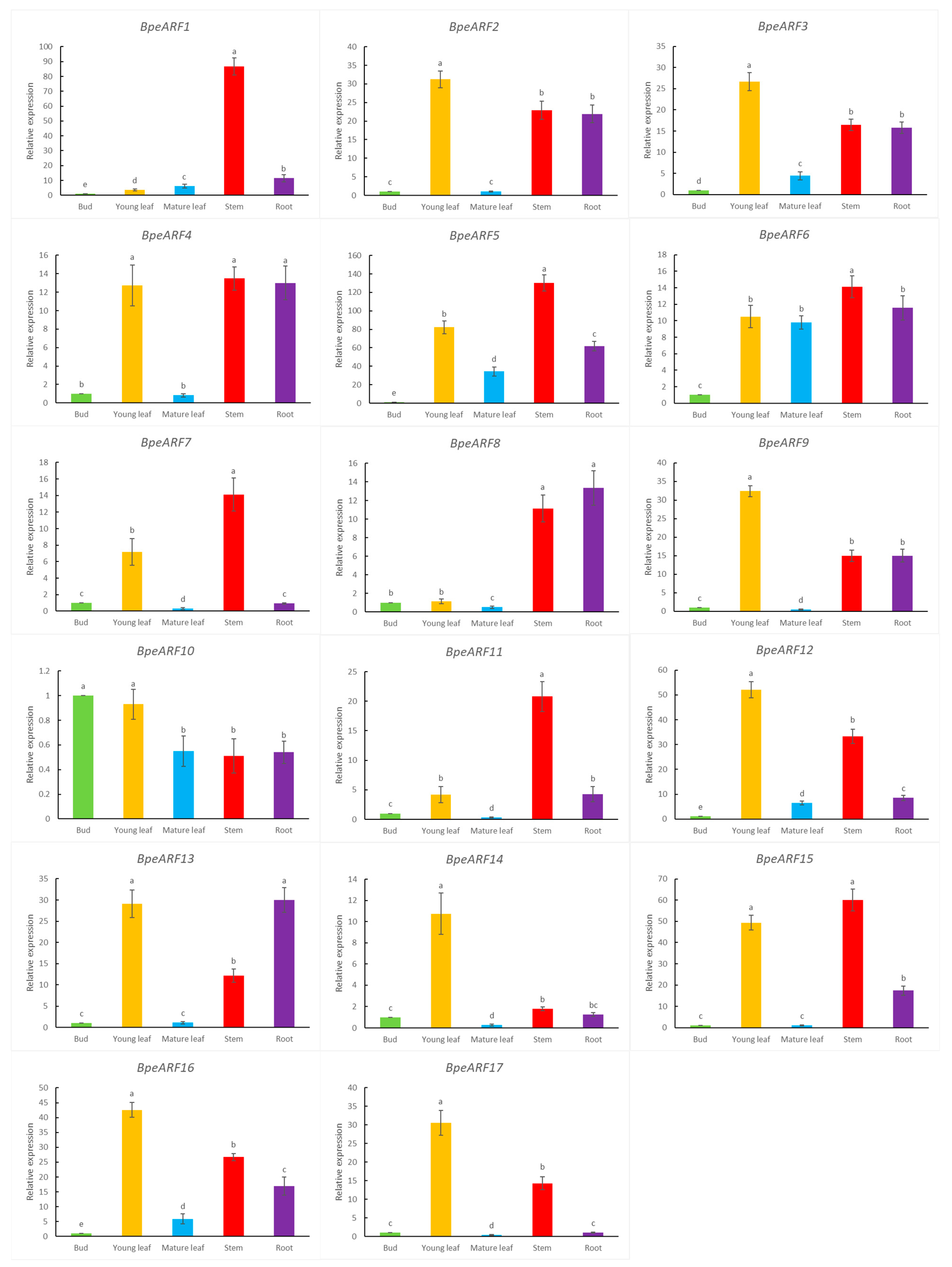
In order to explore the putative functions of BpeARF genes, qRT-PCR was employed to detect the expression levels of BpeARF genes in buds, young leaves, mature leaves, stems, and roots (Figure 6). The buds contain a large number of meristem cells, which eventually develop into various tissues. Therefore, the expression level of buds is set to 1 for normalize the expression level in other tissues. The results indicated that the 17 BpeARFs exhibited divergent expression patterns in different tissues, which showed that these BpeARFs genes performed distinct functions during birch growth and development. The expressions of BpeARF2, -3, -4, -9, -12, -13, -14, -15, -16, and -17 were the high levels in young leaves, implying these genes played an important role in forming leaves. Similar expression patterns were found in BpeARF1, -4, -5, -6, -7, -11, and -15, which had the highest level in stems, suggesting these genes might participate in early development of wood formation. The expression of BpeARF4, -8, and -13 was the most abundant in roots, indicating these genes might be involved in root formation. Interestingly, BpeARF10 exhibited a higher expression in buds compared to other tissues, and most BpeARF genes had a relatively low level in mature leaves except BpeARF6 and BpeARF10.
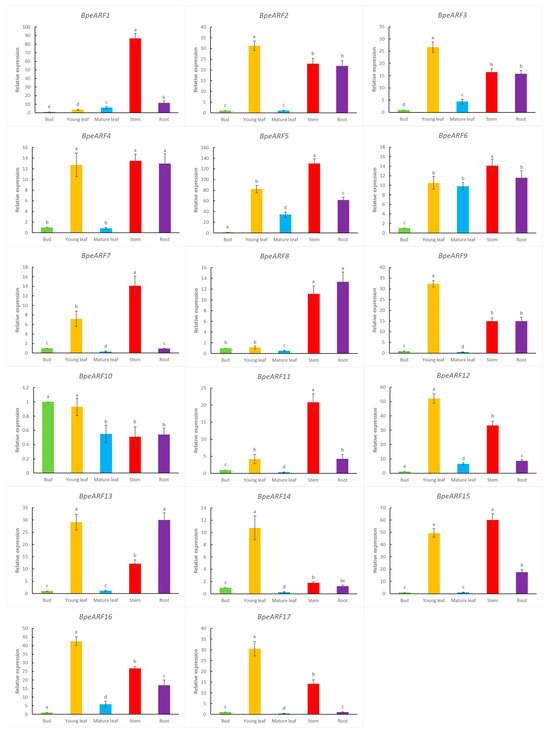
Figure 6.
Expression patterns of BpeARF genes in different tissues of B. pendula. The comparison of means was analyzed by Duncan test. Different lower-case letters indicated significantly differences at 0.05 levels. The error bar meant standard deviation.
