Exploring Genomic Differences between a Pair of Vitis vinifera Clones Using WGS Data: A Preliminary Study
Abstract
1. Introduction
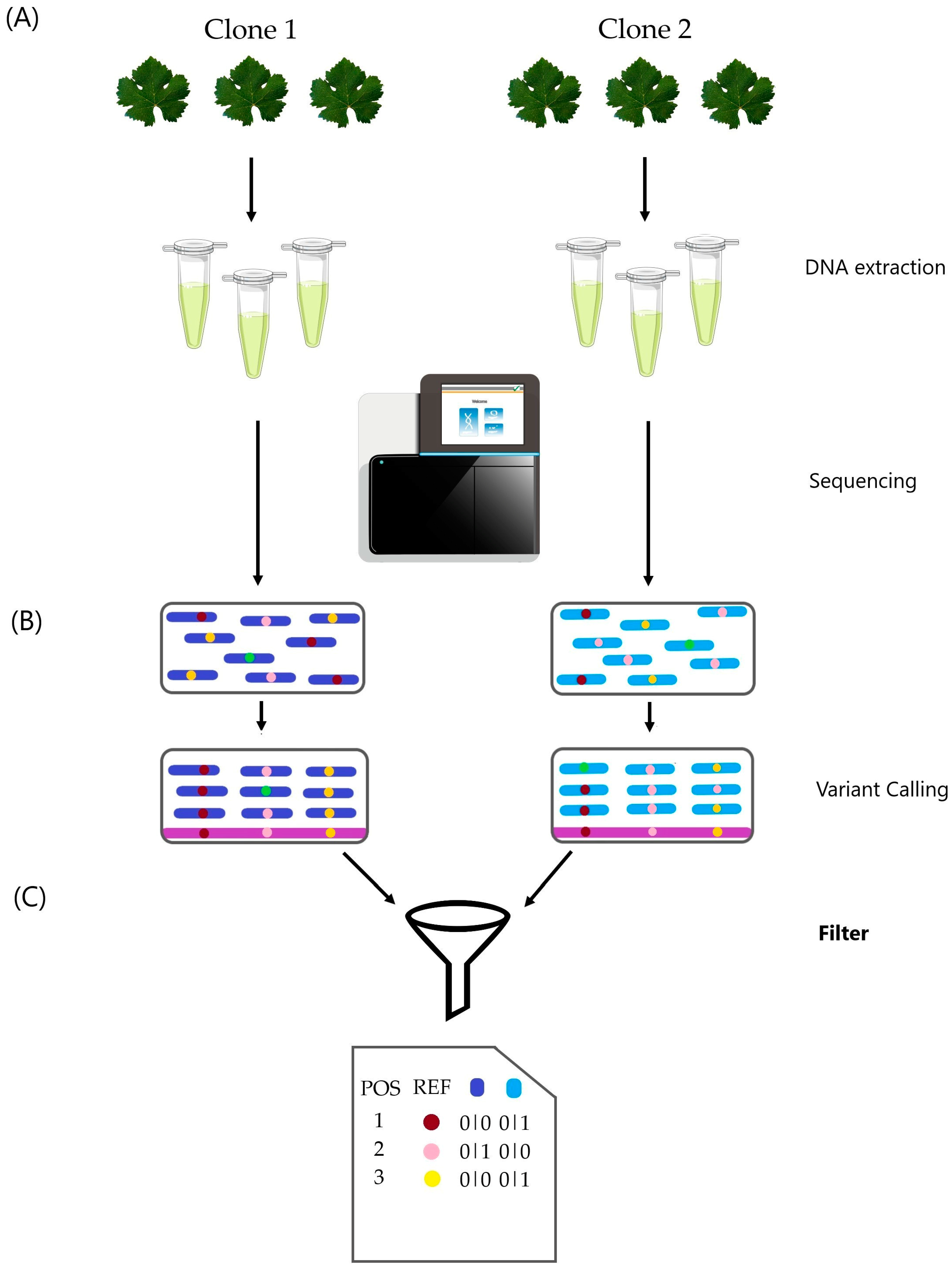
2. Materials and Methods
2.1. Experiments
2.1.1. DNA Extraction
2.1.2. Library Construction and Sequencing
2.2. Bioinformatic Analyses
2.2.1. Quality Control and Trimming of Sequencing Reads
2.2.2. Mapping Reads to the Reference Genome
2.2.3. SNP and InDel Calling
2.2.3.1. Filters
2.2.4. Annotations and Functional Effects of InDels and SNPs
3. Results
3.1. Variants Calling: Variations between Individuals vs. Variations That Differentiate Clones in Carménère and Merlot
3.1.1. Variant Calling of Carménère Clones
3.1.2. Variant Calling of Merlot Clones
3.2. Location of Variations within the Genome and Functional Impact on Transcriptional Sequences
4. Discussion
4.1. Clonal Differentiation Method
4.1.1. Filters
4.1.2. The Variations That Distinguished a Pair of Clones
4.1.3. Limitations of Our Study
- Only two cultivars, only two clones per cultivar, and only three individuals or biological replicates per clone were used. As a preliminary study to find variations in clones, this may be considered the bare minimum amount of data to assess the variations. Despite using what could be considered the bare minimum, we found around 5000 variations per pair of clones per cultivar, out of which between 600 and 1000 variations (per cultivar) were estimated to be validated by visual inspection of the read alignments over the reference genome. However, more sequenced individuals [27] from a larger number of cultivars would be desirable and will be needed to confirm or improve our current estimations.
