Genome-Wide Characterization of Shi-Related Sequence Gene Family and Its Roles in Response to Zn2+ Stress in Cucumber
Abstract
1. Introduction
2. Materials and Methods
2.1. Isolation of SRS Family Genes in Cucumber
2.2. Protein Sequence and Phylogenetic Analysis of the SRS Family Genes in Cucumber
2.3. Gene Duplication, Structure, and Conservation Analysis
2.4. Subcellular Location Prediction, Cis-Element and GO Analysis
2.5. Plant Materials and Zn2+ Treatment
2.6. RNA Isolation and Quantitative Real-Time PCR (qRT-PCR) Analysis
2.7. Zn2+ Tolerance Assay
3. Results
3.1. Eight Genes from SRS Family Were Identified in Cucumber
3.2. The SRS Proteins Are Relatively Conserved in Plants
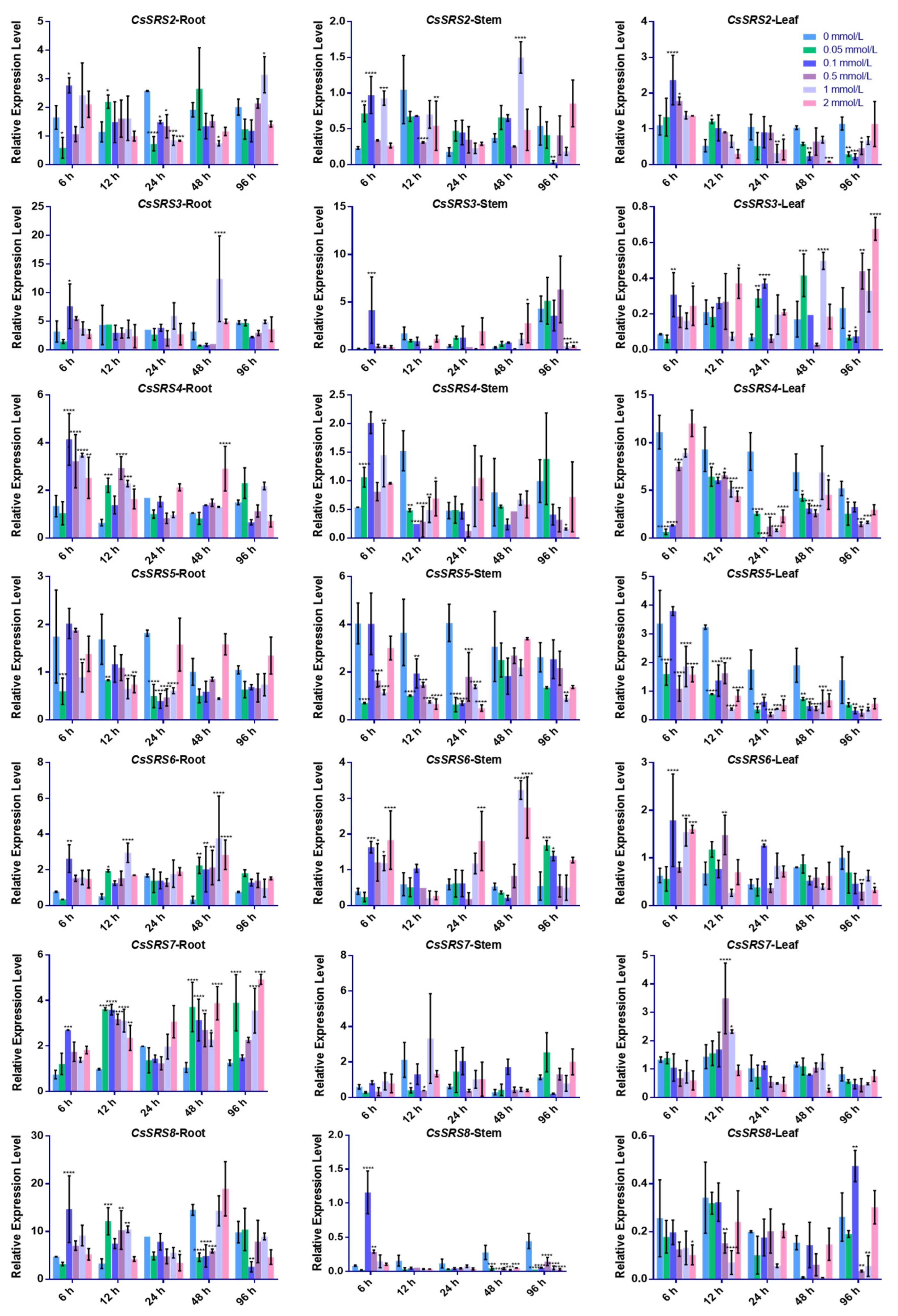
3.3. Expression Pattern of SRS Genes in Cucumber Under Zn2+ Stress
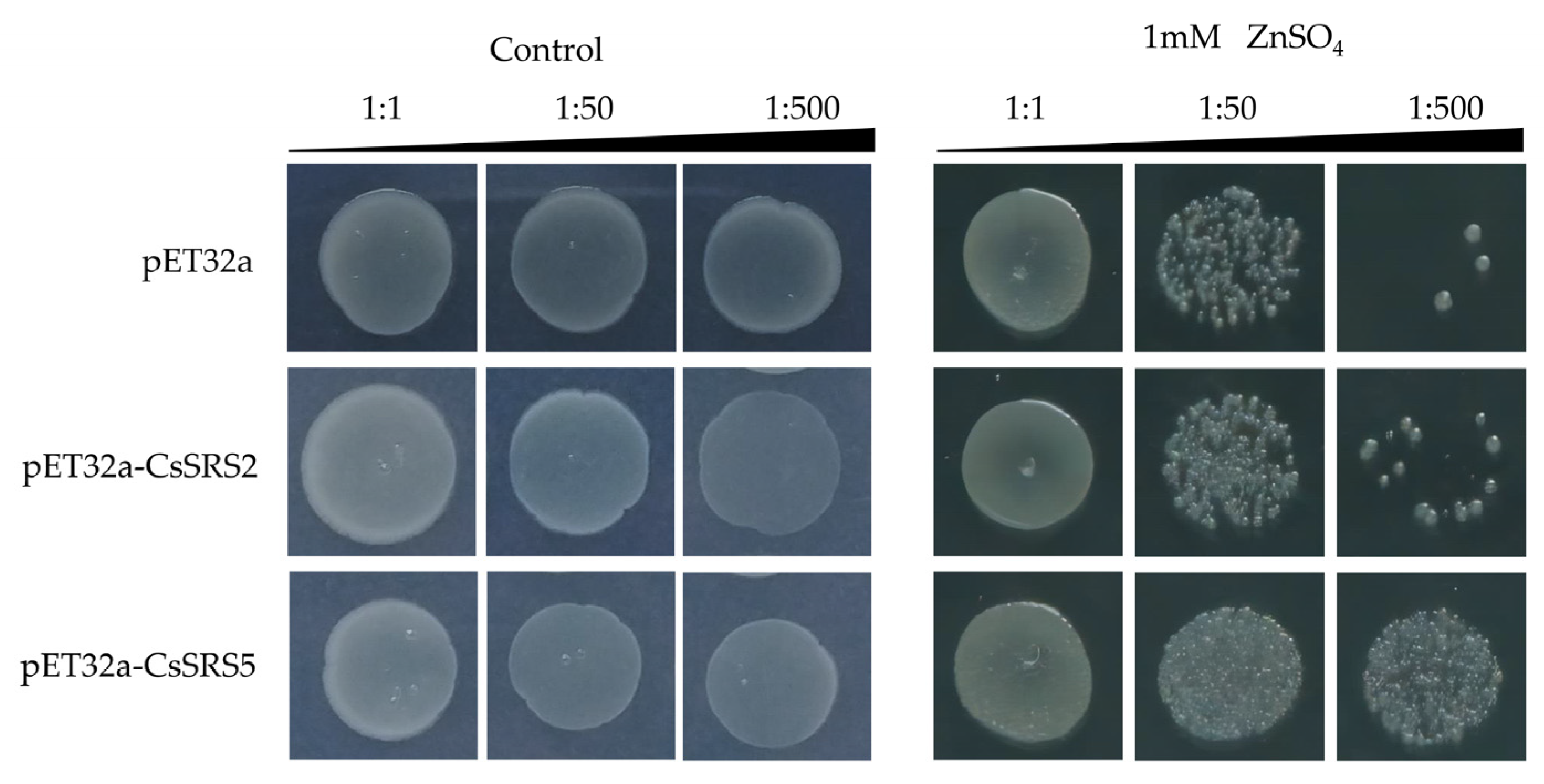
3.4. Overexpressing CsSRS Genes Enhanced Zn2+ Tolerance in E. coli
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Fridborg, I.; Kuusk, S.; Robertson, M.; Sundberg, E. The Arabidopsis Protein SHI Represses Gibberellin Responses in Arabidopsis and Barley. Plant Physiol. 2001, 127, 937–948. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.G.; Lee, S.; Kim, Y.S.; Yun, D.J.; Woo, J.C.; Park, C.M. Activation Tagging of an Arabidopsis SHI-RELATED SEQUENCE Gene Produces Abnormal Anther Dehiscence and Floral Development. Plant Mol. Biol. 2010, 74, 337–351. [Google Scholar] [CrossRef]
- Kuusk, S.; Sohlberg, J.J.; Magnus Eklund, D.; Sundberg, E. Functionally Redundant SHI Family Genes Regulate Arabidopsis Gynoecium Development in a Dose-dependent Manner. Plant J. 2006, 47, 99–111. [Google Scholar] [CrossRef] [PubMed]
- Kaulen, H.; Pognonec, P.; Gregor, P.D.; Roeder, R.G. The Xenopus B1 Factor is Closely Related to The Mammalian Activator USF and Is Implicated in The Developmental Regulation of TFIIIA Gene Expression. Mol. Cell. Biol. 1991, 11, 412–424. [Google Scholar] [CrossRef] [PubMed]
