Genome-Wide Identification of UGT Genes and Analysis of Their Expression Profiles During Fruit Development in Walnut (Juglans regia L.)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Identification of UGT Gene Family in Walnut
2.2. Physical and Chemical Properties, as Well as Cis-Acting Element Analysis
2.3. Phylogenetic and Chromosomal Localization Analysis
2.4. Gene Replication and Collinearity Analysis
2.5. Conserved Sequence and Gene Structure Analysis
2.6. Expression Pattern Analysis of 13 JrUGTs
2.7. Determination of Phenolics
2.8. Data Processing
3. Results
3.1. Identification of UGT Gene Family Members in Walnut
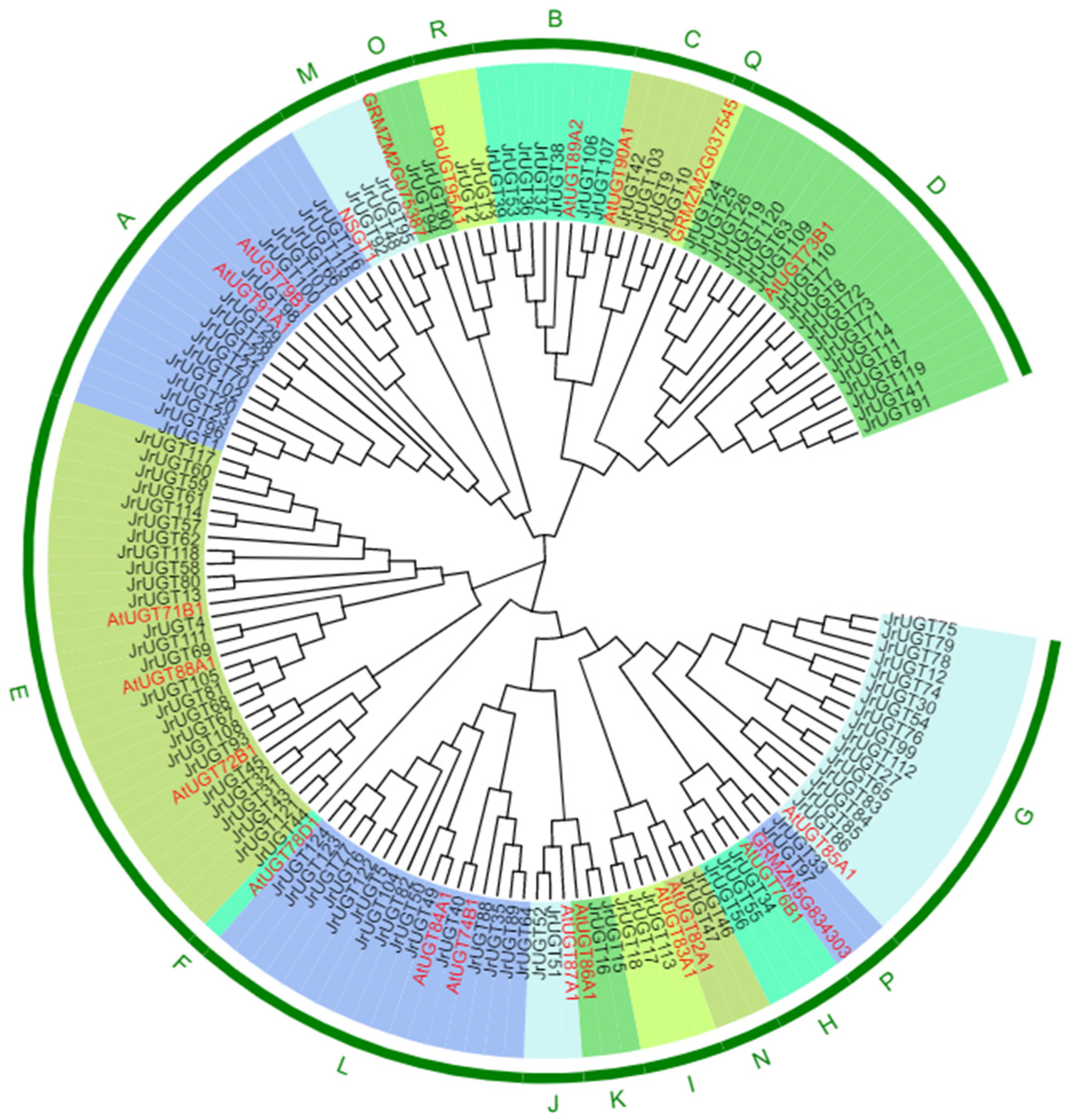
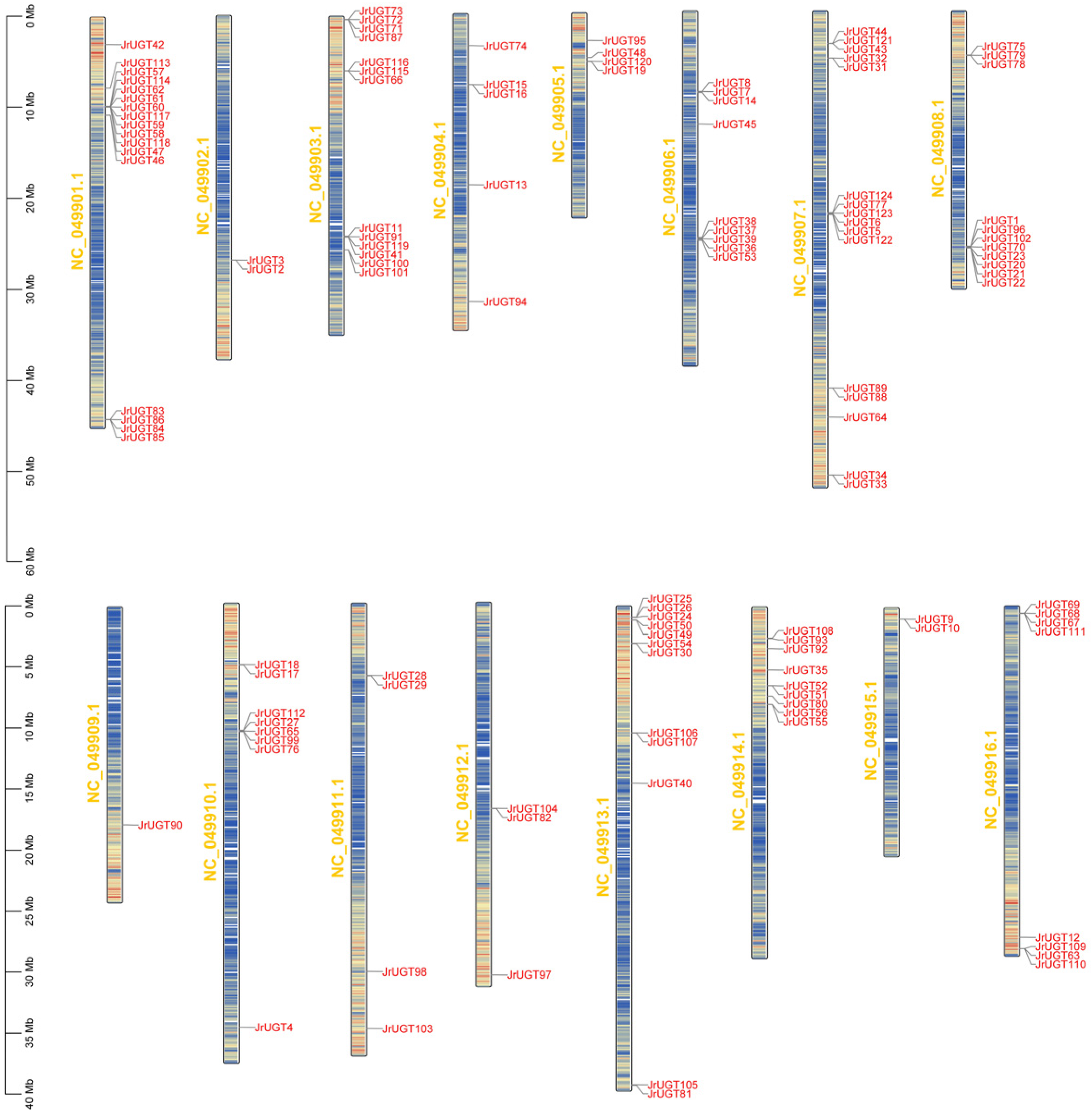
3.2. Phylogenetic and Chromosomal Localization Analysis
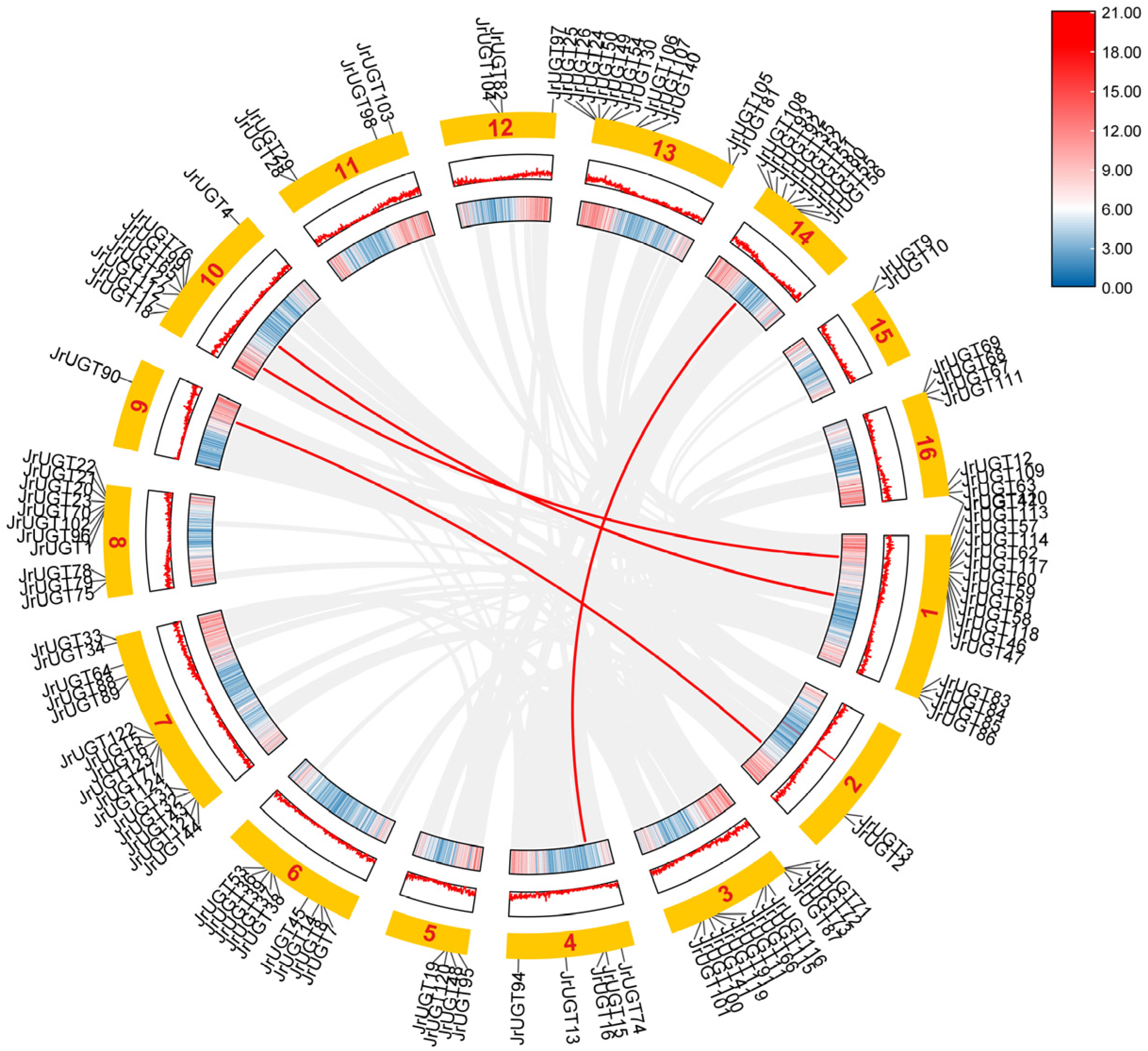
3.3. Gene Replication and Collinearity Analysis
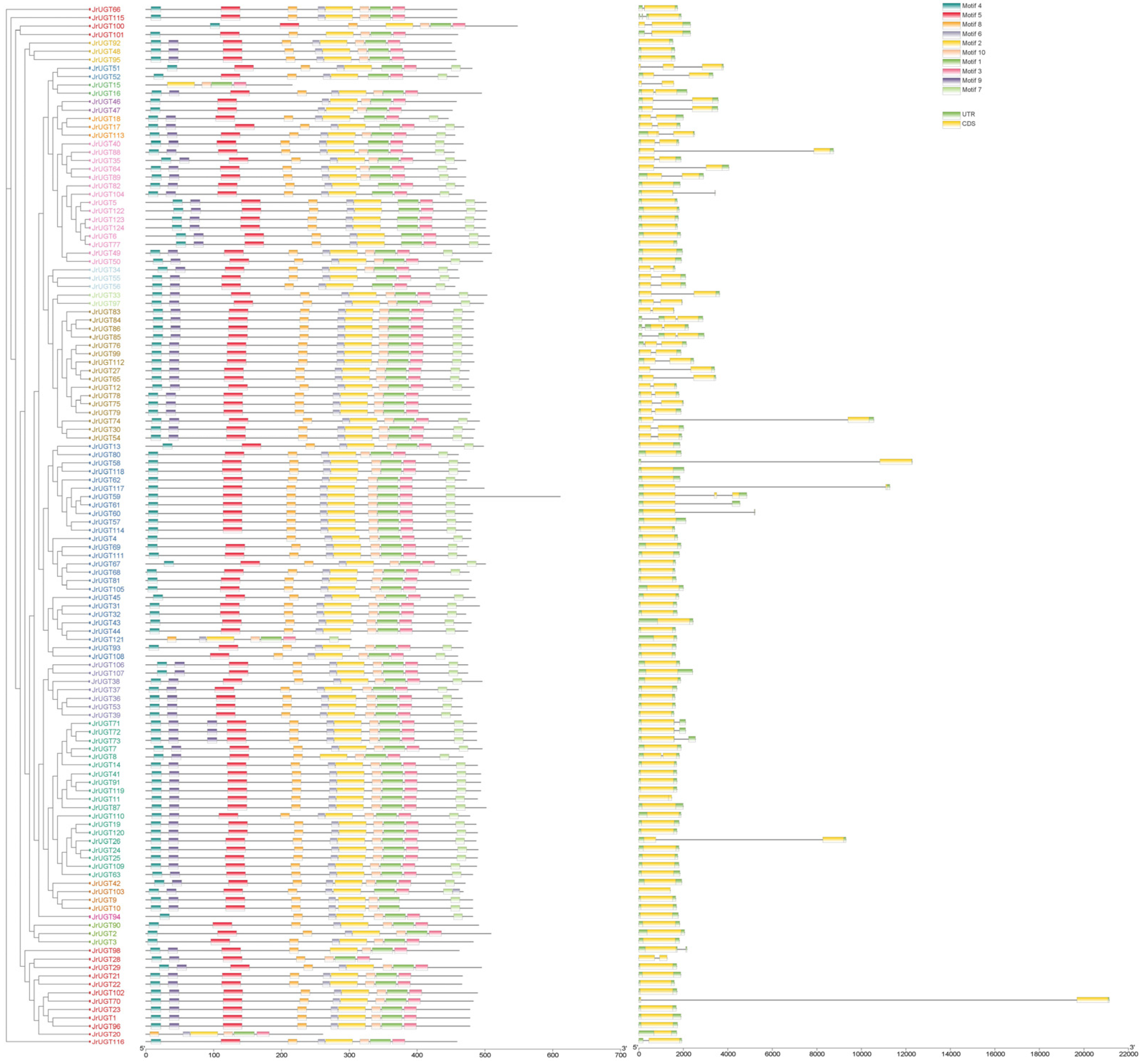
3.4. Conserved Sequence and Gene Structure Analysis
3.5. Promoter Cis-Acting Element Analysis
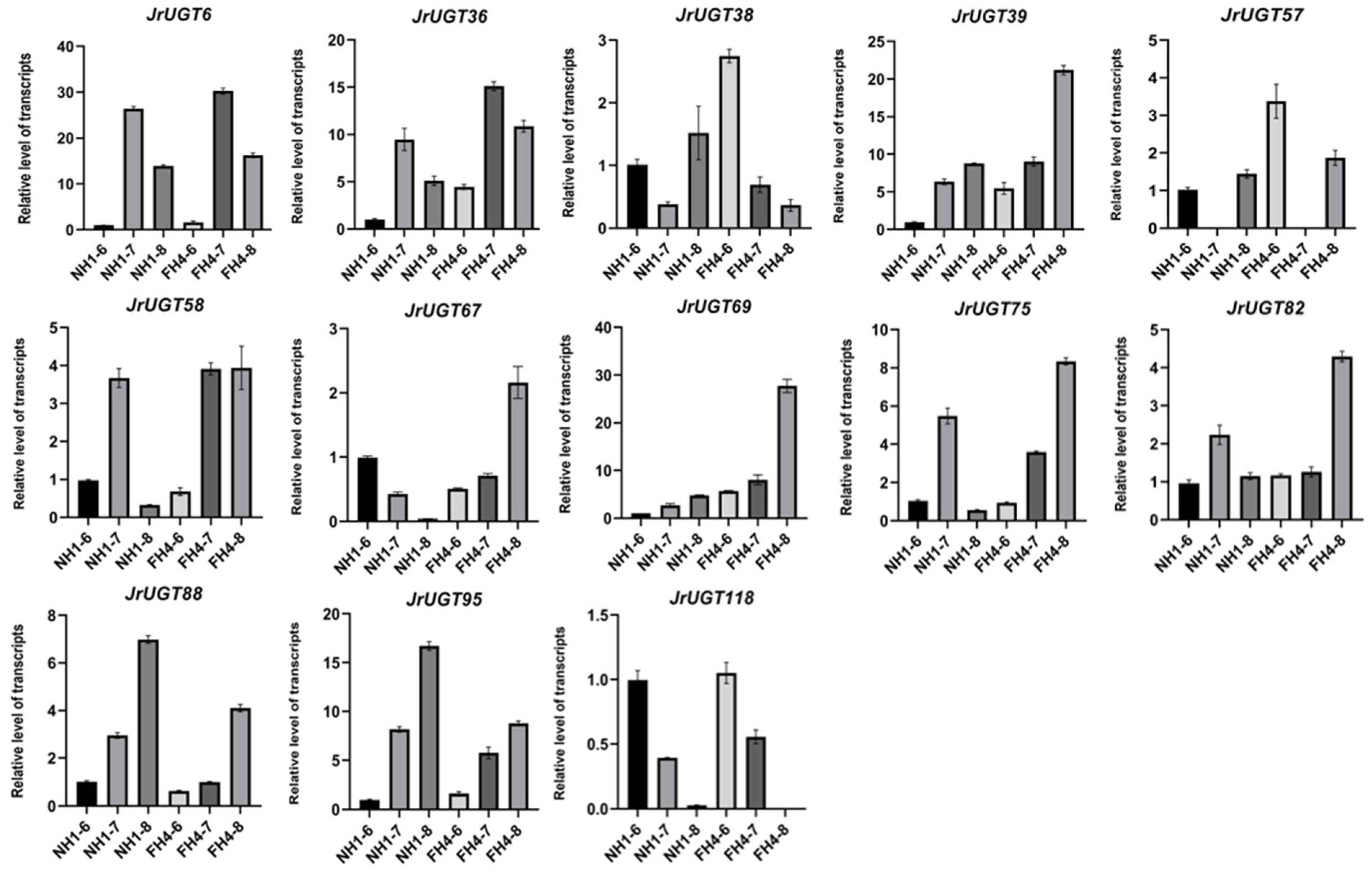
3.6. Expression Patterns of UGT Genes
3.7. Analysis of Phenolic Substances in Walnut Kernel Pellicle at Different Periods
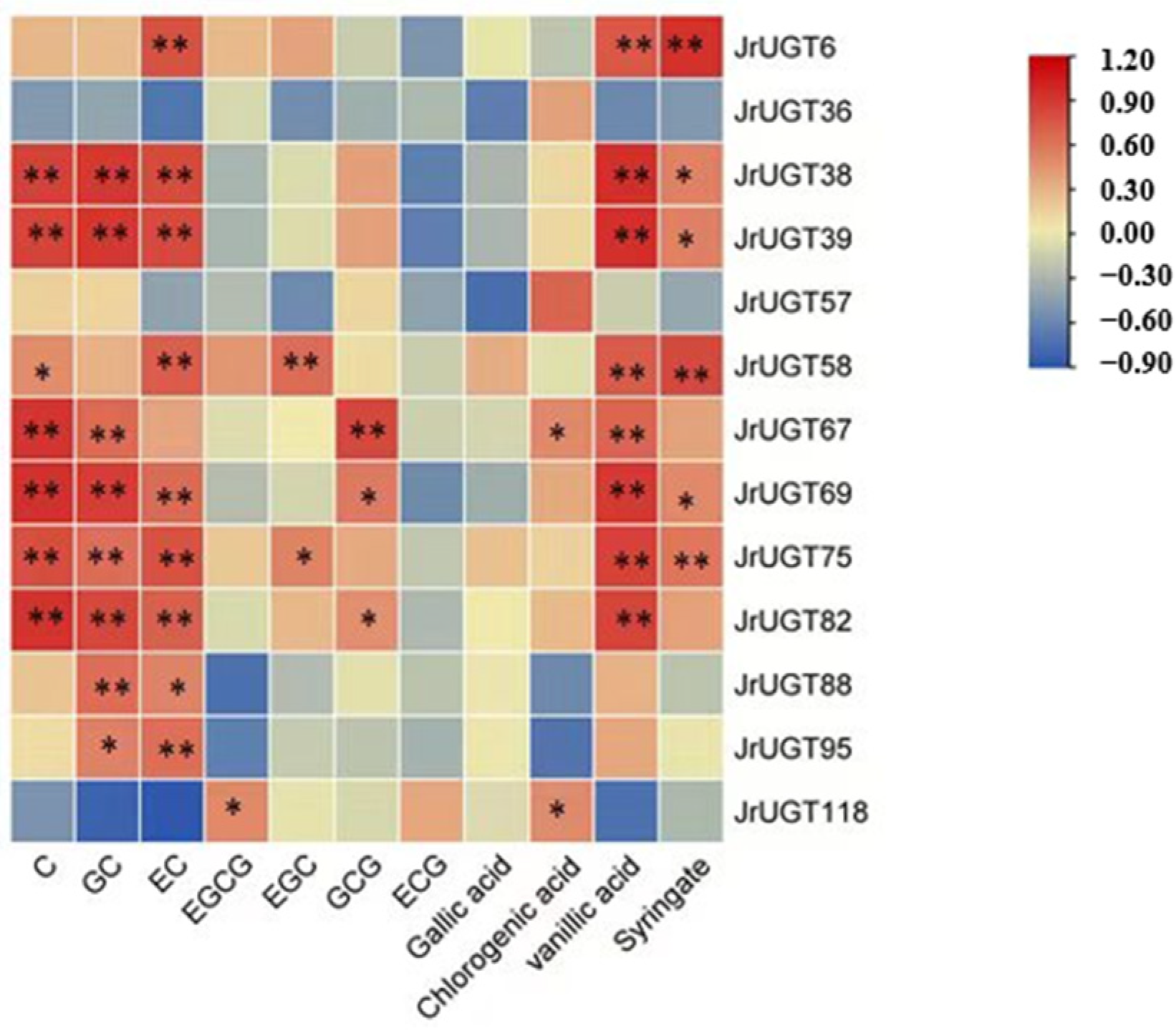
3.8. Correlation Analysis of UGT Gene Expression and Phenolic Content in Walnut
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Feldman, E.B. The scientific evidence for a beneficial health relationship between walnuts and coronary heart disease. J. Nutr. 2002, 132, 1062s–1101s. [Google Scholar] [CrossRef] [PubMed]
- McGranahan, G.; Leslie, C. Walnut. In Fruit Breed; Springer: Boston, MA, USA, 2012; pp. 827–846. [Google Scholar]
- Winkel-Shirley, B. Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol. 2001, 126, 485–493. [Google Scholar] [CrossRef]