3.7. Expression Patterns of BpeARF Genes with Exogenous NAA Treatment
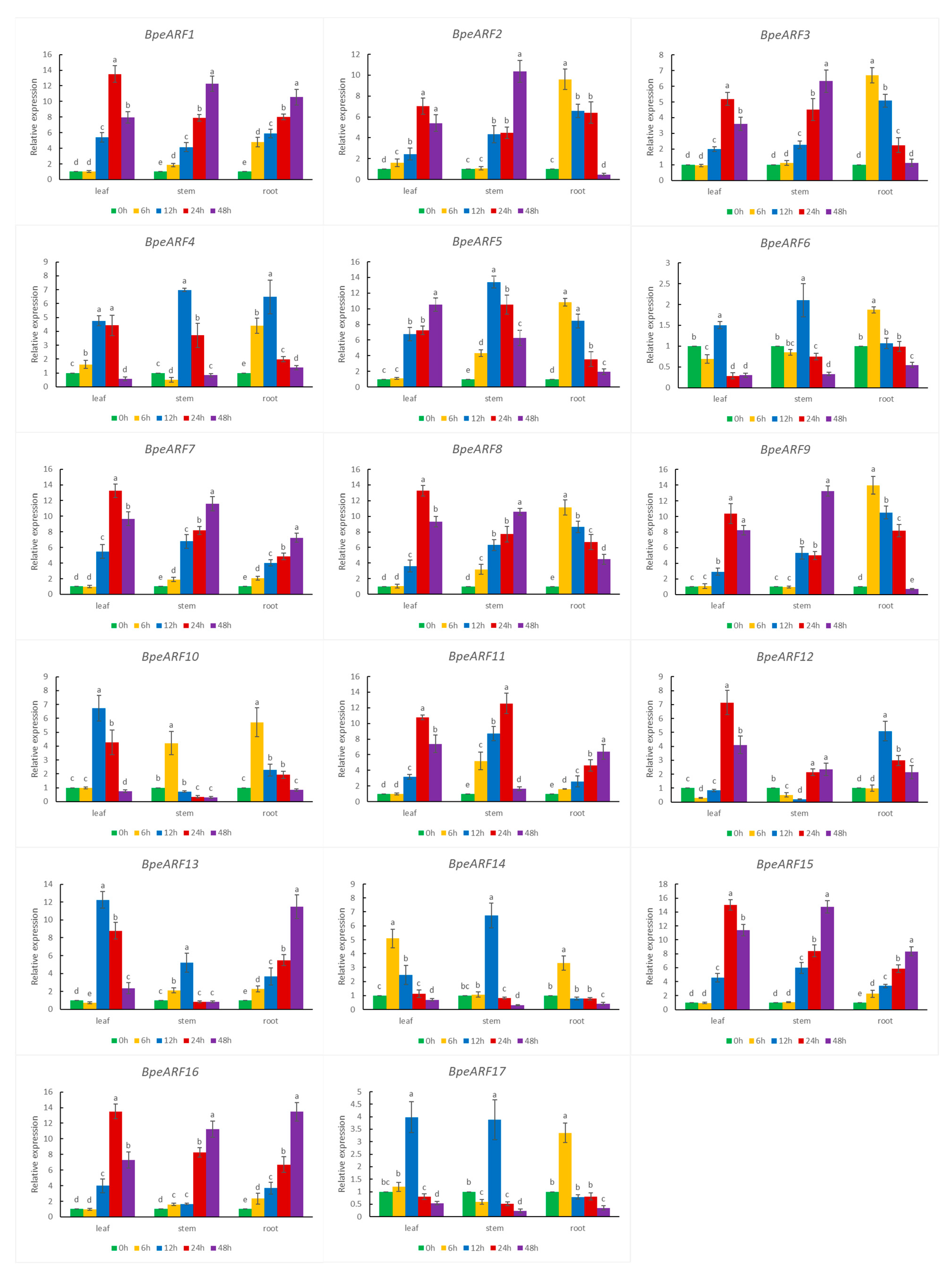
As an important regulator of the auxin signaling pathway, ARF play a key role in auxin response. Under exogenous NAA treatment, the 17 BpeARF genes exhibited different expression patterns among leaves, stems, and roots (Figure 7). In leaves, five BpeARF genes (BpeARF4, -6, -10, -13, and -17) and ten BpeARF genes (BpeARF1, -2, -3, -7, -8, -9, -11, -12, -15, and -16) were the highest expression level at 12 h and 24 h, respectively. In stems, the expression of six BpeARF genes (BpeARF4, -5, -6, -13, -14, and -17) peaked at 12 h and nine BpeARF genes (BpeARF1, -2, -3, -7, -8, -9, -12, -15, and -16) peaked at 48 h. In roots, the expressions of eight BpeARF genes (BpeARF2, -3, -5, -6, -8, -9, -10, and -14) and six BpeARF genes (BpeARF1, -7, -11, -13, -15, and -16) were maximum at 6 h and 48 h, respectively. Additionally, BpeARF1, -5, -7, -8, -11, and -16 in stems and roots were upregulated expression at all timepoints. BpeARF14 was highest expression at 6 h in leaves, and BpeARF10 and BpeARF11 in stems had highest level at 6 h and 24 h, respectively. These results suggested that BpeARF genes might have different roles in auxin signal transduction among different tissues in birch.
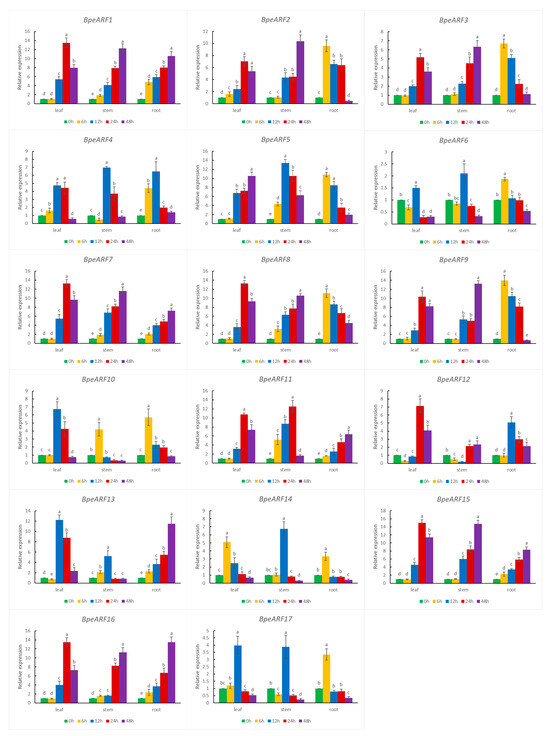
Figure 7.