- Stone, S.L.; Hauksdottir, H.; Troy, A.; Herschleb, J.; Kraft, E.; Callis, J. Functional Analysis of the RING-type Ubiquitin Ligase Family of Arabidopsis. Plant Physiol. 2005, 137, 13–30. [Google Scholar] [CrossRef]
- Yuan, X.; Zhang, S.; Liu, S.; Yu, M.; Su, H.; Shu, H.; Li, X. Global Analysis of Ankyrin Repeat Domain C3HC4-type RING Finger Gene Family in Plants. PLoS ONE 2013, 8, e58003. [Google Scholar] [CrossRef]
- Miller, J.C.; Holmes, M.C.; Wang, J.; Guschin, D.Y.; Lee, Y.L.; Rupniewski, I.; Beausejour, C.M.; Waite, A.J.; Wang, N.S.; Kim, K.A.; et al. An improved Zinc-finger Nuclease Architecture for Highly Specific Genome Editing. Nat. Biotechnol. 2007, 25, 778–785. [Google Scholar] [CrossRef]
- Kim, Y.G.; Cha, J.; Chandrasegaran, S. Hybrid Restriction Enzymes: Zinc Finger Fusions to Fok I Cleavage Domain. Proc. Natl. Acad. Sci. USA 1996, 93, 1156–1160. [Google Scholar] [CrossRef]
- Eklund, D.M.; Staldal, V.; Valsecchi, I.; Cierlik, I.; Eriksson, C.; Hiratsu, K.; Ohme-Takagi, M.; Sundstrom, J.F.; Thelander, M.; Ezcurra, I.; et al. The Arabidopsis thaliana STYLISH1 Protein Acts as a Transcriptional Activator Regulating Auxin Biosynthesis. Plant Cell 2010, 22, 349–363. [Google Scholar] [CrossRef]
- Jung, Y.J.; Lee, I.H.; Nou, I.S.; Lee, K.D.; Rashotte, A.M.; Kang, K.K. BrRZFP1 a Brassica rapa C3HC4-type RING Zinc Finger Protein Involved in Cold, Salt and Dehydration Stress. Plant Biol. 2013, 15, 274–283. [Google Scholar] [CrossRef]
- Singh, S.; Yadav, S.; Singh, A.; Mahima, M.; Singh, A.; Gautam, V.; Sarkar, A.K. Auxin Signaling Modulates LATERAL ROOT PRIMORDIUM1 (LRP1) Expression During Lateral Root Development in Arabidopsis. Plant J. 2020, 101, 87–100. [Google Scholar] [CrossRef] [PubMed]