- Vogt, T.; Jones, P. Glycosyltransferases in plant natural product synthesis: Characterization of a supergene family. Trends Plant Sci. 2000, 5, 380–386. [Google Scholar] [CrossRef] [PubMed]
- Bowles, D.; Lim, E.K.; Poppenberger, B.; Vaistij, F.E. Glycosyltransferases of lipophilic small molecules. Annu. Rev. Plant Biol. 2006, 57, 567–597. [Google Scholar] [CrossRef]
- Yang, B.; Liu, H.L.; Yang, J.L.; Gupta, V.K.; Jiang, Y.M. New insights on bioactivities and biosynthesis of flavonoid glycosides. Trends Food Sci. Technol. 2018, 79, 116–124. [Google Scholar] [CrossRef]
- Naeem, A.; Ming, Y.; Pengyi, H.; Jie, K.Y.; Yali, L.; Haiyan, Z.; Shuai, X.; Wenjing, L.; Ling, W.; Xia, Z.M.; et al. The fate of flavonoids after oral administration: A comprehensive overview of its bioavailability. Crit. Rev. Food Sci. Nutr. 2022, 62, 6169–6186. [Google Scholar] [CrossRef]
- Mackenzie, P.I.; Owens, I.S.; Burchell, B.; Bock, K.W.; Bairoch, A.; Bélanger, A.; Fournel-Gigleux, S.; Green, M.; Hum, D.W.; Iyanagi, T.; et al. The UDP glycosyltransferase gene superfamily: Recommended nomenclature update based on evolutionary divergence. Pharmacogenetics 1997, 7, 255–269. [Google Scholar] [CrossRef]
- Chen, Y.; Fu, M.; Li, H.; Wang, L.; Liu, R.; Liu, Z. Genome-wide characterization of the UDP-glycosyltransferase gene family reveals their potential roles in leaf senescence in cotton. Int. J. Biol. Macromol. 2022, 222 Pt B, 2648–2660. [Google Scholar] [CrossRef]
- Yonekura-Sakakibara, K.; Hanada, K. An evolutionary view of functional diversity in family 1 glycosyltransferases. Plant J. Cell Mol. Biol. 2011, 66, 182–193. [Google Scholar] [CrossRef]
- Caputi, L.; Malnoy, M.; Goremykin, V.; Nikiforova, S.; Martens, S. A genome-wide phylogenetic reconstruction of family 1 UDP-glycosyltransferases revealed the expansion of the family during the adaptation of plants to life on land. Plant J. Cell Mol. Biol. 2012, 69, 1030–1042. [Google Scholar] [CrossRef]
