Abstract
The xyloglucan endotransglucosylase/hydrolase (XTH) family is an important multigene family in plants that plays a key role in cell wall reconstruction and stress tolerance. However, the specific traits of XTH genes and their expression patterns under different stresses have not been systematically studied in melon. In this study, based on the genomic data of Cucumis melon, 29 XTH genes were identified; most of these genes contain two conserved domains (Glyco_hydro_16 and XET_C domains). Based on neighbor-joining phylogenetic analysis, the CmXTHs were divided into four subfamilies, I/II, IIIA, and IIIB, which are distributed across nine chromosomes of melon. Collinearity analysis showed that the melon XTH genes have an evolutionary history consistent with three species: Arabidopsis, tomato, and cucumber. The promoter regions of the CmXTH genes contain numerous cis-acting elements, which are associated with plant growth, hormonal response, and stress responses. RNA-Seq analysis indicated that CmXTH genes exhibit different expression patterns under drought and salt stress treatments, suggesting that this gene family plays an important role under abiotic stress. This study provides a theoretical basis for further studies on the molecular function of XTH genes in melon.
1. Introduction
The cell wall serves as the primary barrier for plants against external stresses and also plays a crucial role in signal transduction [1]. The main components of plant cell walls are cellulose, hemicellulose, lignin, and pectin. Xyloglucan (XG) constitutes the largest proportion of the primary cell wall in dicotyledonous plants [2], and it regulates cell wall tension by cross-linking with cellulose, pectin polysaccharides, and stretch proteins, thus forming a load-bearing network of the primary cell wall [3]. Xyloglucan endotransglucosylase/hydrolase (XTH) regulates cell wall elasticity and ductility by catalyzing molecular grafting between xyloglucan chains or the hydrolysis of xyloglucan, thereby influencing plant growth and development [4]. XTHs are a multigene family that encodes proteins belonging to the GH16 subfamily (Glycoside hydrolase family 16) [5]. XTH enzymes exhibit two distinct catalytic activities: xyloglucan endotransglucosylase (XET) activity and xyloglucan endo-hydrolase (XEH) activity. Based on their protein structural features, XTH proteins are classified into three subfamilies, I, II, and III [6,7,8]. However, with further research, subfamily III was subdivided into two classes: IIIA and IIIB. Classes I and II, which exhibit similar structures and functions, possess both XEH and XET enzymatic activities. In contrast, class IIIA exhibits only XEH activity, while class IIIB displays a more pronounced xyloglucan endotransglucosylase (XET) activity [9]. Currently, XTHs have been identified in a variety of plants, including Arabidopsis (Arabidopsis thaliana, 33), rice (Oryza sativa L., 29), poplar (Populus simonii × Populus nigra, 41), wheat (Triticum aestivum L., 57), tomato (Solanum lycopersicum, 25), kiwi (Actinidia chinensis Planch, 14), and apple (Malus domestica, 11) [10,11,12,13,14,15].
Studies have demonstrated that XTHs are closely related to plant growth and development. For instance, in Arabidopsis, at least 10 genes are significantly expressed in the root differentiation zone, 5 genes are expressed in green angiosperms, and 2 genes are expressed in the stems, promoting cell wall synthesis, as well as cell elongation and expansion [10,16,17]. The overexpression of PtxtXET16-34 affected xyloglucan content by increasing its level in the primary wall xylem while decreasing xyloglucan levels in the secondary wall xylem [18]. The overexpression of cotton (Gossypium hirsutum L.) GhXTH28 increased the length of cotton fibers and promoted the elongation of cotton leaves [19]. In addition, the maize ZmXTH1 gene has been demonstrated to regulate cell wall composition and structure [20]. The heterologous expression of the pepper CaXTH3 gene in Arabidopsis revealed that the leaves exhibited varying degrees of twisting and bending along the margins during early leaf development, resulting in severely wrinkled leaf shapes [21]. Additionally, two XTH genes (DcXTH2 and DcXTH3) in carnation were associated with petal growth and development during the flower opening process [22].
In addition, XTHs are also associated with abiotic stresses. The overexpression of PeXTH enhanced salt tolerance in tobacco plants by promoting leaf succulent development [23]. Mutant Arabidopsis plants lacking the XTH31, XTH17, and XTH15 genes exhibited lower xyloglucan content compared to wild-type plants, resulting in a significant reduction in the accumulation of Al3+ in the root tip and cell wall, which increased tolerance to aluminum stress [24]. The overexpression of the peppers CaXTH15 and CaXTH3 in Arabidopsis enhanced both salt and drought tolerance [21,25]. Additionally, the overexpression of DkXTH1 in persimmon enhanced tolerance to salt and drought stress, as well as increasing ABA (abscisic acid) content in Arabidopsis [26]. The expression of the cucumber CsXTH1 and CsXTH3 genes was affected by mechanical stimuli [27]. However, a recent study indicated that AtXTH19 may act as a negative regulator under low-temperature stress [28], suggesting that XTH plays different roles in responding to abiotic stresses in plants.
Melon (Cucumis melo L.) is an annual herbaceous plant that belongs to the genus Cucumis and the family Cucurbitaceae. It is one of the most widely grown fruit and vegetable cash crops in the world [29]. In addition to its superior flavor, melon is rich in nutrients and health-promoting compounds, including vitamins, antioxidant compounds, and fiber [30,31]. However, melon is sensitive to water availability due to its shallow root system and large, thin leaves [32]. Salt stress is closely related to water scarcity. Thus, the growth of melon seedlings is highly susceptible to drought and salt stress. These factors adversely affect the physiological and biochemical processes of melon growth and development, leading to a significant reduction in yield [33,34,35]. In the present study, 29 XTH family genes in melon were identified, and the evolutionary relationships of the melon XTH family were analyzed through an analysis of gene structure, conserved structural domains, and homologous collinearity. Additionally, the gene expression patterns of XTH family genes under drought and salt stress were profiled. This study provides a theoretical basis for a deeper understanding and analysis of the functions of melon XTH genes.
2. Materials and Methods
2.1. Genome-Wide Characterization of Melon XTH Genes
The melon genome file (DHL92_genome_v4) and the genome annotation total protein sequence file (DHL92_pep_v4.0) for the melon database were downloaded [36]. The hidden Markov model (HMM) files for two XTH family protein structures (PF00722 and PF06955) [37] were downloaded from the Pfam database (https://pfam.xfam.org (accessed on 21 July 2024)). TBtools-II was used to search for and filter all potential XTH proteins in melon, with the identity match threshold set at ≥50% [38]. Protein sequences were submitted to three databases, SMART (http://smart.embl.de/ (accessed on 21 July 2024)), Pfam, and CDD (Conserved Domains Database, https://www.ncbi.nlm.nih.gov/cdd (accessed on 21 July 2024)) [39], to verify all potential XTH proteins.