Expression patterns of BpeARF genes with exogenous NAA treatment in B. pendula. The comparison of means was analyzed by Duncan test. Different lower-case letters indicated significantly differences at 0.05 levels. The error bar meant standard deviation.
3.8. Expression Patterns of BpeARF Genes under Drought Stress
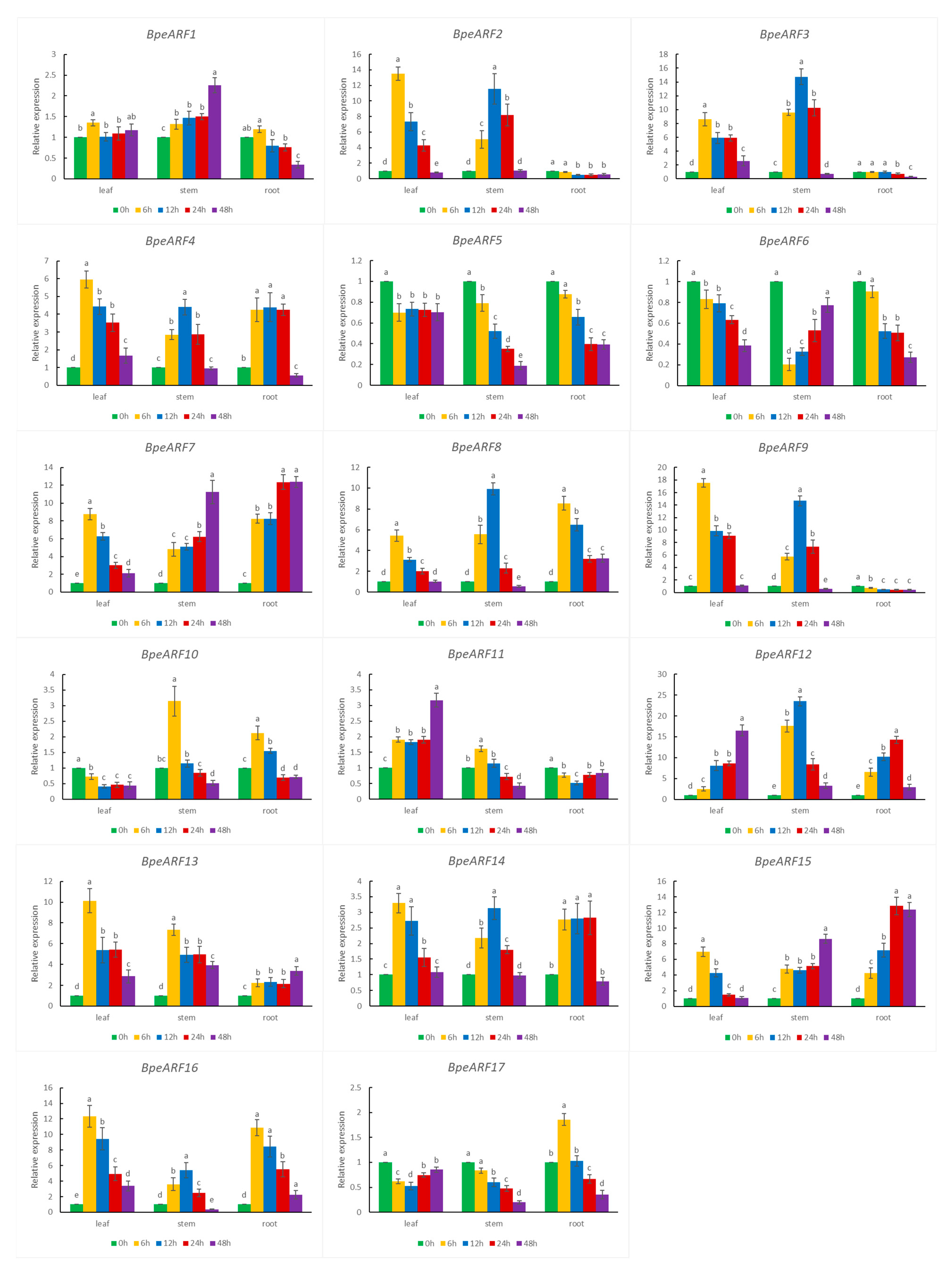
According to cis-acting element data, all the BpeARF genes contained at least one cis-element related to abiotic stress in their promoters. The expression levels of 17 BpeARF genes under drought stress were investigated by qRT-PCR (Figure 8). The results showed that 17 BpeARF genes expressions were highly diverse in different tissues. For instance, most of BpeARF genes (BpeARF1, -2, -3, -4, -7, -8, -9, -13, -14, -15, and -16) exhibited the highest expression at 6 h in leaves, and the expression levels of eight BpeARF genes (BpeARF2, -3, -4, -8, -9, -12, -14, and -16) peaked at 12 h in stems. BpeARF7 and BpeARF15 were upregulated in stems and roots. BpeARF5 and BpeARF6 were downregulated in leaves, stems, and roots, and BpeARF17 was downregulated in leaves and stems. The qRT-PCR results demonstrated that BpeARF genes played different roles in response to drought stress.
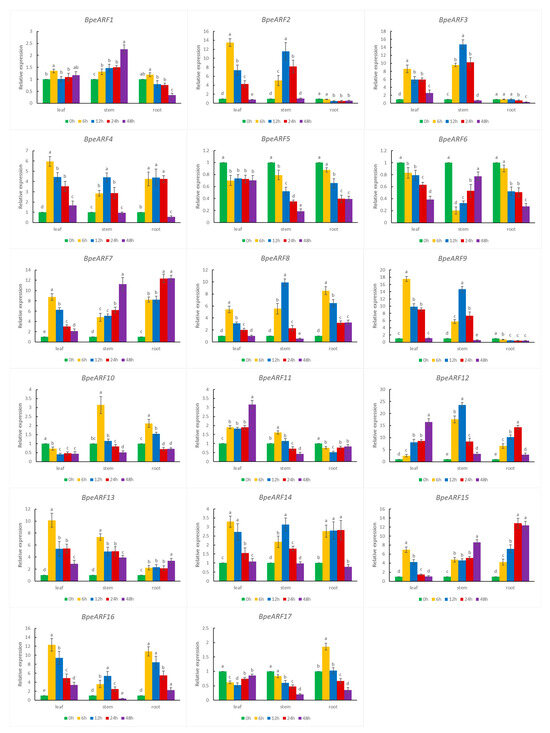
Figure 8.
Expression patterns of BpeARF genes under drought stress in B. pendula. The comparison of means was analyzed by Duncan test. Different lower-case letters indicated significantly differences at 0.05 levels. The error bar meant standard deviation.