- Zhang, P.; Zhang, Z.; Zhang, L.; Wang, J.; Wu, C. Glycosyltransferase GT1 family: Phylogenetic distribution, substrates coverage, and representative structural features. Comput. Struct. Biotechnol. J. 2020, 18, 1383–1390. [Google Scholar] [CrossRef] [PubMed]
- Taguchi, G.; Imura, H.; Maeda, Y.; Kodaira, R.; Hayashida, N.; Shimosaka, M.; Okazaki, M. Purification and characterization of UDP-glucose: Hydroxycoumarin 7-O-glucosyltransferase, with broad substrate specificity from tobacco cultured cells. Plant Sci. Int. J. Exp. Plant Biol. 2000, 157, 105–112. [Google Scholar] [CrossRef]
- Lim, E.K.; Baldauf, S.; Li, Y.; Elias, L.; Worrall, D.; Spencer, S.P.; Jackson, R.G.; Taguchi, G.; Ross, J.; Bowles, D.J. Evolution of substrate recognition across a multigene family of glycosyltransferases in Arabidopsis. Glycobiology 2003, 13, 139–145. [Google Scholar] [CrossRef]
- Li, Y.; Baldauf, S.; Lim, E.K.; Bowles, D.J. Phylogenetic analysis of the UDP-glycosyltransferase multigene family of Arabidopsis thaliana. J. Biol. Chem. 2001, 276, 4338–4343. [Google Scholar] [CrossRef]
- Velasco, R.; Zharkikh, A.; Affourtit, J.; Dhingra, A.; Cestaro, A.; Kalyanaraman, A.; Fontana, P.; Bhatnagar, S.K.; Troggio, M.; Pruss, D.; et al. The genome of the domesticated apple (Malus × domestica Borkh.). Nat. Genet. 2010, 42, 833–839. [Google Scholar] [CrossRef]
- Li, Y.; Li, P.; Wang, Y.; Dong, R.; Yu, H.; Hou, B. Genome-wide identification and phylogenetic analysis of Family-1 UDP glycosyltransferases in maize (Zea mays). Planta 2014, 239, 1265–1279. [Google Scholar] [CrossRef]