2.2. Physicochemical Property Analysis of CmXTH Proteins
The basic physicochemical properties of 29 XTH proteins—including the number of amino acids, molecular weight (MW), protein length, isoelectric point (PI), and instability index—were predicted using the online tool ProtProm in ExPASy [40]. The identified CmXTH genes were named according to their chromosomal positions.
2.3. Phylogenetic Tree of CmXTH Proteins
The amino acid sequences of 33 AtXTHs in Arabidopsis thaliana were downloaded from the TAIR database (https://www.arabidopsis.org/ (accessed on 21 July 2024)). Clustal Muscle was used to align the protein sequences of Arabidopsis thaliana and melon (Cucumis melon L.) [41]. MEGA 7.0 was used to construct phylogenetic trees using the neighbor-joining (NJ) method, with bootstrap analysis conducted for 1000 repetitions [42]. A phylogenetic tree was visualized and beautified using the Evolview online tool (https://evolgenius.info//evolview/ (accessed on 22 July 2024)). The melon XTH gene subfamily was classified based on the Arabidopsis XTH gene subfamily.
2.4. Chromosomal Localization of CmXTH Genes
The genomic data of melon were downloaded from the CuGenDBv2 database [36], and each gene was mapped to the corresponding chromosome position based on the positional information of the melon XTH genes. Chromosome localization and visualization were performed using TBtools-II [38].
2.5. Gene Structure, Protein Structural Domain, and Conserved Motif Analysis of CmXTHs
The melon genomic sequences were downloaded from the CuGenDBv2 database [36]. The online software Pfam 37.0 (https://pfam.xfam.org/search/sequence (accessed on 24 July 2024)) was used to analyze the CmXTH protein family for the presence of conserved structural domains (Glyco_hydro_16 domain, XET_C domain). The conserved motifs in CmXTH proteins were predicted and analyzed using the online software MEME 5.5.6 database (https://meme-suite.org/meme/ (accessed on 24 July 2024)), and their functions were predicted by Pfam. Subsequently, TBtools-II software was used for visualization [38].
2.6. Cis-Acting Element Analysis of CmXTH Genes
The upstream 2000 bp sequences of the XTH genes were obtained from the CuGenDBv2 database [36]. Cis-acting elements in CmXTH genes were predicted using the online tool PlantCARE (https://bioinformatiCs.psb.ugent.be/ (accessed on 24 July 2024)), and cis-elements were visualized with TBtools-II [38].
2.7. Synteny and Duplation Analysis of CmXTH Genes
The amino acid sequences of tomato and cucumber were downloaded from NCBI (https://www.ncbi.nlm.nih.gov/ (accessed on 26 July 2024)). The duplication types of and intraspecific variability in XTH family members were analyzed using the One Step MCsanX-Super Fast function in TBtools-II [38]. The Advanced Circle function in TBtools-II [38] was employed to plot the collinearity relationship of CmXTH genes, as well as the collinearity relationship between melon and three other species, including S. lycopersicum, C. sativus, and A thaliana. To analyze and calculate the gene duplication patterns of CmXTH genes, nonsynonymous (Ka) and synonymous substitutions (Ks) in their paralogous and orthologous homologs and evolutionary rates (Ka/Ks) were evaluated using the Simple Ka/Ks calculator (NJ) in TBtools-II software [38]. In general, Ka/Ks < 1, Ka/Ks = 1, and Ka/Ks > 1 usually indicate purifying, neutral, and positive selection, respectively [43].
2.8. Plant Material and Abiotic Stresses
The experimental material used in this study was melon (“Emerald”), which was planted into plastic containers (10 cm × 10 cm × 10 cm; one seed per container) filled with vermiculite and germinated in a growth chamber (150 μmol·m−2·s−1,12 h light/12 h dark, 25 °C/18 °C). When the cotyledons were fully expanded, the plants were transplanted to a cultivation frame (200 μmol·m−2·s−1, 12 h light/12 h dark, 25 °C/18°). The plants were treated when they had grown to about 3 leaves and 1 heart. Drought and salt stress were simulated using 200 mmol/L NaCl and a 1/2 Hoagland solution containing 20% PEG6000 (mass/volume fraction), respectively. Leaves were collected after 0 h, 3 h, 6 h, 9 h, 12 h, and 24 h. Three biological replicates were performed for each set of experiments.
Total RNA was extracted from approximately 200 mg of the frozen leaf tissue using the SPARKeasy Plant RNA Kit (SparkJade, Qingdao, China) according to the manufacturer’s instructions. The Reverse Transcription Kit (Cofitt, Chengdu, China) was used to reverse transcribe total RNA into cDNA while also removing any genomic DNA present in the sample, following the manufacturer’s protocol. Primers were designed using Primer Premier 5.0 (supplementary file S1). The accession number of the actin gene is MELO3C023264 (LOC103485254). The RT-qPCR assay was conducted using a QuantStudio™ 5 Real-Time PCR instrument (ABI) and SYBR dye (Dr. Di, Shanghai, China). Three independent biological replicates were performed, and the 2−∆∆CT method was employed to calculate the results [44]. The RT-qPCR quantitative data were analyzed using SPSS22.0 software with a one-way ANOVA test, where p < 0.05 was considered significant. TBtools-II and Excel 2020 were utilized to calculate the data and plot all graphs [38].
3. Results
3.1. Identification of XTH Family Genes in Melon
A total of 30 CmXTH genes were identified from the melon genome. These 30 candidate genes were submitted to the SMART database, and incomplete sequences were manually deleted, resulting in 29 XTH genes. According to their positions on the chromosome, the 29 CmXTH genes were sequentially named CmXTH1-CmXTH29.
The identification results are shown in Table 1. The relative molecular weights ranged from 12.58 kD to 40.42 kD. The lengths of the proteins were different, indicating that there was large variability among the CmXTH proteins. The isoelectric points ranged from 4.55 to 9.53. The pI values of 16 out of 29 XTH proteins in melon were higher than 7.0, while 13 out of 29 XTH proteins had values lower than 7.0.

Table 1.
Physicochemical properties of 29 XTH genes in melon.