- Smith, D.L.; Fedoroff, N.V. LRP1, a Gene Expressed in Lateral and Adventitious Root Primordia of Arabidopsis. Plant Cell 1995, 7, 735–745. [Google Scholar] [CrossRef] [PubMed]
- Kuusk, S.; Sohlberg, J.J.; Long, J.A.; Fridborg, I.; Sundberg, E. STY1 and STY2 Promote the Formation of Apical Tissues During Arabidopsis Gynoecium Development. Development 2002, 129, 4707–4717. [Google Scholar] [CrossRef]
- De Rybel, B.; Audenaert, D.; Xuan, W.; Overvoorde, P.; Strader, L.C.; Kepinski, S.; Hoye, R.; Brisbois, R.; Parizot, B.; Vanneste, S.; et al. A Role for the Root Cap in Root Branching Revealed by the Non-auxin Probe Naxillin. Nat. Chem. Biol. 2012, 8, 798–805. [Google Scholar] [CrossRef]
- Fridborg, I.; Kuusk, S.; Moritz, T.; Sundberg, E. The Arabidopsis Dwarf Mutant shi Exhibits Reduced Gibberellin Responses Conferred by Overexpression of a New Putative Zinc Finger Protein. Plant Cell 1999, 11, 1019–1031. [Google Scholar] [CrossRef]
- Zeng, H.; Wu, H.; Wang, G.; Dai, S.; Zhu, Q.; Chen, H.; Yi, K.; Du, L. Arabidopsis CAMTA3/SR1 Is Involved in Drought Stress Tolerance And ABA Signaling. Plant Sci. 2022, 319, 111250. [Google Scholar] [CrossRef]
- Zhao, S.P.; Song, X.Y.; Guo, L.L.; Zhang, X.Z.; Zheng, W.J. Genome-Wide Analysis of the Shi-Related Sequence Family and Functional Identification of GmSRS18 Involving in Drought and Salt Stresses in Soybean. Int. J. Mol. Sci. 2020, 21, 1810. [Google Scholar] [CrossRef]
- Sun, C.; Yu, L.; Zhang, S.; Gu, Q.; Wang, M. Genome-wide Characterization of the SHORT INTER-NODES/STYLISH and Shi-Related Sequence Family in Gossypium hirsutum and Functional Identification of GhSRS21 under Salt Stress. Front. Plant Sci. 2023, 13, 1078083. [Google Scholar] [CrossRef] [PubMed]