- Cui, L.; Yao, S.; Dai, X.; Yin, Q.; Liu, Y.; Jiang, X.; Wu, Y.; Qian, Y.; Pang, Y.; Gao, L.; et al. Identification of UDP-glycosyltransferases involved in the biosynthesis of astringent taste compounds in tea (Camellia sinensis). J. Exp. Bot. 2016, 67, 2285–2297. [Google Scholar] [CrossRef]
- Wu, B.; Liu, X.; Xu, K.; Zhang, B. Genome-wide characterization, evolution and expression profiling of UDP-glycosyltransferase family in pomelo (Citrus grandis) fruit. BMC Plant Biol. 2020, 20, 459–470. [Google Scholar] [CrossRef]
- Xu, C.; Wei, L.; Huang, S.; Yang, C.; Wang, Y.; Yuan, H.; Xu, Q.; Zhang, W.; Wang, M.; Zeng, X.; et al. Drought Resistance in Qingke Involves a Reprogramming of the Phenylpropanoid Pathway and UDP-Glucosyltransferase Regulation of Abiotic Stress Tolerance Targeting Flavonoid Biosynthesis. J. Agric. Food Chem. 2021, 69, 3992–4005. [Google Scholar] [CrossRef]
- Chen, L.; Huang, X.X.; Zhao, S.M.; Xiao, D.W.; Xiao, L.T.; Tong, J.H.; Wang, W.S.; Li, Y.J.; Ding, Z.; Hou, B.K. IPyA glucosylation mediates light and temperature signaling to regulate auxin-dependent hypocotyl elongation in Arabidopsis. Proc. Natl. Acad. Sci. USA 2020, 117, 6910–6917. [Google Scholar] [CrossRef]
- Knoch, E.; Sugawara, S.; Mori, T.; Nakabayashi, R.; Saito, K.; Yonekura-Sakakibara, K. UGT79B31 is responsible for the final modification step of pollen-specific flavonoid biosynthesis in Petunia hybrida. Planta 2018, 247, 779–790. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Huang, R.; Liu, L.; Li, Y.; Wang, W.; Xu, Q.; Yu, Y.; Zhou, T. CsUGT78A15 catalyzes the anthocyanidin 3-O-galactoside biosynthesis in tea plants. Plant Physiol. Biochem. PPB 2021, 166, 738–749. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Zhang, Y.; Qu, X.; Wu, F.; Li, X.; Ren, M.; Tong, Y.; Wu, X.; Yang, A.; Chen, Y.; et al. Genome-wide analysis of UDP-glycosyltransferases family and identification of UGT genes involved in abiotic stress and flavonol biosynthesis in Nicotiana tabacum. BMC Plant Biol. 2023, 23, 204–221. [Google Scholar] [CrossRef]