4. Discussion
4.1. Duplication Events of BpeARF Gene Family
It is known that ARFs were important transcriptional regulators involved in auxin signaling transduction. In the past few decades, ARF gene families have been identified in some model plants, crops, fruits, and trees which have been sequenced genomes [47,48,49,50]. In this study, 17 BpeARF genes were identified in B. pendula genome, suggesting that the number of BpeARF genes was the same with Eucalyptus grandis [50], Prunus persica [51], and Dimocarpus longan [52]. The genome sizes of P. persica, B. pendula, D. longan, and E. grandis were 265 Mb, 440 Mb, 455 Mb, and 640 Mb, respectively [28,53,54,55], indicating that the genome size was not correlated with the number of ARF genes in these plants. Although the number of BpeARF genes was fewer compared to A. thaliana and P. trichocarpa, gene duplication events still occurred in BpeARF gene family. Tandem duplication played an important role in the expansion of ARF genes in some plants [11,56], and chromosomal regions within the 200 kb range of two or more genes are defined as tandem duplication events [57]. In B. pendula, BpeARF2 and BpeARF3, BpeARF6 and BpeARF7, BpeARF8 and BpeARF9, and BpeARF15 and BpeARF16 were located in Chromosome 2, -5, -6, and -13, respectively, and these genes were separated by at least several megabases in the same chromosome. Therefore, no tandem duplication was identified among the BpeARF gene pairs. Collinearity analysis revealed that three BpeARF pairs were formed by segmental duplication, and the Ka/Ks values of these gene pairs were less than 1, demonstrating that these BpeARF genes undergone negative purifying selection to maintain their function [58]. Similar results were also shown in Cicer arietinum [59] and Fagopyrum tataricum [38].
4.2. Phylogenetic Relationship and Diversity of BpeARF Gene Family
The phylogenetic tree was constructed to analyze the relationship of ARF families between B. pendula, B. platyphylla, P. trichocarpa, and A. thaliana. The BpeARF genes were divided into four classes, which was similar to the ARFs classification in Musa acuminata [60], Ananas comosus [61], and Corylus heterophylla [62]. There were 79 ARF gene pairs were identified between B. pendula and other plant species, and the Ka/Ks values of most ARF gene pairs were less than 1, suggesting these BpeARFs were highly homologous and conservative to BpARFs, PtrARFs, and AtARFs.
The BpeARF proteins were ranged in size from 65.209 kDa to 127.009 kDa and were defined by their intron/exon structure, conserved domains and motif composition. The exon numbers of BpeARF genes varied from 2 to 15, which was the same with BpARF genes in B. platyphylla, suggesting ARF genes might have a similar structure in birch [25]. A total of ten motifs was identified in BpeARF proteins, was fewer compared to P. trichocarpa (20) [24] and O. fragrans (12) [26]. Among the 17 BpeARF proteins, BpeARF5 in Class I lacked DBD domain correspond to Motif 1, -2, and -5, indicating it could not recognize the auxin response elements on the promoters of the target genes [63]. The DBD-truncated ARF proteins were also observed in other plant species, such as StARF18c in Solanum tuberosum [64], and PavARF3 in Prunus avium [65]. BpeARF proteins lacking CTD domain in Class IV, should be regulated other transcription factors instead of Aux/IAA [66]. The MR domain was present in all the BpeARF proteins, which existing in Class II was an activator, and in Class I, III, and IV was a repressor. The value of activator/repressor in BpeARF proteins was 0.55, which was higher than that in P. trichocarpa (0.52) [24] and E. grandis (0.42) [50], and lower than that in A. thaliana (0.59) [47] and O. sativa (0.56) [12]. The mechanism of activator/repressor value in plant species is still unclear [24,52]. The protein domain analysis could provide useful information for speculating the putative biological functions of BpeARF genes.
4.3. Expression Characteristics of BpeARF Gene Family
Cis-acting element analysis revealed that BpeARF promoters contained a great number of cis-acting elements associated with tissue development, hormone response, and abiotic stress, suggesting that BpeARF genes might be involved in these biological processes. The 17 BpeARF genes had diverse expression profiles in different tissues. The buds contained a large number of meristem cells, and various tissues were formed by differentiation from meristem cells. BpeARF10 exhibited a lower expression in roots, stems, and leaves compared to buds, indicating BpeARF10 might play a negative regulation on tissue differentiation. BpeARF2, -3, -4, -9, -12, and -13 showed high expression in young leaves, suggesting these genes participated in leaf development like AtARF5, -7, and -19 which were clustered into Class II [67,68]. Compared to young leaves, most BpeARF genes were downregulated expression in mature leaves, demonstrating BpeARF genes might play roles in leaf maturity and senescence. This result was identical to PpARF16 and PpARF17 in P. persica [51]. BpeARF1, -6, -7, -11, and -15 were high expression in stems, illustrating these genes were involved in wood formation, and the result was similar to the function of their homologous genes in P. trichocarpa, such as PtrARF6, -10, -18, -20, -22, and -32 [24]. AtARF7 and AtARF19 induced adventitious root formation by directly regulating the auxin-mediated transcription of LBD/ASL genes in roots [67,69]. Our synteny analysis showed that BpeARF8 was homologous to AtARF7 and AtARF19, and had high expression level in roots, implying BpeARF8 might have some potential functions during root formation. These results demonstrated that BpeARF genes were greatly involved in birch growth and development.
It is significant to determine the response of BpeARF genes to exogenous auxin treatment because the ARF activity is regulated by auxin concentration in plants [70]. In Nicotiana tabacum, the expression patterns of NtARF genes could be divided into two groups with exogenous NAA treatment. The first group comprised six NtARF genes (NtARF1, -10, -14, -26b, -45 and -46), whose expression levels decreased at 1 h and 2 h, and increased at 5 h and 12 h. The second group comprised four NtARF genes (NtARF6, -13, -23 and -29), whose expression gradually increased from 1 h to 12 h, and decreased at 24 h and 48 h [71]. In the present study, all the BpeARF genes was responsive to exogenous NAA treatment and exhibited diverse expression patterns in different tissues. The expressions of three BpeARF genes (BpeARF1, -7, and -8) in stems and four BpeARF genes (BpeARF1, -7, -11, and -13) in roots gradually increased with the prolongation of treatment time, suggesting their expressions were positively regulated by NAA, consistent with their homologs (PtrARF4, -18, -23, -30, -32, and -33) in P. trichocarpa [24]. In contrast, the expression of BpeARF14 in leaves, BpeARF10 in stems, and nine BpeARF genes (BpeARF2, -3, -5, -6, -8, -9, -10, -14, and -17) in roots peaked at 6 h and then decreased at later time, demonstrating NAA induced these genes in a short period, as PvARF3, -4, -23, -24, -25, and -26 in Panicum virgatum, whose expression levels peaked at 1 h and decreased at 2 h and 3 h under NAA treatment [72]. In addition, the expression levels of the BpeARF genes were generally increased in response to exogenous NAA treatment, but the auxin-responsive elements were only detected in the promoters of nine BpeARF genes (BpeARF1, -5, -6, -7, -10, -11, -13, -14, and -17). This result was identical to ZmARF genes in Zea mays [48], and the underlying mechanism needs to be elucidated in future.