- Büyük, İ.; Okay, A.; Aras, S. Identification and Characterization of SRS Genes in Phaseolus vulgaris Genome and Their Responses Under Salt Stress. Biochem. Genet. 2022, 60, 482–503. [Google Scholar] [CrossRef]
- He, B.; Shi, P.; Lv, Y.; Gao, Z.; Chen, G. Gene Coexpression Network Analysis Reveals the Role of SRS Genes in Senescence Leaf of Maize (Zea mays L.). J. Genet. 2020, 99, 3. [Google Scholar] [CrossRef]
- Duan, E.; Wang, Y.; Li, X.; Lin, Q.; Zhang, T.; Wang, Y.; Zhou, C.; Zhang, H.; Jiang, L.; Wang, J.; et al. OsSHI1 Regulates Plant Architecture Through Modulating the Transcriptional Activity of IPA1 in Rice. Plant Cell 2019, 31, 1026–1042. [Google Scholar] [CrossRef] [PubMed]
- Yuo, T.; Yamashita, Y.; Kanamori, H.; Matsumoto, T.; Lundqvist, U.; Sato, K.; Ichii, M.; Jobling, S.A.; Taketa, S. A SHORT INTERNODES (SHI) Family Transcription Factor Gene Regulates Awn Elongation and Pistil Morphology in Barley. J. Exp. Bot. 2012, 63, 5223–5232. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, X.; Han, J.; Lu, W.; Ren, Z. Genome-wide Analysis of the WRKY Gene Family in the Cucumber Genome and Transcriptome-wide Identification of WRKY Transcription Factors that Respond to Biotic and Abiotic Stresses. BMC Plant Biol. 2020, 20, 443. [Google Scholar] [CrossRef]
- Luo, S.; Liu, Z.; Wan, Z.; He, X.; Lv, J.; Yu, J.; Zhang, G. Foliar Spraying of NaHS Alleviates Cucumber Salt Stress by Maintaining N+/K+ Balance and Activating Salt Tolerance Signaling Pathways. Plants 2023, 12, 2450. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, W.; Zhang, D.; Zheng, Y.; Xu, Y.; Liu, B.; Li, Q. Mechanism of [CO2] Enrichment Alleviated Drought Stress in the Roots of Cucumber Seedlings Revealed via Proteomic and Biochemical Analysis. Int. J. Mol. Sci. 2022, 23, 14911. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Pan, Y.; Zhu, M.; Wang, S.; Ma, G.; Huang, X.; Qiao, C.; Wang, R.; Xu, X.; Liang, Y.; Lu, K.; et al. Genome-Wide Characterization and Analysis of Metallothionein Family Genes That Function in Metal Stress Tolerance in Brassica napus L. Int. J. Mol. Sci. 2018, 19, 2181. [Google Scholar] [CrossRef]
- Yu, C.S.; Cheng, C.W.; Su, W.C.; Chang, K.C.; Huang, S.W.; Hwang, J.K.; Lu, C.H. CELLO2GO: A Web Server for Protein Subcellular Localization Prediction with Functional Gene Ontology Annotation. PLoS ONE 2014, 9, e99368. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing Real-time PCR Data by the Comparative C(T) Method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
- Baillo, E.H.; Kimotho, R.N.; Zhang, Z.; Xu, P. Transcription Factors Associated with Abiotic and Biotic Stress Tolerance and Their Potential for Crops Improvement. Genes 2019, 10, 771. [Google Scholar] [CrossRef] [PubMed]
- Talanova, V.V.; Titov, A.F.; Topchieva, L.V.; Malysheva, I.E. Effect of Stress Factors on Expression of the Gene Encoding a CBF Transcription Factor in Cucumber Plants. Dokl. Biol. Sci. 2008, 423, 419–421. [Google Scholar] [CrossRef] [PubMed]
- Qi, C.; Dong, D.; Li, Y.; Wang, X.; Guo, L.; Liu, L.; Dong, X.; Li, X.; Yuan, X.; Ren, S.; et al. Heat Shock-Induced Cold Acclimation in Cucumber Through CsHSFA1d-activated JA Biosynthesis and Signaling. Plant J. 2022, 111, 85–102. [Google Scholar] [CrossRef]
- Liu, X.; Wang, T.; Bartholomew, E.; Black, K.; Dong, M.; Zhang, Y.; Yang, S.; Cai, Y.; Xue, S.; Weng, Y.; et al. Comprehensive Analysis of NAC Transcription Factors and Their Expression During Fruit Spine Development in Cucumber (Cucumis sativus L.). Hortic. Res. 2018, 5, 31. [Google Scholar] [CrossRef]