3.2. Phylogenetic Tree Analysis of Melon XTH Family Genes
A phylogenetic tree was constructed using the XTH protein sequences from both melon and Arabidopsis. As shown in Figure 1, similar to the subfamily classification of the Arabidopsis XTH proteins, the CmXTH proteins were classified into four subgroups, in which CmXTH1, CmXTH12, CmXTH21, and CmXTH26-29 belong to subgroup I; CmXTH2-10 and CmXTH14-20 belong to subgroup II; and CmXTH11, CmXTH13, and CmXTH22-25 belong to subgroup III. CmXTH13 and CmXTH25 belong to III-A, while the rest of the genes belong to III-B. In terms of gene function classification, subgroups I, II, and III-B have XET activity, whereas subgroup III-A exhibits XEH activity. The specific functions of CmXTH proteins can be predicted based on their grouping characteristics.
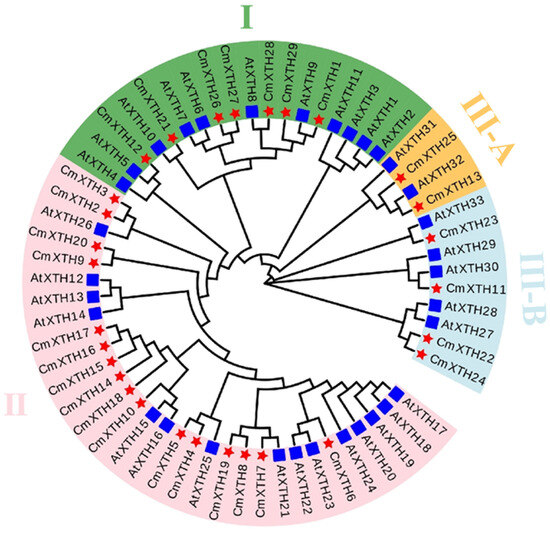
Figure 1.
A phylogenetic tree analysis of XTH proteins in melon and Arabidopsis thaliana. (The evolutionary tree was divided into four subfamilies with different colors. The blue square represents Arabidopsis. The red five-pointed star represents the melon).
3.3. Chromosomal Localization of Melon XTH Family Genes
A total of 29 CmXTH genes were unequally mapped on the 9 chromosomes, with the exception of chromosomes 6, 8, and 11. For instance, chromosome 2 contained the largest number of CmXTHs (8); chromosome 12 had five CmXTH genes, while chromosomes 1, 3, 7, and 9 each contained only one CmXTH gene (Figure 2).
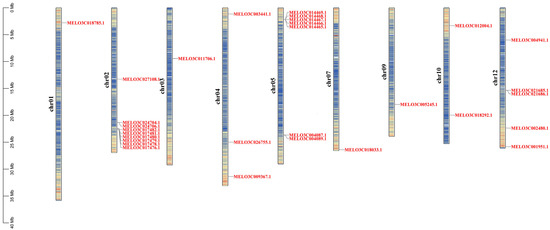
Figure 2.
Chromosomal localization of XTH gene in melon.
Gene amplification refers to the appearance of one or more copies of a DNA segment during genome replication; this can involve a small segment of the genome, an entire chromosome, or the entire genome [45]. Based on the XTH chromosome localization in melon, we identified five gene clusters containing a total of 17 tandem repeat genes. Chromosome 2 has two gene clusters: one of which contains two XTH genes, CmXTH2/CmXTH3, and the other contains six genes, CmXTH4-CmXTH9. Chromosome 5 has two gene clusters, one containing five genes (CmXTH14-CmXTH18) and another containing two genes (CmXTH19/CmXTH20). Chromosome 12 has one gene cluster that contains two genes (CmXTH26/CmXTH27). This suggests that tandem duplication may be an important mode of XTH gene expansion in melon.
3.4. Gene Structure, Protein Structural Domain, and Conserved Motif Analysis of XTH Family in Melon
The number and location of introns are very important in the process of gene expression and regulation. The gene structure analysis of the melon XTH genes showed that each gene contains introns and exons, with varying numbers of each. The CmXTH18 gene contains up to four introns, while the number of introns in other genes does not exceed three (Figure 3c). The results of the conserved motif analysis showed that the CmXTH protein family contained 20 conserved motifs, but there were differences in the types and numbers of conserved motifs among the members of the CmXTH protein family (Figure 3b). The 20 conserved motifs were functionally annotated using the online Pfam software, and the results are shown in Table 2. The functional annotation results showed that motifs 1, 2, 3, 4, and 12 belonged to the GH16 structural domain, and motif 5 belonged to the XET structural domain, while other functions were not understood.
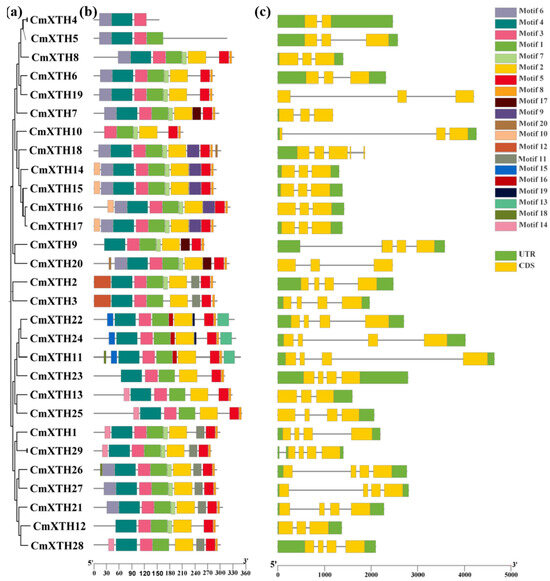
Figure 3.
Phylogenetic tree, gene structure, and conserved motifs of melon XTHs. (a) Phylogenetic relationships of 29 CmXTH genes, (b) protein motifs, and (c) gene structures. Horizontal coordinates denote lengths of genes and amino acid sequences.

Table 2.
Analysis and annotation of conserved motifs of melon XTH proteins.
As shown in Figure 4, the analysis of the conserved protein domain revealed that almost all the genes have both GH16 and XET structural domains, while CmXTH4 and CmXTH5 have only GH16 structural domains. These structurally specialized proteins may have unique functions.
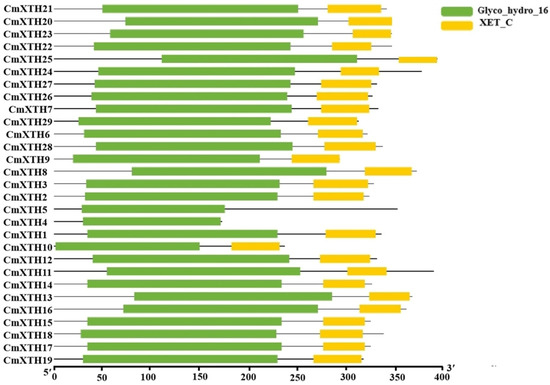
Figure 4.