- Lim, E.K.; Ashford, D.A.; Hou, B.; Jackson, R.G.; Bowles, D.J. Arabidopsis glycosyltransferases as biocatalysts in fermentation for regioselective synthesis of diverse quercetin glucosides. Biotechnol. Bioeng. 2004, 87, 623–631. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.; Gu, J.; Luo, Y.; Wang, Y.; Pang, Y.; Shen, G.; Guo, B. Genome-wide analysis of UGT gene family identified key gene for the biosynthesis of bioactive flavonol glycosides in Epimedium pubescens Maxim. Synth. Syst. Biotechnol. 2022, 7, 1095–1107. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome Biol. 2010, 11, R106. [Google Scholar] [CrossRef]
- Feng, C.; Xu, C.J.; Wang, Y.; Liu, W.L.; Yin, X.R.; Li, X.; Chen, M.; Chen, K.S. Codon usage patterns in Chinese bayberry (Myrica rubra) based on RNA-Seq data. BMC Genom. 2013, 14, 732. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Lu, W.Q.; Zhou, X.M.; Zhang, H.Y.; Jiang, J.S.; Fu, L. Simultaneous determination of 9 polyphenols in fruits by high performance liquid chromatography. Chin. J. Health Lab. Technol. 2017, 2, 317–320. [Google Scholar]
- Yang, J.C.; Bai, X.M. Simultaneous Determination of catechins and caffeine content in tea by 359 HPLC. Guizhou Agric. Sci 2020, 48, 99–102. [Google Scholar]
- Paquette, S.; Møller, B.L.; Bak, S. On the origin of family 1 plant glycosyltransferases. Phytochemistry 2003, 62, 399–413. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Ahmad, D.; Zhang, X.; Zhang, Y.; Wu, L.; Jiang, P.; Ma, H. Genome-wide analysis of family-1 UDP glycosyltransferases (UGT) and identification of UGT genes for FHB resistance in wheat (Triticum aestivum L.). BMC Plant Biol. 2018, 18, 67. [Google Scholar] [CrossRef]