- Zhou, Y.; Ge, L.; Li, G.; Jiang, L.; Yang, Y. Characterization and Expression Analysis of Growth Regulating Factor (GRF) Family Genes in Cucumber. Arch. Biol. Sci. 2018, 70, 629–637. [Google Scholar] [CrossRef]
- Yuan, T.T.; Xu, H.H.; Li, J.; Lu, Y.T. Auxin Abolishes SHI-RELATED SEQUENCE5-Mediated Inhibition of Lateral Root Development in Arabidopsis. New Phytol. 2020, 225, 297–309. [Google Scholar] [CrossRef]
- Eklund, D.M.; Cierlik, I.; Ståldal, V.; Claes, A.R.; Vestman, D.; Chandler, J.; Sundberg, E. Expression of Arabidopsis SHORT INTERNODES/STYLISH Family Genes in Auxin Biosynthesis Zones of Aerial Organs is Dependent on a GCC Box-like Regulatory Element. Plant Physiol. 2011, 157, 2069–2080. [Google Scholar] [CrossRef]
- Sohlberg, J.J.; Myrenas, M.; Kuusk, S.; Lagercrantz, U.; Kowalczyk, M.; Sandberg, G.; Sundberg, E. STY1 Regulates Auxin Homeostasis and Affects Apical-basal Patterning of the Arabidopsis Gynoecium. Plant J. 2006, 47, 112–123. [Google Scholar] [CrossRef] [PubMed]
- Youssef, H.M.; Eggert, K.; Koppolu, R.; Alqudah, A.M.; Poursarebani, N.; Fazeli, A.; Sakuma, S.; Tagiri, A.; Rutten, T.; Govind, G.; et al. VRS2 Regulates Hormone-mediated Inflorescence Patterning in Barley. Nat. Genet. 2017, 49, 157–161. [Google Scholar] [CrossRef]
- Yang, J.; Xu, P.; Yu, D. Genome-Wide Identification and Characterization of the SHI-Related Sequence Gene Family in Rice. Evol. Bioinform. Online 2020, 16, 1176934320941495. [Google Scholar] [CrossRef]
- Huang, H.; Song, J.; Feng, Y.; Zheng, L.; Chen, Y.; Luo, K. Genome-Wide Identification and Expression Analysis of the SHI-Related Sequence Family in Cassava. Genes 2023, 14, 870. [Google Scholar] [CrossRef] [PubMed]
- Küpper, H.; Andresen, E. Mechanisms of Metal Toxicity in Plants. Metallomic 2016, 8, 269–285. [Google Scholar] [CrossRef] [PubMed]