A conserved domain analysis of CmXTH proteins. The green box represents the Glyco_hydro_16 domain, while the yellow box represents the XET_C domain.
3.5. Cis-Acting Element Analysis of Melon XTH Family Genes
In order to further predict the functions of melon XTH genes, the 2000 bp upstream sequences of the 29 CmXTHs were extracted and submitted to the PlantCARE website for cis-acting element prediction. It was found that the melon XTH family contains 21 types of cis-acting elements, which can be divided into the following five categories: light response-related, hormonal response-related, environmental-stress-related, developmental-related, and binding-site-related elements (Figure 5). Among these, there are many elements related to abiotic stresses, such as drought inducibility, defense and stress responsiveness, low-temperature responsiveness, and wound response. Moreover, there are many elements related to plant hormone responses, including abscisic acid (ABA), gibberellin (GA), salicylic acid (SA), methyl jasmonate (MeJA), zein metabolism, and auxin responses. Additionally, many cis-regulatory elements are connected with plant growth and development, including meristem expression, circadian control, cell cycle regulation, and light response.
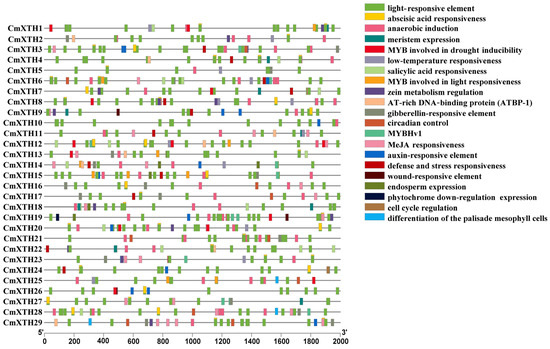
Figure 5.
Cis-element analysis of 29 CmXTH gene promoters. Each box is filled with different colors representing different cis-acting elements.
3.6. Synteny Analysis of Melon XTH Family Genes
In order to further understand the evolutionary relationship of melon XTH genes, an intraspecific covariance analysis of melon XTH genes was performed in this study. As shown in Figure 6, two pairs of tandem repetitive events were identified: CmXTH6 and CmXTH20 and CmXTH13 and CmXTH25. These genes may have been derived from the same intra-chromosomal segment and possess similar characteristics.
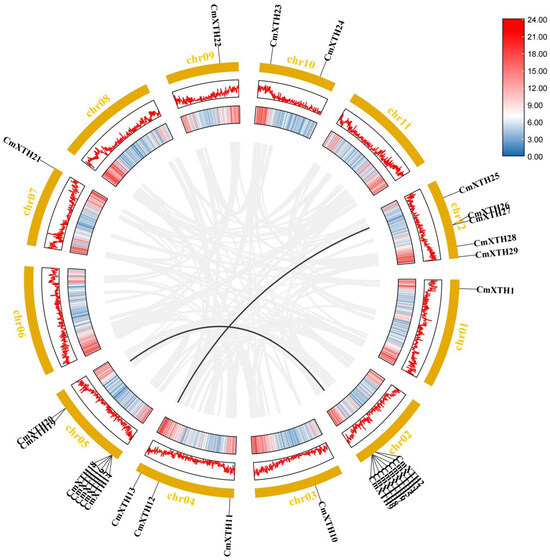
Figure 6.
A fragment duplication analysis of the melon XTHs. The black lines represent the fragmentary duplicated gene pair, and the gray lines represent the synteny blocks of the XTH genes in the melon genome. The line and heat map in the outer circle represent gene density on the chromosome.
In addition, the duplicated genes belonged to the same subfamily, and both pairs of genes were found to have strong collinearity. CmXTH6 and CmXTH20 belonged to subfamily II, while CmXTH13 and CmXTH25 belonged to subfamily III-A. Moreover, Ka/Ks ratios were determined to understand the mechanisms of gene divergence and evolutionary pressures. As seen in Table 3, all Ka/Ks values were less than 0.5, which implies that XTH family genes underwent strong purifying selection during evolution.

Table 3.
Ka/Ks analysis of homologous gene pairs of XTHs in melon.
3.7. Collinearity Analysis of Melon XTH Genes with Three Other Species
In order to further study the evolutionary relationship of the XTH family, this study analyzed the collinearity between melon and Arabidopsis, tomato, and cucumber. The results showed 12 homologous pairs in Arabidopsis, 11 homologous pairs in tomato, and 15 homologous pairs in cucumber (Figure 7). And interestingly, the eight homologous pairs obtained between melon and Arabidopsis also appeared in collinearity relationships with the other three species. This indicates that the evolutionary process of these eight genes is highly conserved.
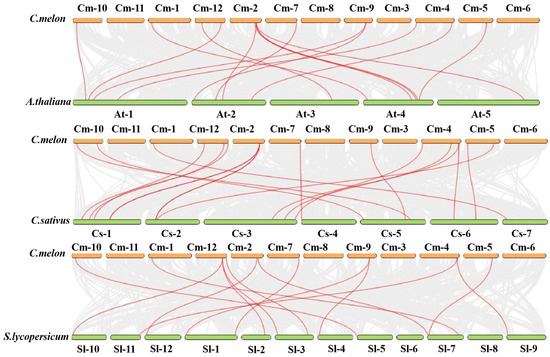
Figure 7.
A collinearity analysis of XTH genes between melon and Arabidopsis thaliana, Solanum lycopersicum, and Cucumis sativus. The red lines represent XTH syntenic gene pairs of melon with three other species, and the gray lines represent orthologous genes of melon with three other species.
3.8. CmXTH Gene Expression in Response to Multiple Stresses
In order to explore the expression patterns of CmXTH genes under abiotic stress, we analyzed the expression levels of CmXTH genes under salt and drought stress using RT-qPCR. The expression of XTHs under drought treatments varied at different time points (Figure 8a). CmXTH26, CmXTH14, and CmXTH4 were up-regulated after 3 h; CmXTH1, CmXTH27, and CmXTH29 were up-regulated after 6 h, while CmXTH11 exhibited the highest expression after 9 h. The number of members under the 12 h drought treatment reached a maximum of 13 (Figure 8a). Conversely, the expression of CmXTH17 and CmXTH24 decreased under drought treatments.