- Ren, C.; Cao, Y.; Xing, M.; Guo, Y.; Li, J.; Xue, L.; Sun, C.; Xu, C.; Chen, K.; Li, X. Genome-wide analysis of UDP-glycosyltransferase gene family and identification of members involved in flavonoid glucosylation in Chinese bayberry (Morella rubra). Front. Plant Sci. 2022, 13, 998985. [Google Scholar] [CrossRef]
- Wu, B.; Gao, L.; Gao, J.; Xu, Y.; Liu, H.; Cao, X.; Zhang, B.; Chen, K. Genome-Wide Identification, Expression Patterns, and Functional Analysis of UDP Glycosyltransferase Family in Peach (Prunus persica L. Batsch). Front. Plant Sci. 2017, 8, 389. [Google Scholar] [CrossRef]
- Barvkar, V.T.; Pardeshi, V.C.; Kale, S.M.; Kadoo, N.Y.; Gupta, V.S. Phylogenomic analysis of UDP glycosyltransferase 1 multigene family in Linum usitatissimum identified genes with varied expression patterns. BMC Genom. 2012, 13, 175. [Google Scholar] [CrossRef]
- Ross, J.; Li, Y.; Lim, E.; Bowles, D.J. Higher plant glycosyltransferases. Genome Biol. 2001, 2, Reviews3004. [Google Scholar] [CrossRef]
- Jaillon, O.; Aury, J.M.; Noel, B.; Policriti, A.; Clepet, C.; Casagrande, A.; Choisne, N.; Aubourg, S.; Vitulo, N.; Jubin, C.; et al. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 2007, 449, 463–467. [Google Scholar]
- Ren, Z.; Ji, X.; Jiao, Z.; Luo, Y.; Zhang, G.Q.; Tao, S.; Lei, Z.; Zhang, J.; Wang, Y.; Liu, Z.J.; et al. Functional analysis of a novel C-glycosyltransferase in the orchid Dendrobium catenatum. Hortic. Res. 2020, 7, 111. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Pang, C.; Fan, S.; Song, M.; Yu, J.; Wei, H.; Ma, Q.; Li, L.; Zhang, C.; Yu, S. Genome-wide analysis of the family 1 glycosyltransferases in cotton. Mol. Genet. Genom. MGG 2015, 290, 1805–1818. [Google Scholar] [CrossRef] [PubMed]