- Whole-genome sequencing of Vitis vinifera clones is an expensive and long-term enterprise. This study aimed to explore variations to see if a pair of clones can be differentiated through genomic variations. The next step is to look for patterns of variation in clones from different cultivars and find a cheaper and faster way of differentiating them.
- According to Ajay, 2011, the naïve approach of variant calling followed by the comparison of samples can often lead to errors [25,26]. As we used this exact approach, the filters step was crucial to obtain a more reliable group of candidate variations. In addition, visual inspection of these variations was necessary. In our case, the results of the visually validated variations, although estimated to be only between 12% (Carménère) and 20% (Merlot) of the total, confirmed the results of the pipeline for both cultivars.
- Additional filters can increase the number of variations that can differentiate a pair of clones. For example, Li remarked that one frequent source of errors in SNP calling is mismapped reads [25]. In our work, we considered all mapped reads, as opposed to considering only the uniquely mapped reads of the reference genome. Uniquely mapped reads might increase the number of variation candidates as a larger proportion of the sites would pass the biallelic sites filter, meaning the number of biallelic sites would increase after the multi-mapped reads were removed.
5. Conclusions
- It was estimated that between 600 (for Carménère) and 1000 (for Merlot) SNPs can differentiate a pair of clones in the Carménère and Merlot cultivars of Vitis vinifera. Three deletions were visually validated in either cultivar.
- It was found that all visually validated variations had a homozygous genotype in one clone and a heterozygous genotype in the other, with one allele of the second clone matching that of the first clone.
- The proportion of SNPs located within genes versus intergenic regions was found to be 32% for Carménère and 12% for Merlot. All SNPs in gene regions were classified as having low, moderate, or modifier impacts, indicating that none of the identified variations was responsible for phenotypic differences between the clones, despite some being missense mutations. However, we speculate that many of these variations may be associated with gene regulatory functions.
- Despite the high coverage of some of the visually validated variations from Carménère, only four of them were found in non-nuclear genomes.