- Priya, A.K.; Gnanasekaran, L.; Dutta, K.; Rajendran, S.; Balakrishnan, D.; Soto-Moscoso, M. Biosorption of Heavy Metals by Microorganisms: Evaluation of Different Underlying Mechanisms. Chemosphere 2022, 307, 135957. [Google Scholar] [CrossRef]
- Wang, Y.; Shi, J.; Wang, H.; Lin, Q.; Chen, X.; Chen, Y. The influence of Soil Heavy Metals Pollution on Soil Microbial Biomass, Enzyme Activity, and Community Composition Near a Copper Smelter. Ecotoxicol. Environ. Saf. 2007, 67, 75–81. [Google Scholar] [CrossRef] [PubMed]
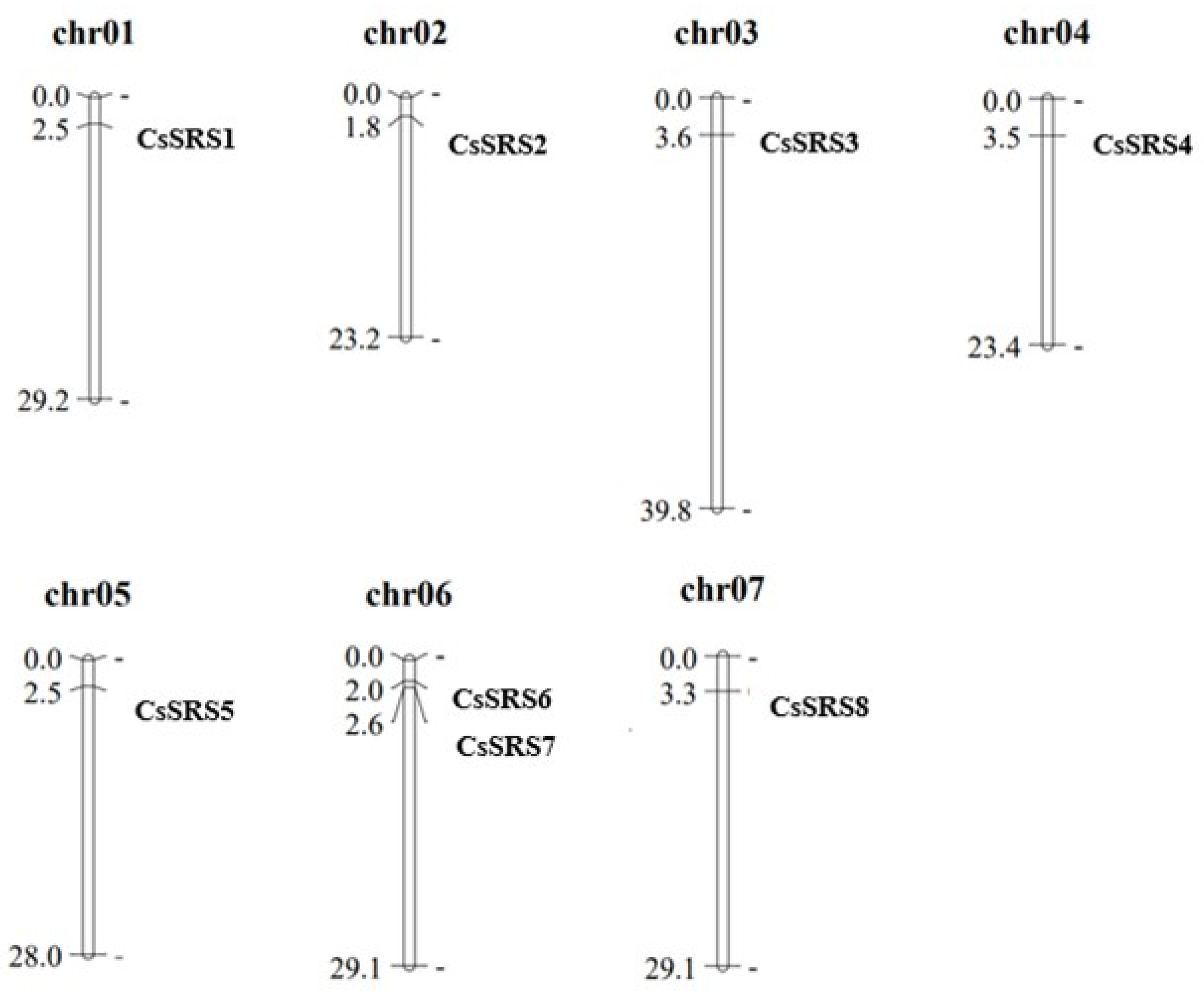
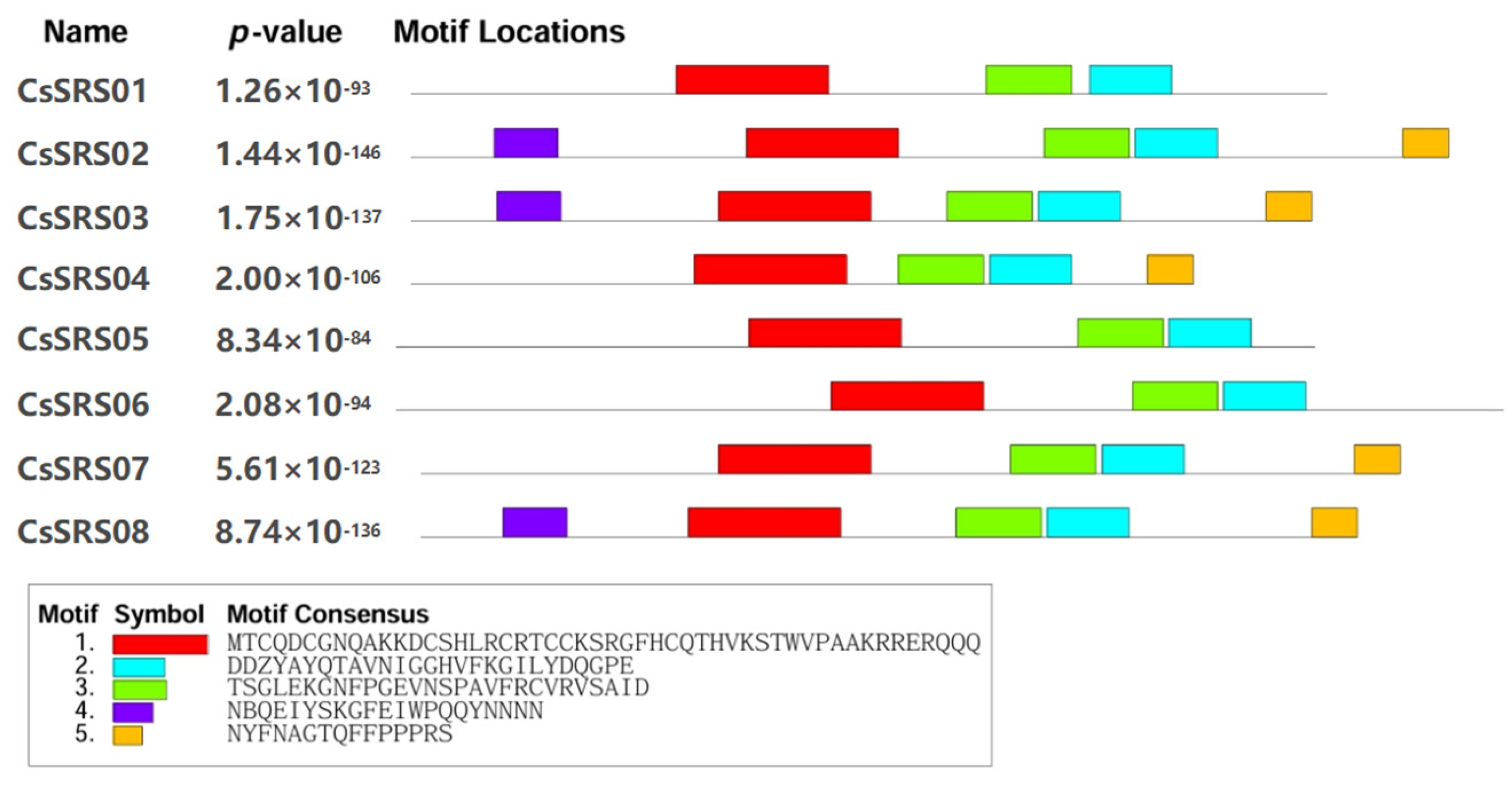
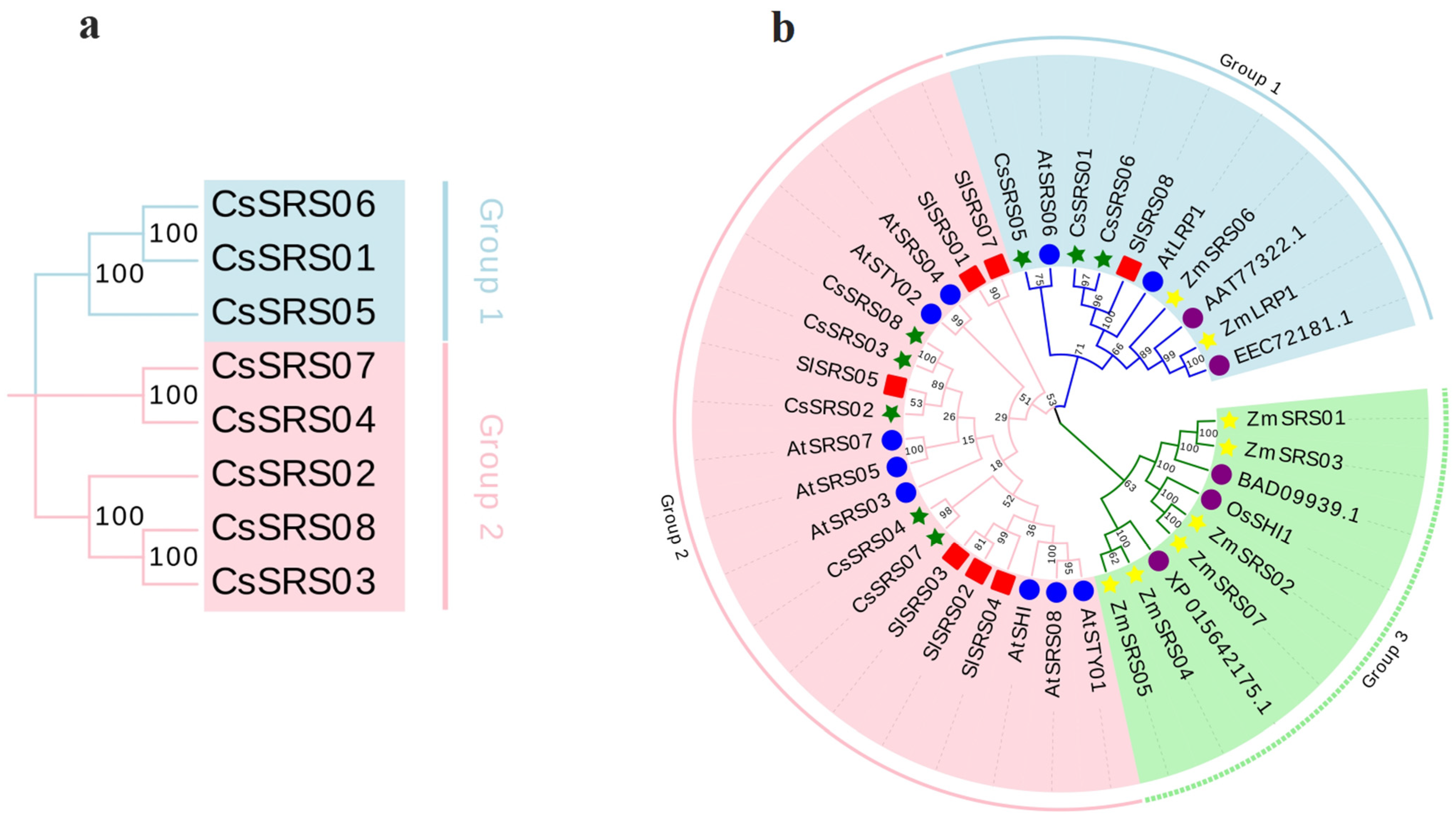
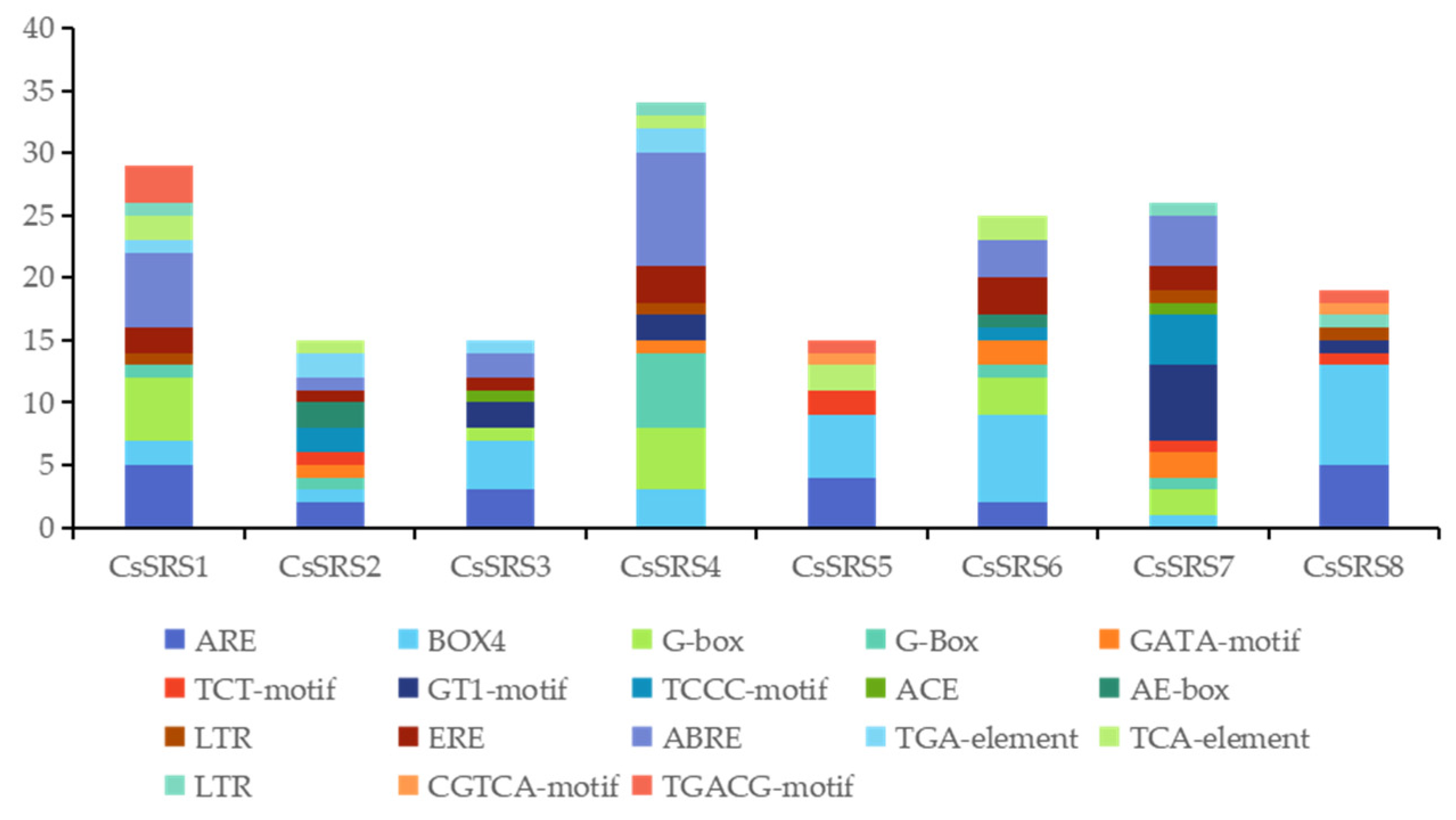
| Gene Locus ID | Gene Number | Start | Stop | Gene Bases (bp) | CDS (bp) | Amino Acids (aa) | MW (Da) | pI |
|---|---|---|---|---|---|---|---|---|
| Csa1G628000.1 | CsSRS1 | 24,761,047 | 24,763,733 | 2687 | 924 | 307 | 31,146.62 | 8.90 |
| Csa2G364580.1 | CsSRS2 | 17,580,604 | 17,582,096 | 1493 | 1044 | 347 | 37,386.30 | 6.45 |
| Csa3G865400.1 | CsSRS3 | 36,176,110 | 36,177,603 | 1494 | 909 | 302 | 33,877.29 | 7.65 |
| Csa4G043960.1 | CsSRS4 | 3,461,111 | 3,462,133 | 1023 | 792 | 263 | 28,682.78 | 6.24 |
| Csa5G623840.1 | CsSRS5 | 25,062,878 | 25,064,891 | 2014 | 912 | 303 | 32,073.74 | 8.89 |
| Csa6G432270.1 | CsSRS6 | 20,428,913 | 20,430,800 | 1888 | 1098 | 365 | 37,662.42 | 7.15 |
| Csa6G504650.2 | CsSRS7 | 25,599,240 | 25,601,517 | 1560 | 996 | 331 | 35,897.29 | 7.00 |
| Csa7G051450.1 | CsSRS8 | 3,302,327 | 3,303,550 | 1134 | 954 | 317 | 35,221.94 | 7.13 |
| Protein Name | Localization Prediction | Molecular Function | Biological Process |
|---|---|---|---|
| CsSRS1 | Extracellular and nuclear | DNA and protein binding | Anatomical structure development Reproduction |
| CsSRS2 | Nuclear | DNA and protein binding | Anatomical structure development |