- Qiao, X.; Li, Q.; Yin, H.; Qi, K.; Li, L.; Wang, R.; Zhang, S.; Paterson, A.H. Gene duplication and evolution in recurring polyploidization-diploidization cycles in plants. Genome Biol. 2019, 20, 38. [Google Scholar] [CrossRef]
- Wang, F.; Su, Y.; Chen, N.; Shen, S. Genome-Wide Analysis of the UGT Gene Family and Identification of Flavonoids in Broussonetia papyrifera. Molecules 2021, 26, 3449. [Google Scholar] [CrossRef]
- Dong, L.; Tang, Z.; Yang, T.; Hao, F.; Deng, X. Genome-Wide Analysis of UGT Genes in Petunia and Identification of PhUGT51 Involved in the Regulation of Salt Resistance. Plants 2022, 11, 2434. [Google Scholar] [CrossRef]
- Sun, L.; Zhao, L.; Huang, H.; Zhang, Y.; Wang, J.; Lu, X.; Wang, S.; Wang, D.; Chen, X.; Chen, C.; et al. Genome-wide identification, evolution and function analysis of UGTs superfamily in cotton. Front. Mol. Biosci. 2022, 9, 965403. [Google Scholar] [CrossRef]


| Developmental Stage | NH1-6 | NH1-7 | NH1-8 | FH4-6 | FH4-7 | FH4-8 | |
|---|---|---|---|---|---|---|---|
| Substance Name | |||||||
| C | 866.71 ± 2.71 b | 822.45 ± 16.30 c | 746.36 ± 12.40 e | 733.57 ± 5.98 e | 785.20 ± 11.82 d | 1797.77 ± 13.64 a | |
| GC | 168.22 ± 2.31 e | 203.57 ± 2.49 c | 355.09 ± 2.61 b | 144.95 ± 4.70 f | 185.72 ± 4.39 d | 591.43 ± 4.24 a | |
| EC | 56.97 ± 4.37 e | 230.58 ± 2.91 c | 213.56 ± 2.17 d | 57.44 ± 2.91 e | 243.31 ± 6.38 b | 315.93 ± 2.13 a | |
| EGCG | 209.25 ± 6.12 bc | 254.38 ± 28.44 a | 152.46 ± 10.50 d | 231.61 ± 1.67 ab | 232.77 ± 4.42 ab | 191.65 ± 2.40 c | |
| EGC | 318.98 ± 7.78 d | 646.48 ± 13.98 a | 186.41 ± 12.01 f | 282.21 ± 15.76 e | 369.06 ± 7.51 b | 344.71 ± 4.98 c | |
| GCG | 39.93 ± 4.05 b | 8.18 ± 0.40 e | 15.21 ± 0.30 cd | 13.07 ± 1.69 d | 17.52 ± 0.58 c | 45.02 ± 2.73 a | |
| ECG | 359.08 ± 25.51 a | 296.73 ± 24.67 b | 177.76 ± 4.80 c | 151.43 ± 0.80 cd | 154.68 ± 7.26 cd | 145.33 ± 1.43 d | |