Previous researches have shown that ARF genes are widely participated in the regulation of plant tolerance to drought stress. In Cocos nucifera, six CnARF genes were significantly downregulated under mannitol and PEG treatment [73]. SlARF8A and SlARF10A were upregulated in response to drought stress in Solanum lycopersicum [74]. Our results suggested that the expressions of BpeARF genes were changed under drought stress. For example, eleven BpeARF genes (BpeARF1, -2, -3, -4, -7, -8, -9, -13, -14, -15, and -16) in leaves and five BpeARF genes (BpeARF1, -8, -10, -16, and -17) in roots exhibited the highest expression at 6 h, respectively, and the expressions of eight BpeARF genes (BpeARF2, -3, -4, -8, -9, -12, -14, and -16) peaked at 12 h in stems. BpeARF5 and BpeARF6 were downregulated in leaves, stems, and roots. In particular, BpeARF11 and BpeARF12 in leaves, together with BpeARF7 and BpeARF15 in stems and roots, showed increasing expression levels with the prolongation of PEG treatment, as their homologous genes in B. platyphylla, such as BpARF1 and BpARF4 [25]. Conversely, BpeARF5 was continuously downregulated in leaves, stems, and roots, similar to CsARF4, -11, and -19 in Camellia sinensis and EgARF1, -14, -17, and -20 in Elaeis guineensis were inhibited under drought condition [27,75]. These genes could be considered as candidate genes for breeding drought-resistant birch.
5. Conclusions
In the current study, 17 BpeARF genes were identified from B. pendula genome, which were unevenly distributed on 13 chromosomes. Among them, three BpeARF gene pairs were formed by segmental duplication, and undergone negative purifying selection to maintain their function. The BpeARF genes could be divided into four classes using phylogenetic analysis, and the intron/exon structure, conserved domain, and motif composition were similar among the BpeARF genes within the same class. The cis-acting elements in the promoter regions of BpeARF genes were related to tissue development, hormone response, and stress resistance. The expression data revealed BpeARF genes involved in birch growth and development, and responded to exogenous auxin treatment and drought stress. The expressions of one (BpeARF10), ten (BpeARF2, -3, -4, -9, -12, -13, -14, -15, -16, and -17), seven (BpeARF1, -4, -5, -6, -7, -11, and -15), and three BpeARF genes (BpeARF4, -8, and -13) were the high levels in buds, young leaves, stems, and roots, respectively. Under exogenous NAA treatment, six BpeARF genes in stems and roots were upregulated expression at all timepoints. Under drought stress, BpeARF7 and BpeARF15 were upregulated in stems and roots, and BpeARF5 and BpeARF6 were downregulated in leaves, stems, and roots. This study provided valuable information for the classification and putative functions of BpeARF gene family. Further research in the selection and functional verification of BpeARF genes should be performed based on the present study, which may be helpful for the genetic engineering of birch trees.
Author Contributions
Conceptualization, H.M. and L.L.; Formal analysis, X.J., S.L. (Songtong Lv) and S.L. (Sheng Long); Funding acquisition, H.M. and L.L.; Investigation, Y.L. and L.C.; Methodology, H.M.; Resources, L.L.; Supervision, L.L.; Writing—original draft, X.J. and S.L. (Songtong Lv); Writing—review and editing, H.M. and L.L. All authors have read and agreed to the published version of the manuscript.
Funding
This work was financially supported by a grant from the National Natural Science Foundation of China (32001333), the Central Finance Forestry Science and Technology Promotion Demonstration Program of China (JLT2021-35), and the National Key Research and Development Program of China (2023YFF1304004-05).
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Acknowledgments
We thank Xinshan Nursery Stock Cooperative for support related to the plant material and greenhouse facilities.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Koski, V.; Rousi, M. A review of the promises and constraints of breeding silver birch (Betula pendula Roth) in Finland. Forestry 2005, 78, 187–198. [Google Scholar] [CrossRef]
- Gomes, G.L.B.; Scortecci, K.C. Auxin and its role in plant development: Structure, signalling, regulation and response mechanisms. Plant Biol. 2021, 23, 894–904. [Google Scholar] [CrossRef] [PubMed]
- Gu, C.R.; Han, R.; Liu, C.Y.; Fang, G.G.; Yuan, Q.H.; Zheng, Z.M.; Yu, Q.B.; Jiang, J.; Liu, S.Z.; Xie, L.N.; et al. Heritable epigenetic modification of BpPIN1 is associated with leaf shapes in Betula pendula. Tree Physiol. 2023, 43, 1811–1824. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.; Chen, S.; Wang, S.; Liu, G.F.; Li, H.Y.; Huang, H.J.; Jiang, J. BpGH3.5, an early auxin-response gene, regulates root elongation in Betula platyphylla × Betula pendula. Plant Cell Tissue Organ Cult. 2015, 120, 239–250. [Google Scholar] [CrossRef]
- Yu, Z.P.; Zhang, F.; Friml, J.; Ding, Z.J. Auxin signaling: Research advances over the past 30 years. J. Integr. Plant Biol. 2022, 64, 371–392. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Zhou, J.J.; Zhang, J.Z. Aux/IAA gene family in plants: Molecular structure, regulation, and function. Int. J. Mol. Sci. 2018, 19, 259. [Google Scholar] [CrossRef] [PubMed]