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Franks, T.; Botta, R.; Thomas, M.R.; Franks, J. Chimerism in Grapevines: Implications for Cultivar Identity, Ancestry and Genetic Improvement. Theor. Appl. Genet. 2002, 104, 192–199. [Google Scholar] [CrossRef]
- Imazio, S.; Labra, M.; Grassi, F.; Winfield, M.; Bardini, M.; Scienza, A. Molecular Tools for Clone Identification: The Case of the Grapevine Cultivar ‘Traminer’. Plant Breed. 2002, 121, 531–535. [Google Scholar] [CrossRef]
- Regner, F.; Stadlbauer, A.; Eisenheld, C.; Kaserer, H. Genetic Relationships Among Pinots and Related Cultivars. Am. J. Enol. Vitic. 2000, 51, 7–14. [Google Scholar] [CrossRef]
- Konradi, J.; Blaich, R.; Forneck, A. Variation among Clones and Sports of “Pinot Noir” (Vitis Vinifera L.). Eur. J. Hortic. Sci. 2007, 72, 275–279. [Google Scholar]
- Moncada, X.; Muñoz, L.; Merdinoglu, D.; Castro, M.H.; Hinrichsen, P. Clonal Polymorphism in the Red Wine Cultivars ’Carmenére´ And ´Cabernet Sauvignon´. Acta Hortic. 2004, 689, 513–520. [Google Scholar] [CrossRef]
- Moncada, X.; Hinrichsen, P. Limited Genetic Diversity among Clones of Red Wine Cultivar “Carmenère” as Revealed by Microsatellite and AFLP Markers. VITIS J. Grapevine Res. 2007, 46, 174–180. [Google Scholar] [CrossRef]
- Adam-Blondon, A.-F.; Roux, C.; Claux, D.; Butterlin, G.; Merdinoglu, D.; This, P. Mapping 245 SSR Markers on the Vitis Vinifera Genome: A Tool for Grape Genetics. Theor. Appl. Genet. 2004, 109, 1017–1027. [Google Scholar] [CrossRef]
- Calderón, L.; Mauri, N.; Muñoz, C.; Carbonell-Bejerano, P.; Bree, L.; Bergamin, D.; Sola, C.; Gomez-Talquenca, S.; Royo, C.; Ibáñez, J.; et al. Whole Genome Resequencing and Custom Genotyping Unveil Clonal Lineages in ‘Malbec’ Grapevines (Vitis Vinifera L.). Sci. Rep. 2021, 11, 7775. [Google Scholar] [CrossRef]
- Urra, C.; Sanhueza, D.; Pavez, C.; Tapia, P.; Núñez-Lillo, G.; Minio, A.; Miossec, M.; Blanco-Herrera, F.; Gainza, F.; Castro, A.; et al. Identification of Grapevine Clones via High-Throughput Amplicon Sequencing: A Proof-of-Concept Study. G3 Genes. Genomes Genet. 2023, 13, jkad145. [Google Scholar] [CrossRef]
- Gambino, G.; Dal Molin, A.; Boccacci, P.; Minio, A.; Chitarra, W.; Avanzato, C.G.; Tononi, P.; Perrone, I.; Raimondi, S.; Schneider, A.; et al. Whole-Genome Sequencing and SNV Genotyping of ‘Nebbiolo’ (Vitis vinifera L.). Clones. Sci. Rep. 2017, 7, 17294. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool. for High. Throughput Sequence Data. 2010. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 8 September 2024).
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and Accurate Short Read Alignment with Burrows–Wheeler Transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map Format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
- DePristo, M.A.; Banks, E.; Poplin, R.; Garimella, K.V.; Maguire, J.R.; Hartl, C.; Philippakis, A.A.; Del Angel, G.; Rivas, M.A.; Hanna, M.; et al. A Framework for Variation Discovery and Genotyping Using Next-Generation DNA Sequencing Data. Nat. Genet. 2011, 43, 491–498. [Google Scholar] [CrossRef]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M.; et al. Twelve Years of SAMtools and BCFtools. GigaScience 2021, 10, giab008. [Google Scholar] [CrossRef]
- Robinson, J.T.; Thorvaldsdóttir, H.; Winckler, W.; Guttman, M.; Lander, E.S.; Getz, G.; Mesirov, J.P. Integrative Genomics Viewer. Nat. Biotechnol. 2011, 29, 24–26. [Google Scholar] [CrossRef]
- Jombart, T. Adegenet: A R Package for the Multivariate Analysis of Genetic Markers. Bioinformatics 2008, 24, 1403–1405. [Google Scholar] [CrossRef]
- Quinlan, A.R.; Hall, I.M. BEDTools: A Flexible Suite of Utilities for Comparing Genomic Features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and Applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef]