| Gallic acid | 1390.92 ± 46.78 b | 1535.90 ± 11.41 a | 1223.70 ± 4.08 c | 1086.18 ± 11.21 d | 1243.39 ± 19.68 c | 1218.43 ± 15.23 c | |
| Chlorogenic acid | 366.99 ± 9.65 c | 263.89 ± 7.50 d | 142.40 ± 4.66 e | 547.09 ± 32.01 a | 256.41 ± 6.06 d | 449.65 ± 11.78 b | |
| vanillic acid | 185.66 ± 5.70 f | 333.59 ± 6.35 c | 321.29 ± 2.72 d | 230.55 ± 3.22 e | 451.43 ± 11.37 b | 645.98 ± 2.52 a | |
| Syringate | 55.07 ± 3.76 e | 97.62 ± 7.02 c | 63.65 ± 3.50 e | 76.84 ± 6.60 d | 186.88 ± 9.21 a | 139.95 ± 1.75 b | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shi, D.; Yang, J.; Li, G.; Zhou, Y.; Yao, P.; Shi, Y.; Tian, J.; Zhang, X.; Liu, Q. Genome-Wide Identification of UGT Genes and Analysis of Their Expression Profiles During Fruit Development in Walnut (Juglans regia L.). Horticulturae 2024, 10, 1130. https://doi.org/10.3390/horticulturae10111130
Shi D, Yang J, Li G, Zhou Y, Yao P, Shi Y, Tian J, Zhang X, Liu Q. Genome-Wide Identification of UGT Genes and Analysis of Their Expression Profiles During Fruit Development in Walnut (Juglans regia L.). Horticulturae. 2024; 10(11):1130. https://doi.org/10.3390/horticulturae10111130
Chicago/Turabian StyleShi, Danhua, Jinyu Yang, Gengyang Li, Yuanting Zhou, Pei Yao, Yanyu Shi, Jieyun Tian, Xiaojun Zhang, and Qunlong Liu. 2024. "Genome-Wide Identification of UGT Genes and Analysis of Their Expression Profiles During Fruit Development in Walnut (Juglans regia L.)" Horticulturae 10, no. 11: 1130. https://doi.org/10.3390/horticulturae10111130
APA StyleShi, D., Yang, J., Li, G., Zhou, Y., Yao, P., Shi, Y., Tian, J., Zhang, X., & Liu, Q. (2024). Genome-Wide Identification of UGT Genes and Analysis of Their Expression Profiles During Fruit Development in Walnut (Juglans regia L.). Horticulturae, 10(11), 1130. https://doi.org/10.3390/horticulturae10111130