- Kou, X.H.; Zhao, X.Y.; Wu, B.D.; Wang, C.; Wu, C.E.; Yang, S.; Zhou, J.Q.; Xue, Z.H. Auxin response factors are ubiquitous in plant growth and development, and involved in crosstalk between plant hormones: A review. Appl. Sci. 2022, 12, 1360. [Google Scholar] [CrossRef]
- Mironova, V.V.; Omelyanchuk, N.A.; Wiebe, D.S.; Levitsky, V.G. Computational analysis of auxin responsive elements in the Arabidopsis thaliana L. genome. BMC Genom. 2014, 15, S4. [Google Scholar] [CrossRef]
- Li, S.B.; Xie, Z.Z.; Hu, C.G.; Zhang, J.Z. A review of auxin response factors (ARFs) in plants. Front. Plant Sci. 2016, 7, 47. [Google Scholar] [CrossRef]
- Cancé, C.; Martin-Arevalillo, R.; Boubekeur, K.; Dumas, R. Auxin response factors are keys to the many auxin doors. New Phytol. 2022, 235, 402–419. [Google Scholar] [CrossRef]
- Tombuloglu, H. Genome-wide analysis of the auxin response factors (ARF) gene family in barley (Hordeum vulgare L.). J. Plant Biochem. Biot. 2019, 28, 14–24. [Google Scholar] [CrossRef]
- Wang, D.K.; Pei, K.M.; Fu, Y.P.; Sun, Z.X.; Li, S.J.; Liu, H.Q.; Tang, K.; Han, B.; Tao, Y.Z. Genome-wide analysis of the auxin response factors (ARF) gene family in rice (Oryza sativa). Gene 2007, 394, 13–24. [Google Scholar] [CrossRef]
- Liu, Z.N.; Miao, L.M.; Huo, R.X.; Song, X.Y.; Johnson, C.; Kong, L.J.; Sundaresan, V.; Yu, X.L. ARF2–ARF4 and ARF5 are essential for female and male gametophyte development in Arabidopsis. Plant Cell Physiol. 2018, 59, 179–189. [Google Scholar] [CrossRef]
- Schruff, M.C.; Spielman, M.; Tiwari, S.; Adams, S.; Fenby, N.; Scott, R.J. The AUXIN RESPONSE FACTOR 2 gene of Arabidopsis links auxin signalling, cell division, and the size of seeds and other organs. Development 2006, 133, 251–261. [Google Scholar] [CrossRef]
- Nagpal, P.; Ellis, C.M.; Weber, H.; Ploense, S.E.; Barkawi, L.S.; Guilfoyle, T.J.; Hagen, G.; Alonso, J.M.; Cohen, J.D.; Farmer, E.E.; et al. Auxin response factors ARF6 and ARF8 promote jasmonic acid production and flower maturation. Development 2005, 132, 4107–4118. [Google Scholar] [CrossRef]
- Reed, J.W.; Wu, M.F.; Reeves, P.H.; Hodgens, C.; Yadav, V.; Hayes, S.; Pierik, R. Three auxin response factors promote hypocotyl elongation. Plant Physiol. 2018, 178, 864–875. [Google Scholar] [CrossRef]
- Xiao, G.H.; He, P.; Zhao, P.; Liu, H.; Zhang, L.; Pang, C.Y.; Yu, J.N. Genome-wide identification of the GhARF gene family reveals that GhARF2 and GhARF18 are involved in cotton fibre cell initiation. J. Exp. Bot. 2018, 69, 4323–4337. [Google Scholar] [CrossRef]
- de Jong, M.; Wolters-Arts, M.; García-Martínez, J.L.; Mariani, C.; Vriezen, W.H. The Solanum lycopersicum AUXIN RESPONSE FACTOR 7 (SlARF7) mediates cross-talk between auxin and gibberellin signalling during tomato fruit set and development. J. Exp. Bot. 2011, 62, 617–626. [Google Scholar] [CrossRef] [PubMed]
- Sagar, M.; Chervin, C.; Mila, I.; Hao, Y.W.; Roustan, J.P.; Benichou, M.; Gibon, Y.; Biais, B.; Maury, P.; Latché, A.; et al. SlARF4, an auxin response factor involved in the control of sugar metabolism during tomato fruit development. Plant Physiol. 2013, 161, 1362–1374. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.J.; Mei, L.H.; Wu, M.B.; Wei, W.; Shan, W.; Gong, Z.H.; Zhang, Q.; Yang, F.Q.; Yan, F.; Zhang, Q.; et al. SlARF10, an auxin response factor, is involved in chlorophyll and sugar accumulation during tomato fruit development. J. Exp. Bot. 2018, 69, 5507–5518. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.N.; Dong, L.W.; Deng, X.; Liu, D.M.; Liu, Y.; Li, M.F.; Hu, Y.K.; Yan, Y.M. Genome-wide identification, molecular evolution, and expression analysis of auxin response factor (ARF) gene family in Brachypodium distachyon L. BMC Plant Biol. 2018, 18, 336. [Google Scholar] [CrossRef]
- Tang, Y.Y.; Du, G.N.; Xiang, J.; Hu, C.L.; Li, X.T.; Wang, W.H.; Zhu, H.; Qiao, L.X.; Zhao, C.M.; Wang, J.S.; et al. Genome-wide identification of auxin response factor (ARF) gene family and the miR160-ARF18-mediated response to salt stress in peanut (Arachis hypogaea L.). Genomics 2022, 114, 171–184. [Google Scholar] [CrossRef]
- Pei, Q.Y.; Li, N.; Yang, Q.H.; Wu, T.; Feng, S.Y.; Feng, X.H.; Jing, Z.G.; Zhou, R.; Gong, K.; Yu, T.; et al. Genome-wide identification and comparative analysis of ARF family genes in three Apiaceae species. Front. Genet. 2021, 11, 590535. [Google Scholar] [CrossRef]
- Liu, Y.Y.; Wang, R.Q.; Yu, J.J.; Huang, S.; Zhang, Y.; Wei, H.R.; Wei, Z.G. Genome-wide identification and characterization of auxin response factor (ARF) gene family involved in wood formation and response to exogenous hormone treatment in Populus trichocarpa. Int. J. Mol. Sci. 2023, 24, 740. [Google Scholar] [CrossRef]
- Li, H.Y.; Zhang, X.; Tong, B.T.; Wang, Y.C.; Yang, C.P. Expression analysis of the BpARF genes in Betula platyphylla under drought stress. Plant Physiol. Bioch. 2020, 148, 273–281. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.W.; Yue, Y.Z.; Li, L.; Li, Y.L.; Li, H.Y.; Ding, W.J.; Shi, T.T.; Yang, X.L.; Wang, L.G. Genome-wide identification of the auxin response factor (ARF) gene family and their expression analysis during flower development of Osmanthus fragrans. Forests 2020, 11, 245. [Google Scholar] [CrossRef]