- Cingolani, P.; Platts, A.; Wang, L.L.; Coon, M.; Nguyen, T.; Wang, L.; Land, S.J.; Lu, X.; Ruden, D.M. A Program for Annotating and Predicting the Effects of Single Nucleotide Polymorphisms, SnpEff: SNPs in the Genome of Drosophila Melanogaster Strain W1118; Iso-2; Iso-3. Fly 2012, 6, 80–92. [Google Scholar] [CrossRef]
- Jiang, Y.; Jiang, Y.; Wang, S.; Zhang, Q.; Ding, X. Optimal Sequencing Depth Design for Whole Genome Re-Sequencing in Pigs. BMC Bioinform. 2019, 20, 556. [Google Scholar] [CrossRef]
- Kishikawa, T.; Momozawa, Y.; Ozeki, T.; Mushiroda, T.; Inohara, H.; Kamatani, Y.; Kubo, M.; Okada, Y. Empirical Evaluation of Variant Calling Accuracy Using Ultra-Deep Whole-Genome Sequencing Data. Sci. Rep. 2019, 9, 1784. [Google Scholar] [CrossRef]
- Rieber, N.; Zapatka, M.; Lasitschka, B.; Jones, D.; Northcott, P.; Hutter, B.; Jäger, N.; Kool, M.; Taylor, M.; Lichter, P.; et al. Coverage Bias and Sensitivity of Variant Calling for Four Whole-Genome Sequencing Technologies. PLoS ONE 2013, 8, e66621. [Google Scholar] [CrossRef]
- Li, H. A Statistical Framework for SNP Calling, Mutation Discovery, Association Mapping and Population Genetical Parameter Estimation from Sequencing Data. Bioinformatics 2011, 27, 2987–2993. [Google Scholar] [CrossRef]
- Ajay, S.S.; Parker, S.C.J.; Ozel Abaan, H.; Fuentes Fajardo, K.V.; Margulies, E.H. Accurate and Comprehensive Sequencing of Personal Genomes. Genome Res. 2011, 21, 1498–1505. [Google Scholar] [CrossRef]
- Robasky, K.; Lewis, N.E.; Church, G.M. The Role of Replicates for Error Mitigation in Next-Generation Sequencing. Nat. Rev. Genet. 2014, 15, 56–62. [Google Scholar] [CrossRef]
- Lefouili, M.; Nam, K. The Evaluation of Bcftools Mpileup and GATK HaplotypeCaller for Variant Calling in Non-Human Species. Sci. Rep. 2022, 12, 11331. [Google Scholar] [CrossRef]
- Zhou, Y.; Minio, A.; Massonnet, M.; Solares, E.; Lv, Y.; Beridze, T.; Cantu, D.; Gaut, B.S. The Population Genetics of Structural Variants in Grapevine Domestication. Nat. Plants 2019, 5, 965–979. [Google Scholar] [CrossRef]
- Vondras, A.M.; Minio, A.; Blanco-Ulate, B.; Figueroa-Balderas, R.; Penn, M.A.; Zhou, Y.; Seymour, D.; Ye, Z.; Liang, D.; Espinoza, L.K.; et al. The Genomic Diversification of Grapevine Clones. BMC Genom. 2019, 20, 972. [Google Scholar] [CrossRef]




| Cultivar | N° Variations before Filters | N° Variations after Filters | N° of Variations Assessed for Visual Inspection | Proportion of Visually Inspected Variations |
|---|---|---|---|---|
| Carménère | 2,835,299 | 5718 | 408 | 12% |
| Merlot | 5,718,098 | 5218 | 244 | 20% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Araya-Ortega, D.; Gainza-Cortés, F.; Riadi, G. Exploring Genomic Differences between a Pair of Vitis vinifera Clones Using WGS Data: A Preliminary Study. Horticulturae 2024, 10, 1026. https://doi.org/10.3390/horticulturae10101026
Araya-Ortega D, Gainza-Cortés F, Riadi G. Exploring Genomic Differences between a Pair of Vitis vinifera Clones Using WGS Data: A Preliminary Study. Horticulturae. 2024; 10(10):1026. https://doi.org/10.3390/horticulturae10101026
Chicago/Turabian StyleAraya-Ortega, Daniela, Felipe Gainza-Cortés, and Gonzalo Riadi. 2024. "Exploring Genomic Differences between a Pair of Vitis vinifera Clones Using WGS Data: A Preliminary Study" Horticulturae 10, no. 10: 1026. https://doi.org/10.3390/horticulturae10101026
APA StyleAraya-Ortega, D., Gainza-Cortés, F., & Riadi, G. (2024). Exploring Genomic Differences between a Pair of Vitis vinifera Clones Using WGS Data: A Preliminary Study. Horticulturae, 10(10), 1026. https://doi.org/10.3390/horticulturae10101026