| CsSRS3 | Nuclear | DNA and protein binding | Anatomical structure development |
| CsSRS4 | Nuclear | DNA and protein binding | Anatomical structure development |
| CsSRS5 | Extracellular and nuclear | DNA and protein binding | Anatomical structure development |
| CsSRS6 | Extracellular and nuclear | DNA and protein binding | Anatomical structure development Reproduction |
| CsSRS7 | Nuclear | DNA and protein binding | Anatomical structure development |
| CsSRS8 | Nuclear | DNA and protein binding | Anatomical structure development |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, X.; Ahmad, B.; Zeng, S.; Lan, Y.; Hu, X.; Fu, L.; Hu, T.; Li, J.; Zhang, X.; Pan, Y.; et al. Genome-Wide Characterization of Shi-Related Sequence Gene Family and Its Roles in Response to Zn2+ Stress in Cucumber. Horticulturae 2024, 10, 1154. https://doi.org/10.3390/horticulturae10111154
Zhang X, Ahmad B, Zeng S, Lan Y, Hu X, Fu L, Hu T, Li J, Zhang X, Pan Y, et al. Genome-Wide Characterization of Shi-Related Sequence Gene Family and Its Roles in Response to Zn2+ Stress in Cucumber. Horticulturae. 2024; 10(11):1154. https://doi.org/10.3390/horticulturae10111154
Chicago/Turabian StyleZhang, Xinhui, Bilal Ahmad, Shuang Zeng, Yuhan Lan, Xin Hu, Lingling Fu, Tian Hu, Jinhua Li, Xingguo Zhang, Yu Pan, and et al. 2024. "Genome-Wide Characterization of Shi-Related Sequence Gene Family and Its Roles in Response to Zn2+ Stress in Cucumber" Horticulturae 10, no. 11: 1154. https://doi.org/10.3390/horticulturae10111154
APA StyleZhang, X., Ahmad, B., Zeng, S., Lan, Y., Hu, X., Fu, L., Hu, T., Li, J., Zhang, X., Pan, Y., & Du, D. (2024). Genome-Wide Characterization of Shi-Related Sequence Gene Family and Its Roles in Response to Zn2+ Stress in Cucumber. Horticulturae, 10(11), 1154. https://doi.org/10.3390/horticulturae10111154