- Jin, L.F.; Yarra, R.; Zhou, L.X.; Cao, H.X. The auxin response factor (ARF) gene family in Oil palm (Elaeis guineensis Jacq.): Genome-wide identification and their expression profiling under abiotic stresses. Protoplasma 2022, 259, 47–60. [Google Scholar] [CrossRef]
- Salojärvi, J.; Smolander, O.P.; Nieminen, K.; Rajaraman, S.; Safronov, O.; Safdari, P.; Lamminmäki, A.; Immanen, J.; Lan, T.Y.; Tanskanen, J.; et al. Genome sequencing and population genomic analyses provide insights into the adaptive landscape of silver birch. Nat. Genet. 2017, 49, 904–912. [Google Scholar] [CrossRef]
- Ye, J.; McGinnis, S.; Madden, T.L. BLAST: Improvements for better sequence analysis. Nucleic Acids Res. 2006, 34, W6–W9. [Google Scholar] [CrossRef] [PubMed]
- Eddy, S.R. Accelerated profile HMM searches. PLoS Comput. Biol. 2011, 7, e1002195. [Google Scholar] [CrossRef] [PubMed]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A.; et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2019, 47, D427–D432. [Google Scholar] [CrossRef]
- Yang, M.Z.; Derbyshire, M.K.; Yamashita, R.A.; Marchler-Bauer, A. NCBI’s conserved domain database and tools for protein domain analysis. Curr. Protoc. Bioinform. 2020, 69, e90. [Google Scholar] [CrossRef]
- Gasteiger, E.; Gattiker, A.; Hoogland, C.; Ivanyi, I.; Appel, R.D.; Bairoch, A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003, 31, 3784–3788. [Google Scholar] [CrossRef]
- Hu, B.; Jin, J.P.; Guo, A.Y.; Zhang, H.; Luo, J.C.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef]
- Horton, P.; Park, K.J.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.J.; Nakai, K. WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 2007, 35, W585–W587. [Google Scholar] [CrossRef]
- Chao, J.T.; Li, Z.Y.; Sun, Y.H.; Aluko, O.O.; Wu, X.R.; Wang, Q.; Liu, G.S. MG2C: A user-friendly online tool for drawing genetic maps. Mol. Hortic. 2021, 1, 16. [Google Scholar] [CrossRef]
- Wang, Y.P.; Tang, H.B.; DeBarry, J.D.; Tan, X.; Li, J.P.; Wang, X.Y.; Lee, T.H.; Jin, H.Z.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Liu, M.Y.; Ma, Z.T.; Wang, A.H.; Zheng, T.R.; Huang, L.; Sun, W.J.; Zhang, Y.J.; Jin, W.Q.; Zhan, J.Y.; Cai, Y.T.; et al. Genome-wide investigation of the auxin response factor gene family in tartary buckwheat (Fagopyrum tataricum). Int. J. Mol. Sci. 2018, 19, 3256. [Google Scholar] [CrossRef]
- Zhang, Z. KaKs_Calculator 3.0: Calculating selective pressure on coding and non-coding sequences. Genom. Proteom. Bioinf. 2022, 20, 536–540. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Chen, C.J.; Wu, Y.; Xia, R. A painless way to customize Circos plot: From data preparation to visualization using TBtools. iMeta 2022, 1, e35. [Google Scholar] [CrossRef]
- Mu, H.Z.; Lin, L.; Zhang, Q.Y.; Tang, X.J.; Zhang, X.; Cheng, G.Y. Growth, proline content and proline-associated gene expression of autotetraploid Betula platyphylla responding to NaHCO3 stress. Dendrobiology 2016, 75, 123–129. [Google Scholar] [CrossRef][Green Version]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
- Okushima, Y.; Overvoorde, P.J.; Arima, K.; Alonso, J.M.; Chan, A.; Chang, C.; Ecker, J.R.; Hughes, B.; Lui, A.; Nguyen, D.; et al. Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana: Unique and overlapping functions of ARF7 and ARF19. Plant Cell 2005, 17, 444–463. [Google Scholar] [CrossRef]
- Xing, H.Y.; Pudake, R.N.; Guo, G.G.; Xing, G.F.; Hu, Z.R.; Zhang, Y.R.; Sun, Q.X.; Ni, Z.F. Genome-wide identification and expression profiling of auxin response factor (ARF) gene family in maize. BMC Genom. 2011, 12, 178. [Google Scholar] [CrossRef]
- Niu, J.; Bi, Q.X.; Deng, S.Y.; Chen, H.P.; Yu, H.Y.; Wang, L.B.; Lin, S.Z. Identification of AUXIN RESPONSE FACTOR gene family from Prunus sibirica and its expression analysis during mesocarp and kernel development. BMC Plant Biol. 2018, 18, 21. [Google Scholar] [CrossRef]
- Yu, H.; Soler, M.; Mila, I.; Clemente, H.S.; Savelli, B.; Dunand, C.; Paiva, J.A.P.; Myburg, A.A.; Bouzayen, M.; Grima-Pettenati, J.; et al. Genome-wide characterization and expression profiling of the AUXIN RESPONSE FACTOR (ARF) gene family in Eucalyptus grandis. PLoS ONE 2014, 9, e108906. [Google Scholar] [CrossRef]
- Diao, D.H.; Hu, X.; Guan, D.; Wang, W.; Yang, H.Q.; Liu, Y.P. Genome-wide identification of the ARF (auxin response factor) gene family in peach and their expression analysis. Mol. Biol. Rep. 2020, 47, 4331–4344. [Google Scholar] [CrossRef]
- Peng, Y.; Fang, T.; Zhang, Y.Y.; Zhang, M.Y.; Zeng, L.H. Genome-wide identification and expression analysis of auxin response factor (ARF) gene family in longan (Dimocarpus longan L.). Plants-Basel 2020, 9, 221. [Google Scholar] [CrossRef]
- Verde, I.; Abbott, A.G.; Scalabrin, S.; Jung, S.; Shu, S.Q.; Marroni, F.; Zhebentyayeva, T.; Dettori, M.T.; Grimwood, J.; Cattonaro, F.; et al. The high-quality draft genome of peach (Prunus persica) identifies unique patterns of genetic diversity, domestication and genome evolution. Nat. Genet. 2013, 45, 487–494. [Google Scholar] [CrossRef]
- Wang, J.; Li, J.G.; Li, Z.R.; Liu, B.; Zhang, L.L.; Guo, D.L.; Huang, S.L.; Qian, W.Q.; Guo, L. Genomic insights into longan evolution from a chromosome-level genome assembly and population genomics of longan accessions. Hortic. Res. 2022, 9, uhac021. [Google Scholar] [CrossRef]
- Myburg, A.A.; Grattapaglia, D.; Tuskan, G.A.; Hellsten, U.; Hayes, R.D.; Grimwood, J.; Jenkins, J.; Lindquist, E.; Tice, H.; Bauer, D.; et al. The genome of Eucalyptus grandis. Nature 2014, 510, 356–362. [Google Scholar] [CrossRef]