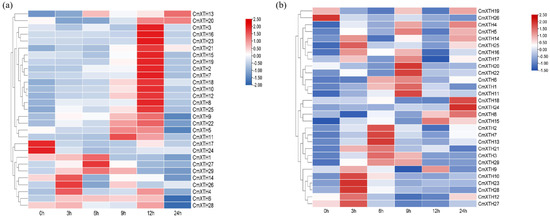
Figure 8.
Expression pattern of melon XTHs under different abiotic stress. (a) Expression pattern of CmXTHs under drought stress. (b) Expression pattern of CmXTHs under salt stress. Samples for expression profiling were collected from melon at 0, 3, 6, 9, 12, and 24 h post drought and salt stress. Red represents high expression, and blue represents low expression. All ratios are log2-transformed so that inductions and repressions of identical magnitude are numerically equal but have opposite signs.
As shown in Figure 8b, the CmXTHs displayed specific expression profiles under different stress times. For example, CmXTH10, CmXTH12, CmXTH23, CmXTH27, and CmXTH28 were mainly expressed after 3 h; CmXTH2, CmXTH7, and CmXTH13 were highly expressed at 6 h, whereas CmXTH11, CmXTH20, and CmXTH22 were up-regulated at 9 h. The expression of CmXTH18 and CmXTH24 reached their maximum at 24 h, while the expression of CmXTH26 decreased under stress treatment. These results indicate that the CmXTH family exhibits different responses to drought and salt stress under different durations.
4. Discussion
The XTH gene family plays an important role in plant growth and development, making it a key focus in melon breeding. In this study, 29 XTH genes were identified from the melon genome and named CmXTH1-CmXTH29 based on their localization on chromosomes (Table 1). Phylogenetic tree analysis showed that, similar to Arabidopsis, the 29 CmXTHs could be classified into four groups (Figure 1). Most XTH genes contain two major conserved structural domains (Glyco_hydro_16 and XET_C). However, we found that CmXTH4 and CmXTH5 lacked the XET_C structural domain (Figure 4). We hypothesize that the loss of the XET_C domain occurred during the evolution of the melon XTH gene family. Plants underwent various whole-genome replication events during evolution, including tandem repeats, fragment repeats, and conversion events [46,47]. Chromosomal localization revealed that the melon XTH gene family has five gene clusters, containing a total of seventeen tandemly repeated genes, suggesting that tandem repeats may be one of the important mechanisms of gene amplification in the CmXTH gene family (Figure 2). Synteny analysis showed that the XTH gene family includes two fragment repeat gene pairs (CmXTH6-CmXTH20, CmXTH13-CmXTH 25) (Figure 6).
Cis-acting regulatory elements function as crucial molecular switches in regulating gene expression [48]. A sequence analysis of the upstream promoters of CmXTH family genes revealed that these promoters contain multiple cis-acting elements, such as MYB, ABRE, and MBS, which are involved in both biotic and abiotic stress responses. MYB, MBS, and other cis-elements serve as binding sites for MYB transcription factors, which regulate hormone signaling and defense responses by binding to MBS elements on target genes [49,50]. Extensive studies have shown that various phytohormones can be involved in the regulation of plant responses to stress [51]. Additionally, several cis-acting elements responsive to phytohormones were identified, including ABRE, GARE-motif, CGTCA-motif, and SARE (Figure 5). Studies have shown that plant hormones induce XTH expression [52,53,54,55]. These regulators ensure that CmXTHs genes respond rapidly to stress conditions.
During the processes of growth and development, plants suffer multiple abiotic stresses such as high temperature, salinity, and drought [56]. The XTH enzyme is an important cell wall-modifying enzyme involved in cell wall construction and degradation, as well as maintaining cell wall integrity and tolerance under both normal and stress conditions [57]. Dhar et al. found that the overexpression of AtXTH22 promoted cell division, elongation, and primary root growth [58]. Similarly, Takeda et al. found that adding xyloglucans to excised pea stems resulted in increased stiffness [59]. Furthermore, numerous studies have shown that the XTH gene family plays a significant role in improving plant stress tolerance [60]. One study found that the overexpression of the CaXTH3 gene in tomato and Arabidopsis thaliana increased tolerance to drought and salt stress by affecting stomatal opening and closing [21,25]. However, Bi et al. found that TaXTH17 plays a negative role in plant resistance to salt and drought [61]. In this study, we observed significant differences in the expression patterns of CmXTHs under different drought and salt stress exposure times. The expression of 13 CmXTHs genes reached a maximum at 12 h with increasing drought stress duration (Figure 8a). CmXTH17 and CmXTH24 were down-regulated at the onset of drought stress, similar to the expression pattern of several XTH genes (AtXTH6, 9, 15, and 16) in Arabidopsis thaliana under drought stress [62], with CmXTH17 being more closely related to AtXTH15 and AtXTH16. These results suggest that the expression of most CmXTH genes is induced by the duration of drought. Expression patterns were also analyzed to show that CmXTHs play an important role in salt stress. With increasing time to salt stress, there were five significantly up-regulated genes at 3 h, three significantly up-regulated genes at 6 h and 9 h each, and two significantly up-regulated genes at 24 h (Figure 8b). Yan et al. found that the knockout of AtXTH30 slowed the decrease in crystalline cellulose content and the depolymerization of cortical microtubules under salt stress, thereby enhancing the resistance of Arabidopsis to salt stress [63]. Therefore, it is reasonable to assume that the 29 CmXTH genes in Cucumis melon could play a role in the adaptation to abiotic stress responses (Figure 8a,b). This is highly significant for improving breeding efficiency, accelerating the development of new varieties, and enhancing crop stress resistance.
5. Conclusions
This study identified 29 XTH genes in melon, which were divided into 4 groups. A phylogenetic analysis, chromosomal localization, and structural analysis of genes and proteins were performed to gain a comprehensive understanding of the CmXTH gene family. We also investigated their promoter regions and collinearity relationships with three other species. The cis-acting element of the CmXTH family members contained a large number of hormonal response- and stress-related elements. In addition, we systematically investigated the expression profiles of CmXTHs under salt and drought stress. All the results of this study provide a theoretical basis for the function identification of melon XTH genes.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/horticulturae10101017/s1, File S1: List of qRT-PCR primers used for expression analysis of CmXTH genes; File S2: gene.position; File S3: protein sequence of CmXTH genes in melon; File S4: Identification of conserved domains in XTH proteins using the CDD tool; File S5: Promoter element prediction analysis of CmXTH genes; File S6: Synteny analysis of XTH genes between melon and other species; File S7: TBtools.kaks.tab.
Author Contributions
Methodology, S.Z.; Data curation, S.Z.; Writing—original draft, S.Z.; formal analysis, X.Z.; Writing—review and editing, Y.W. and Z.Y.; Visualization, Y.K. and Y.L.; Funding acquisition, Z.Y. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Shaanxi Provincial Technological Innovation Guiding Special Project (2021QFY08-02); Shaanxi Province 100 Billion Facility Agriculture Special Project in 2021 (K3030821094); Key Technological Innovation and Integration of Facility Vegetables in the Tibetan Plateau (XZ202202YD0002C); Introduction of Famous Varieties of Facility Vegetables, Melons and Fruits and Construction of Standardised Demonstration Bases (QYXTZX-AL2023-07).
Data Availability Statement
The genome sequences of the cucumis melon, including the predicted gene model annotation for this study, can be found in the CuGenDBv2 database (CuGenDBv2 (http://cucurbitgenomics.org)).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Rui, Y.; Dinneny, J.R. A wall with integrity: Surveillance and maintenance of the plant cell wall under stress. New Phytol. 2020, 225, 1428–1439. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.B.; Cosgrove, D.J. Xyloglucan and its interactions with other components of the growing cell wall. Plant Cell Physiol. 2015, 56, 180–194. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.B.; Cosgrove, D.J. A revised architecture of primary cell walls based on biomechanical changes induced by substrate-specific endoglucanases. Plant Physiol. 2012, 158, 1933–1943. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.E.; Ban, Q.; Li, H.; Hou, Y.; Jin, M.; Han, S.; Rao, J. DkXTH8, a novel xyloglucan endotransglucosylase/hydrolase in persimmon, alters cell wall structure and promotes leaf senescence and fruit postharvest softening. Sci. Rep. 2016, 6, 39155. [Google Scholar] [CrossRef]
- Viborg, A.H.; Terrapon, N.; Lombard, V.; Michel, G.; Czjzek, M.; Henrissat, B.; Brumer, H. A subfamily roadmap of the evolutionarily diverse glycoside hydrolase family 16 (GH16). J. Biol. Chem. 2019, 294, 15973–15986. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Xu, Z.C.; Ding, A.M.; Kong, Y.Z. Genome-wide identification and expression profiling analysis of the Xyloglucan Endotransglucosylase/hydrolase gene family in tobacco (Nicotiana tabacum L.). Genes 2018, 9, 273. [Google Scholar] [CrossRef]
- Li, M.; Xie, F.; He, Q.; Li, J.; Liu, J.L.; Sun, B.; Luo, Y.Y.; Zhang, J.L.; Liu, J.L.; Sun, B.; et al. Expression analysis of XTH in stem swelling of stem mustard and selection of reference genes. Genes 2020, 11, 113. [Google Scholar] [CrossRef]
- Eklöf, J.M.; Brumer, H. The XTH gene family: An update on enzyme structure, function, and phylogeny in xyloglucan remodeling. Plant Physiol. 2010, 153, 456–466. [Google Scholar] [CrossRef]
- Baumann, M.J.; Eklöf, J.M.; Michel, G.; Kallas, A.M.; Teeri, T.T.; Czjzek, M.; Brumer, H. Structural evidence for the evolution of xyloglucanase activity from xyloglucan endo-transglycosylases: Biological implications for cell wall metabolism. Plant Cell 2007, 19, 1947–1963. [Google Scholar] [CrossRef]
- Yokoyama, R.; Nishitani, K. A comprehensive expression analysis of all members of a gene family encoding cell-wall enzymes allowed us to predict cis- regulatory regions involved in cell-wall construction in specific organs of arabidopsis. Plant Cell Physiol. 2001, 42, 1025–1033. [Google Scholar] [CrossRef]
- Yokoyama, R.; Rose, J.K.C.; Nishitani, K. A surprising diversity and abundance of xyloglucan endotransglucosylase/hydrolases in rice. Classification and expression analysis. Plant Physiol. 2004, 134, 1088–1099. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Z.H.; Zhang, X.M.; Yao, W.J.; Gao, Y.; Zhao, K.; Guo, Q.; Zhou, B.R.; Jiang, T.B. Genome-wide identification and expression analysis of the xyloglucan endotransglucosylase/hydrolase gene family in poplar. BMC Genom. 2021, 22, 804. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, D.C.; Zhang, H.Y.; Gao, H.B.; Guo, X.L.; Wang, D.M.; Zhang, X.Q.; Zhang, A.M. The alpha- and beta-expansin and xyloglucan endotransglucosylase/hydrolase gene families of wheat: Molecular cloning, gene expression, and EST data mining. Genomics 2007, 90, 516–529. [Google Scholar] [CrossRef]
- Miedes, E.; Lorences, E.P. Xyloglucan endotransglucosylase/hydrolases (XTHs) during tomato fruit growth and ripening. Plant Physiol. 2009, 166, 489–498. [Google Scholar] [CrossRef] [PubMed]
- Atkinson, R.G.; Johnston, S.L.; Yauk, Y.K.; Sharma, N.N.; Schroder, R. Analysis of xyloglucan endotransglucosylase/hydrolase (XTH) gene families in kiwifruit and apple. Postharvest Biol. Technol. 2009, 51, 149–157. [Google Scholar] [CrossRef]
- Maris, A.; Suslov, D.; Fry, S.C.; Verbelen, J.P.; Vissenberg, K. Enzymic characterization of two recombinant xyloglucan endotransglucosylase/hydrolase (XTH) proteins of Arabidopsis and their effect on root growth and cell wall extension. J. Exp. Bot. 2009, 60, 3959–3972. [Google Scholar] [CrossRef]
- Osato, Y.; Yokoyama, R.; Nishitani, K. A principal role for AtXTH18 in Arabidopsis thaliana root growth: A functional analysis using RNAi plants. J. Plant Res. 2006, 119, 153–162. [Google Scholar] [CrossRef] [PubMed]