- Zong, Y.; Gu, L.L.; Shen, Z.L.; Kang, H.T.; Li, Y.Q.; Liao, F.L.; Xu, L.S.; Guo, W.D. Genome-wide identification and bioinformatics analysis of auxin response factor genes in highbush blueberry. Horticulturae 2021, 7, 403. [Google Scholar] [CrossRef]
- Holub, E.B. The arms race is ancient history in Arabidopsis, the wildflower. Nat. Rev. Genet. 2001, 2, 516–527. [Google Scholar] [CrossRef]
- Li, J.; Zhang, Z.; Vang, S.; Yu, J.; Wong, G.K.S.; Wang, J. Correlation between Ka/Ks and Ks is related to substitution model and evolutionary lineage. J. Mol. Evol. 2009, 68, 414–423. [Google Scholar] [CrossRef]
- Die, J.V.; Gil, J.; Millan, T. Genome-wide identification of the auxin response factor gene family in Cicer arietinum. BMC Genom. 2018, 19, 301. [Google Scholar] [CrossRef]
- Hu, W.; Zuo, J.; Hou, X.W.; Yan, Y.; Wei, Y.X.; Liu, J.H.; Li, M.Y.; Xu, B.Y.; Jin, Z.Q. The auxin response factor gene family in banana: Genome-wide identification and expression analyses during development, ripening, and abiotic stress. Front. Plant Sci. 2015, 6, 742. [Google Scholar] [CrossRef]
- Su, Z.X.; Wang, L.L.; Li, W.M.; Zhao, L.H.; Huang, X.Y.; Azam, S.M.; Qin, Y. Genome-wide identification of auxin response factor (ARF) genes family and its tissue-specific prominent expression in pineapple (Ananas comosus). Trop. Plant Biol. 2017, 10, 86–96. [Google Scholar] [CrossRef]
- Wei, H.; Cheng, Y.Q.; Sun, Y.; Zhang, X.Z.; He, H.L.; Liu, J.F. Genome-wide identification of the ARF gene family and ARF3 target genes regulating ovary initiation in hazel via ChIP sequencing. Front. Plant Sci. 2021, 12, 715820. [Google Scholar] [CrossRef] [PubMed]
- Fontana, M.; Roosjen, M.; García, I.C.; van den Berg, W.; Malfois, M.; Boer, R.; Weijers, D.; Hohlbein, J. Cooperative action of separate interaction domains promotes high-affinity DNA binding of Arabidopsis thaliana ARF transcription factors. Proc. Natl. Acad. Sci. USA 2023, 120, e2219916120. [Google Scholar] [CrossRef] [PubMed]
- Song, S.W.; Hao, L.Y.; Zhao, P.; Xu, Y.; Zhong, N.Q.; Zhang, H.J.; Liu, N. Genome-wide identification, expression profiling and evolutionary analysis of auxin response factor gene family in potato (Solanum tuberosum Group Phureja). Sci. Rep. 2019, 9, 1755. [Google Scholar] [CrossRef] [PubMed]
- Hou, Q.D.; Qiu, Z.L.; Wen, Z.; Zhang, H.M.; Li, Z.C.; Hong, Y.; Qiao, G.; Wen, X.P. Genome-wide identification of ARF gene family suggests a functional expression pattern during fruitlet abscission in Prunus avium L. Int. J. Mol. Sci. 2021, 22, 11968. [Google Scholar] [CrossRef]
- Guilfoyle, T.J.; Hagen, G. Auxin response factors. Curr. Opin. Plant Biol. 2007, 10, 453–460. [Google Scholar] [CrossRef] [PubMed]
- Wilmoth, J.C.; Wang, S.C.; Tiwari, S.B.; Joshi, A.D.; Hagen, G.; Guilfoyle, T.J.; Alonso, J.M.; Ecker, J.R.; Reed, J.W. NPH4/ARF7 and ARF19 promote leaf expansion and auxin-induced lateral root formation. Plant J. 2005, 43, 118–130. [Google Scholar] [CrossRef]
- Krogan, N.T.; Ckurshumova, W.; Marcos, D.; Caragea, A.E.; Berleth, T. Deletion of MP/ARF5 domains III and IV reveals a requirement for Aux/IAA regulation in Arabidopsis leaf vascular patterning. New Phytol. 2012, 194, 391–401. [Google Scholar] [CrossRef]
- Okushima, Y.; Fukaki, H.; Onoda, M.; Theologis, A.; Tasaka, M. ARF7 and ARF19 regulate lateral root formation via direct activation of LBD/ASL genes in Arabidopsis. Plant Cell 2007, 19, 118–130. [Google Scholar] [CrossRef]
- Caumon, H.; Vernoux, T. A matter of time: Auxin signaling dynamics and the regulation of auxin responses during plant development. J. Exp. Bot. 2023, 74, 3887–3902. [Google Scholar] [CrossRef]
- Zhang, J.; Khan, R.; Zhou, L.; Wu, X.Y.; Xu, N.; Ma, X.H.; Zhang, Y. Genome-wide identification analysis of the auxin response factors family in Nicotiana tabacum and the function of NtARF10 in leaf size regulation. J. Plant Biol. 2021, 64, 281–297. [Google Scholar] [CrossRef]
- Wang, J.L.; Wu, Z.Y.; Shen, Z.B.; Bai, Z.T.; Zhong, P.; Ma, L.C.; Pan, D.F.; Zhang, R.B.; Li, D.M.; Zhang, H.L.; et al. Genome-wide identification, phylogeny, and expression analysis of ARF genes involved in vegetative organs development in switchgrass. Int. J. Genom. 2018, 2018, 7658910. [Google Scholar] [CrossRef] [PubMed]
- Santhi, C.K.V.; Rajesh, M.K.; Ramesh, S.V.; Muralikrishna, K.S.; Gangaraj, K.P.; Gupta, P.; Dash, P.K. Genome-wide exploration of auxin response factors (ARFs) and their expression dynamics in response to abiotic stresses and growth regulators in coconut (Cocos nucifera L.). Plant Gene 2021, 28, 100344. [Google Scholar]
- Bouzroud, S.; Gouiaa, S.; Hu, N.; Bernadac, A.; Mila, I.; Bendaou, N.; Smouni, A.; Bouzayen, M.; Zouine, M. Auxin response factors (ARFs) are potential mediators of auxin action in tomato response to biotic and abiotic stress (Solanum lycopersicum). PLoS ONE 2018, 13, e0193517. [Google Scholar] [CrossRef]
- Xu, Y.X.; Mao, J.; Chen, W.; Qian, T.T.; Liu, S.C.; Hao, W.J.; Li, C.F.; Chen, L. Identification and expression profiling of the auxin response factors (ARFs) in the tea plant (Camellia sinensis (L.) O. Kuntze) under various abiotic stresses. Plant Physiol. Biochem. 2016, 98, 46–56. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).