- Nishikubo, N.; Takahashi, J.; Roos, A.A.; Derba-Maceluch, M.; Piens, K.; Brumer, H.; Teeri, T.T.; Stålbrand, H.; Mellerowicz, E.J. Xyloglucan endo- transglycosylase-mediated xyloglucan rearrangements in developing wood of hybrid aspen. Plant Physiol. 2011, 155, 399–413. [Google Scholar] [CrossRef]
- Li, Q.Y.; Li, H.Y.; Yin, C.Y.; Wang, X.T.; Jiang, Q.; Zhang, R.; Ge, F.F.; Chen, Y.D.; Yang, L. Genome-wide identification and characterization of xyloglucan endotransglycosylase/hydrolase in ananas comosus during development. Genes 2019, 10, 537. [Google Scholar] [CrossRef]
- Du, H.; Hu, X.; Yang, W.; Hu, W.; Yan, W.; Li, Y.; He, W.; Cao, M.; Zhang, X.; Luo, B. ZmXTH, a xyloglucan endotransglucosylase/hydrolase gene of maize, conferred aluminum tolerance in Arabidopsis. Plant Physiol. 2021, 266, 153520. [Google Scholar] [CrossRef]
- Cho, S.K.; Kim, J.E.; Park, J.A.; Eom, T.J.; Kim, W.T. Constitutive expression of abiotic stress-inducible hot pepper CaXTH3, which encodes a xyloglucan endotransglucosylase/hydrolase homolog, improves drought and salt tolerance in transgenic Arabidopsis plants. FEBS Lett. 2006, 580, 3136–3144. [Google Scholar] [CrossRef] [PubMed]
- Harada, T.; Torii, Y.; Morita, S.; Onodera, R.; Hara, Y.; Yokoyama, R.; Nishitani, K.; Satoh, S. Cloning, characterization, and expression of xyloglucan endotransglucosylase/hydrolase and expansin genes associated with petal growth and development during carnation flower opening. J. Exp. Bot. 2011, 62, 815–823. [Google Scholar] [CrossRef]
- Han, Y.S.; Sa, G.; Sun, J.; Shen, Z.D.; Zhao, R.; Ding, M.Q.; Deng, S.R.; Lu, Y.J.; Zhang, Y.H.; Shen, X.; et al. Overexpression of Populus euphratica xyloglucan endotransglucosylase/hydrolase gene confers enhanced cadmium tolerance by the restriction of root cadmium uptake in transgenic tobacco. Environ. Exp. Bot. 2014, 100, 74–83. [Google Scholar] [CrossRef]
- Zhu, X.F.; Shi, Y.Z.; Lei, G.J.; Fry, S.C.; Zhang, B.C.; Zhou, Y.H.; Braam, J.; Jiang, T.; Xu, X.Y.; Mao, C.Z.; et al. XTH31, Encoding an in vitro XEH/XET-active enzyme, regulates aluminum sensitivity by modulating in vivo XET action, cell wall xyloglucan content, and aluminum binding capacity in arabidopsis. aluminum binding capacity in arabidopsis. Plant Cell 2012, 24, 4731–4747. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.Y.; Seo, Y.S.; Kim, S.J.; Kim, W.T.; Shin, J.S. Constitutive expression of CaXTH3, a hot pepper xyloglucan endotransglucosylase/hydrolase, enhanced tolerance to salt and drought stresses in tomato plants (Solanum lycopersicum cv. Dotaerang). Plant Cell Rep. 2011, 30, 867–877. [Google Scholar] [CrossRef]
- Han, Y.; Han, S.K.; Ban, Q.Y.; He, Y.H.; Jin, M.J.; Rao, J.P. Overexpression of persimmon DkXTH1 enhanced tolerance to abiotic stress and delayed fruit softening in transgenic plants. Plant Cell Rep. 2017, 36, 583–596. [Google Scholar] [CrossRef] [PubMed]
- Malinowski, R.; Fry, S.C.; Zuzga, S.; Wiśniewska, A.; Godlewski, M.; Noyszewski, A. Developmental expression of the cucumber Cs-XTH1 and Cs-XTH3 genes, encoding xyloglucan endotransglucosylase/hydrolases, can be influenced by mechanical stimuli. Acta Physiol. Plant. 2018, 40, 130–140. [Google Scholar] [CrossRef]
- Takahashi, D.; Johnson, K.; Hao, P.F.; Tuong, T.; Erban, A.; Sampathkumar, A.; Bacic, A.; Livingston, D.P.; Kopka, J.; Kuroha, T.; et al. Cell wall modification by the xyloglucan endotransglucosylase/hydrolase XTH19 influences freezing tolerance after cold and sub-zero acclimation. Plant Cell Environ. 2021, 44, 915–930. [Google Scholar] [CrossRef]
- Chikh-Rouhou, H.; Abdedayem, W.; Solmaz, I.; Sari, N.; Garcés-Claver, N. Melon (Cucumis melo L.): Genomics and breeding. In Smart Plant Breeding for Vegetable Crops in Post-Genomics Era; Springer Nature: Singapore, 2023; pp. 25–52. [Google Scholar]
- DellaPenna, D.; Pogson, B.J. Vitamin synthesis in plants: Tocopherols and Carotenoids. Annu. Rev. Plant Biol. 2006, 57, 711–738. [Google Scholar] [CrossRef]
- Nguyen, P.D.T.; Tran, T.; Thieu, H.H.; Lao, T.; Le, T.; Nguyen, N. Hybridization Between the canary Melon and a Vietnamese Non-sweet Melon Cultivar. Aiming to Improve the Growth Performance and Fruit Quality in Melon (Cucumis melo L.). Mol. Biotechnol. 2024, 66, 1673–1683. [Google Scholar] [CrossRef]
- Cabello, M.J.; Castellanos, M.T.; Romojaro, F.; Martínez-Madrid, C.; Ribas, F. Yield and quality of melon grown under different irrigation and nitrogen rates. Agric. Water Manag. 2009, 96, 866–874. [Google Scholar] [CrossRef]
- Akrami, M.; Arzani, A. Physiological alterations due to field salinity stress in melon (Cucumis melo L.). Acta Physiol. Plant. 2018, 40, 91. [Google Scholar] [CrossRef]
- Chevilly, S.; Dolz-Edo, L.; Martínez-Sánchez, G.; Morcillo, L.; Vilagrosa, A.; López-Nicolás, J.M.; Blanca, J.; Yenush, L.; Mulet, J.M. Distinctive traits for drought and salt stress tolerance in melon (Cucumis melo L.). Front. Plant Sci. 2021, 12, 777060. [Google Scholar] [CrossRef]
- Akrami, M.; Arzani, A.; Majnoun, Z. Leaf ion content, yield and fruit quality of field-grown melon under saline conditions. Exp. Agric. 2019, 55, 707–722. [Google Scholar] [CrossRef]
- Yu, J.; Wu, S.; Sun, H.; Wang, X.; Tang, X.; Guo, S.; Zhang, Z.; Huang, S.; Xu, Y.; Weng, Y.; et al. CuGenDBv2: An updated database for cucurbit genomics. Nucleic Acids Res. 2022, 51, D1457–D1464. [Google Scholar] [CrossRef]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Lisanna, P.; Shriya, R.; Richardson, L.J. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef]
- Lu, S.N.; Wang, J.Y.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; Hurwitz, D.I.; Marchler, G.H.; Song, J.S. CDD/SPARCLE: The conserved domain database in 2020. Nucleic Acids Res. 2020, 48, D265–D268. [Google Scholar] [CrossRef]
- Wilkins, M.R.; Gasteiger, E.; Bairoch, A.; Sanchez, J.C.; Williams, K.L.; Appel, R.D.; Hochstrasser, D.F. Protein identification and analysis tools in the ExPASy server. Methods Mol. Biol. 1999, 112, 531–552. [Google Scholar]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [PubMed]
- Zhang, Z.; Li, J.; Zhao, X.Q.; Wang, J.; Wong, G.K.S.; Yu, J. KaKs_calculator: Calculating Ka and Ks through model selection and model averaging. Genom. Proteom. Bioinform. 2006, 4, 259–263. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Zheng, L.; Wu, H.H.; Qanmber, G.; Ali, F.; Wang, L.L.; Liu, Z.D.; Yu, Q.; Wang, Q.; Xu, A.X.; Yang, Z.R. Genome-wide study of the GATL gene family in Gossypium hirsutum L. reveals that GhGATL genes act on pectin synthesis to regulate plant growth and fiber elongation. Genes 2020, 11, 64. [Google Scholar] [CrossRef]
- Tuskan, G.A.; DiFazio, S.; Jansson, S.; Bohlmann, J.; Grigoriev, I.; Hellsten, U.; Putnam, N.; Ralph, S.; Rombauts, S.; Salamov, A.; et al. The genome of black cottonwood, Populus trichocarpa (Torr. & gray). Science 2006, 313, 1596–1604. [Google Scholar]
- Flagel, L.E.; Wendel, J.F. Gene duplication and evolutionary novelty in plants. New Phytol. 2009, 183, 557–564. [Google Scholar] [CrossRef] [PubMed]
- Ding, X.; Li, J.H.; Pan, Y.; Zhang, Y.; Ni, L.; Wang, Y.L. Genome-wide identification and expression analysis of the UGlcAE gene family in tomato. Int. J. Mol. Sci. 2018, 19, 1583. [Google Scholar] [CrossRef]
- Shan, T.; Rong, W.; Xu, H.; Du, L.; Liu, X.; Zhang, Z. The wheat R2R3-MYB transcription factor TaRIM1 participates in resistance response against the pathogen Rhizoctonia cerealis infection through regulating defense genes. Sci. Rep. 2016, 6, 28777. [Google Scholar] [CrossRef]
- Ma, R.; Liu, B.; Geng, X.; Ding, X.; Yan, N.; Sun, X.; Wang, W.; Sun, X.; Zheng, C. Biological function and stress response mechanism of MYB transcription factor family genes. J. Plant Growth Regul. 2023, 42, 83–95. [Google Scholar] [CrossRef]
- Wani, S.H.; Kumar, V.; Shriram, V.; Sah, S.K. Phytohormones and their metabolic engineering for abiotic stress tolerance in crop plants. Crop J. 2016, 4, 162–176. [Google Scholar] [CrossRef]
- Liu, Y.B.; Lu, S.M.; Zhang, J.F.; Liu, S.; Lu, Y.T. A xyloglucanendotransglucosylase/hydrolase involves in the growth of primaryroot and alters the deposition of cellulose in Arabidopsis. Planta 2007, 226, 1547–1560. [Google Scholar] [CrossRef]
- Jan, A. Characterization of a xyloglucan endotransglucosylase gene that is up-regulated by gibberellin in rice. Plant Physiol. 2004, 136, 3670–3681. [Google Scholar] [CrossRef]
- Catala, C.; Rose, J.K.C.; York, W.S.; Albersheim, P.; Darvill, A.G.; Bennett, A.B. Characterization of a Tomato Xyloglucan Endotransglycosylase Gene That Is Down-Regulated by Auxin in Etiolated Hypocotyls. Plant Physiol. 2001, 127, 1180–1192. [Google Scholar] [CrossRef]
- Becnel, J.; Natarajan, M.; Kipp, A.; Braam, J. Developmental expression patterns of Arabidopsis XTH genes reported by transgenes and Genevestigator. Plant Mol. Biol. 2006, 61, 451–467. [Google Scholar] [CrossRef]
- Ahuja, I.; de Vos, R.C.H.; Bones, A.M.; Hall, R.D. Plant molecular stress responses face climate change. Trends Plant Sci. 2010, 15, 664–674. [Google Scholar] [CrossRef]
- Cosgrove, D.J. Growth of the plant cell wall. Nat. Rev. Mol. Cell Biol. 2005, 6, 850–861. [Google Scholar] [CrossRef]
- Dhar, S.; Kim, J.; Yoon, E.K.; Jang, S.; Ko, K.; Lim, J. SHORT-ROOT controls cell elongation in the etiolated arabidopsis hypocotyl. Mol. Cells 2002, 45, 243–256. [Google Scholar] [CrossRef]
- Takeda, T.; Furuta, Y.; Awano, T.; Mizuno, K.; Mitsuishi, Y.; Hayashi, T. Suppression and acceleration of cell elongation by integration of xyloglucans in pea stem segments. Proc. Natl. Acad. Sci. USA 2002, 99, 9055–9060. [Google Scholar] [CrossRef]
- Soga, K.; Wakabayashi, K.; Kamisaka, S.; Hoson, T. Effects of hypergravity on expression of XTH genes in azuki bean epicotyls. Physiol. Plant. 2007, 131, 332–340. [Google Scholar] [CrossRef]
- Bi, H.; Liu, Z.; Liu, S.; Qiao, W.; Zhang, K.; Zhao, M.; Wang, D. Genome-wide analysis of wheat xyloglucan endotransglucosylase/hydrolase (XTH) gene family revealed TaXTH17 involved in abiotic stress responses. BMC Plant Biol. 2024, 24, 640. [Google Scholar] [CrossRef]
- Clauw, P.; Coppens, F.; De Beuf, K.; Dhondt, S.; Van Daele, T.; Maleux, K.; Veronique, S.; Lieven, C.; Nathalie, G.; Inzé, D. Leaf responses to mild drought stress in natural variants of Arabidopsis. Plant Physiol. 2015, 167, 800–816. [Google Scholar] [CrossRef]
- Yan, J.; Huang, Y.; He, H.; Han, T.; Di, P.; Sechet, J.; Fang, L.; Liang, Y.; Scheller, H.V.; Mortimer, J.C.; et al. Xyloglucan endotransglucosylase-hydrolase30 negatively affects salt tolerance in Arabidopsis. J. Exp. Bot. 2019, 70, 5495–5506. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).