Genome-Wide Identification of the Heat Shock Transcription Factor Gene Family in Rosemary (Salvia rosmarinus)
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Resources
2.2. Identification of SrHSF Gene Family Members
2.3. Chromosome Localization and Numbering of SrHSF
2.4. Physicochemical Properties of SrHSF Proteins
2.5. Phylogenetic Tree Construction of SrHSF Protein Sequences
2.6. Analysis of SrHSF Gene Structure and Conserved Motifs
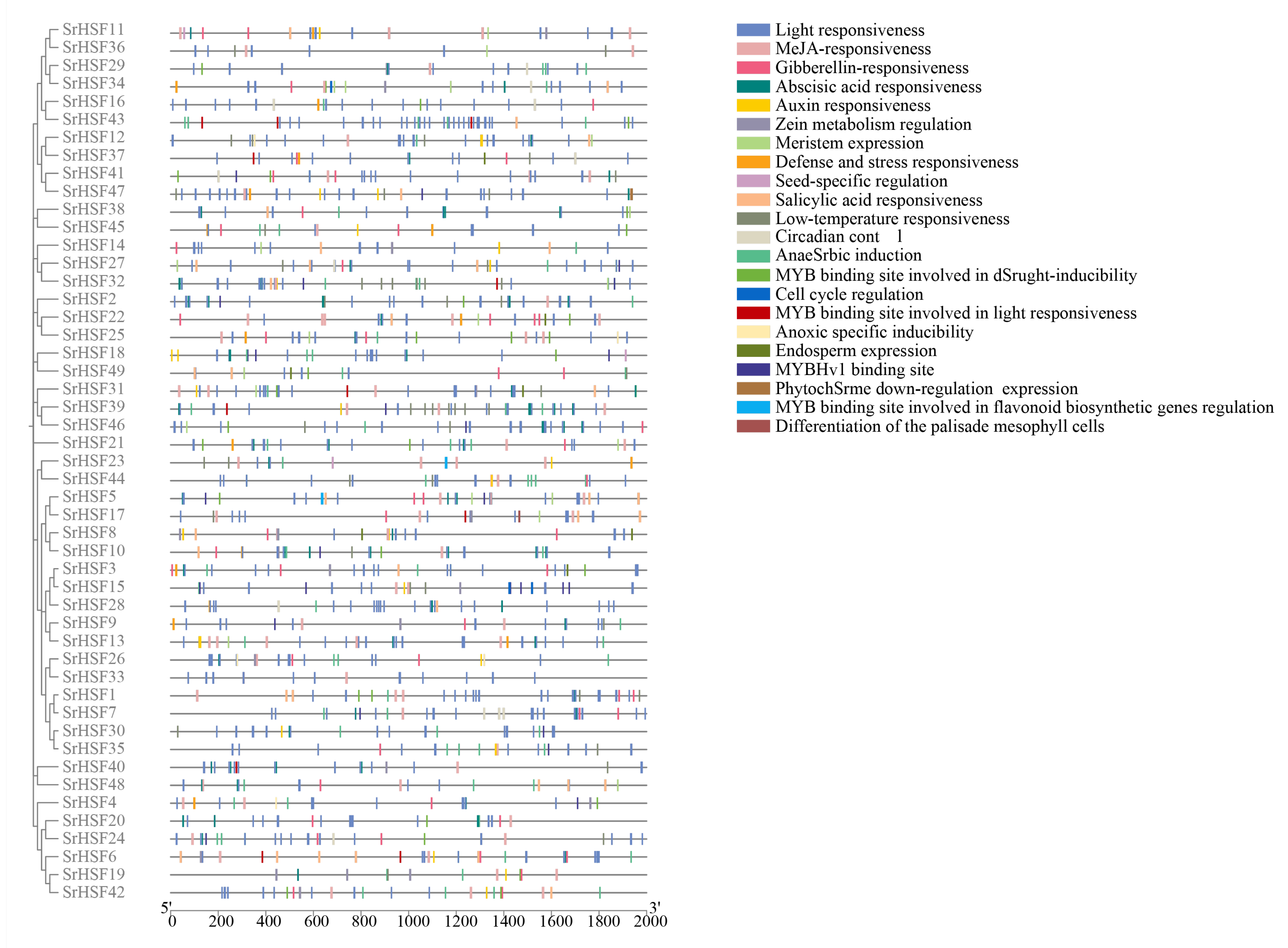
2.7. Prediction of SrHSF Promoter Functions
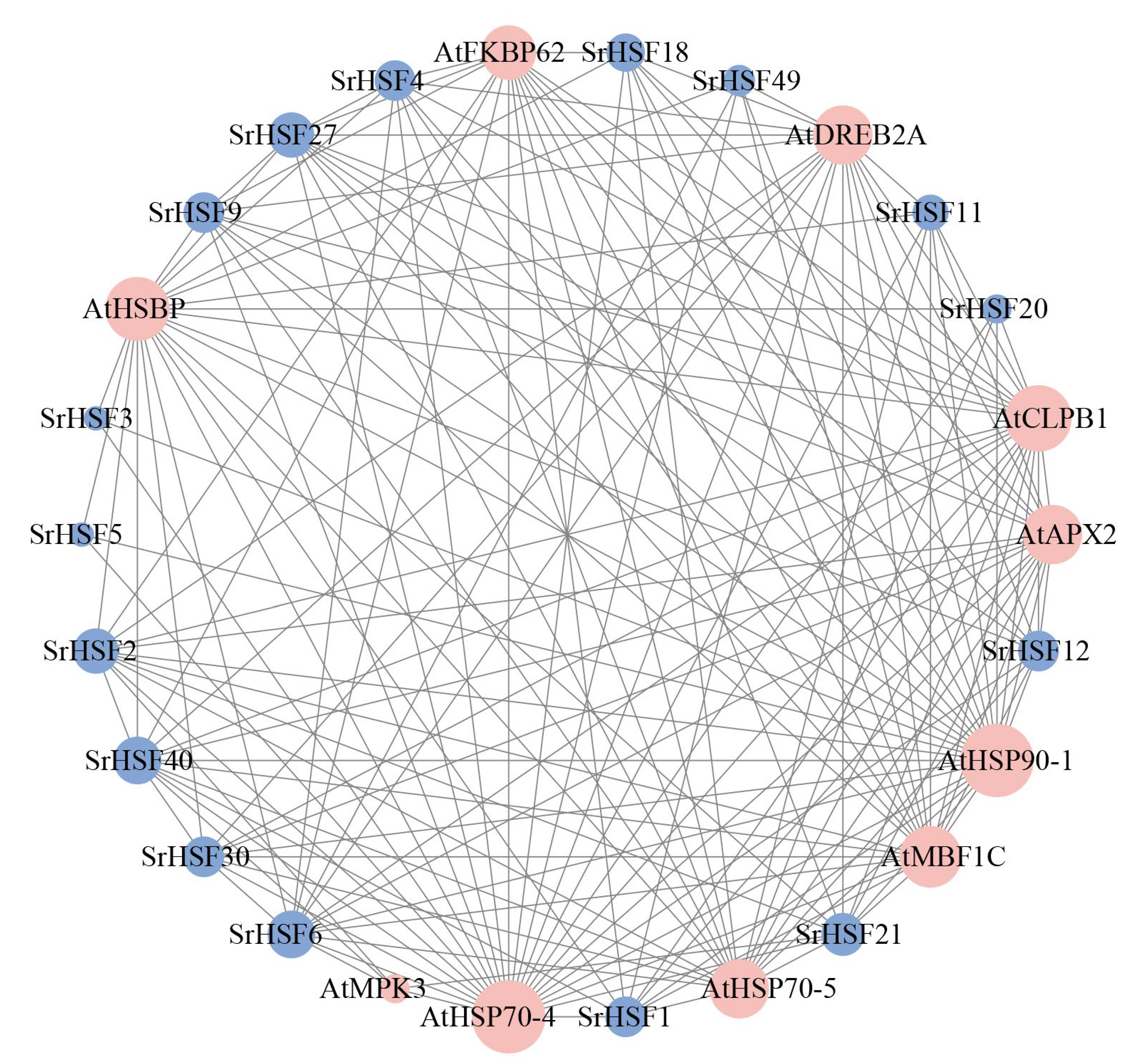
2.8. Structure and Interaction of SrHSF Proteins
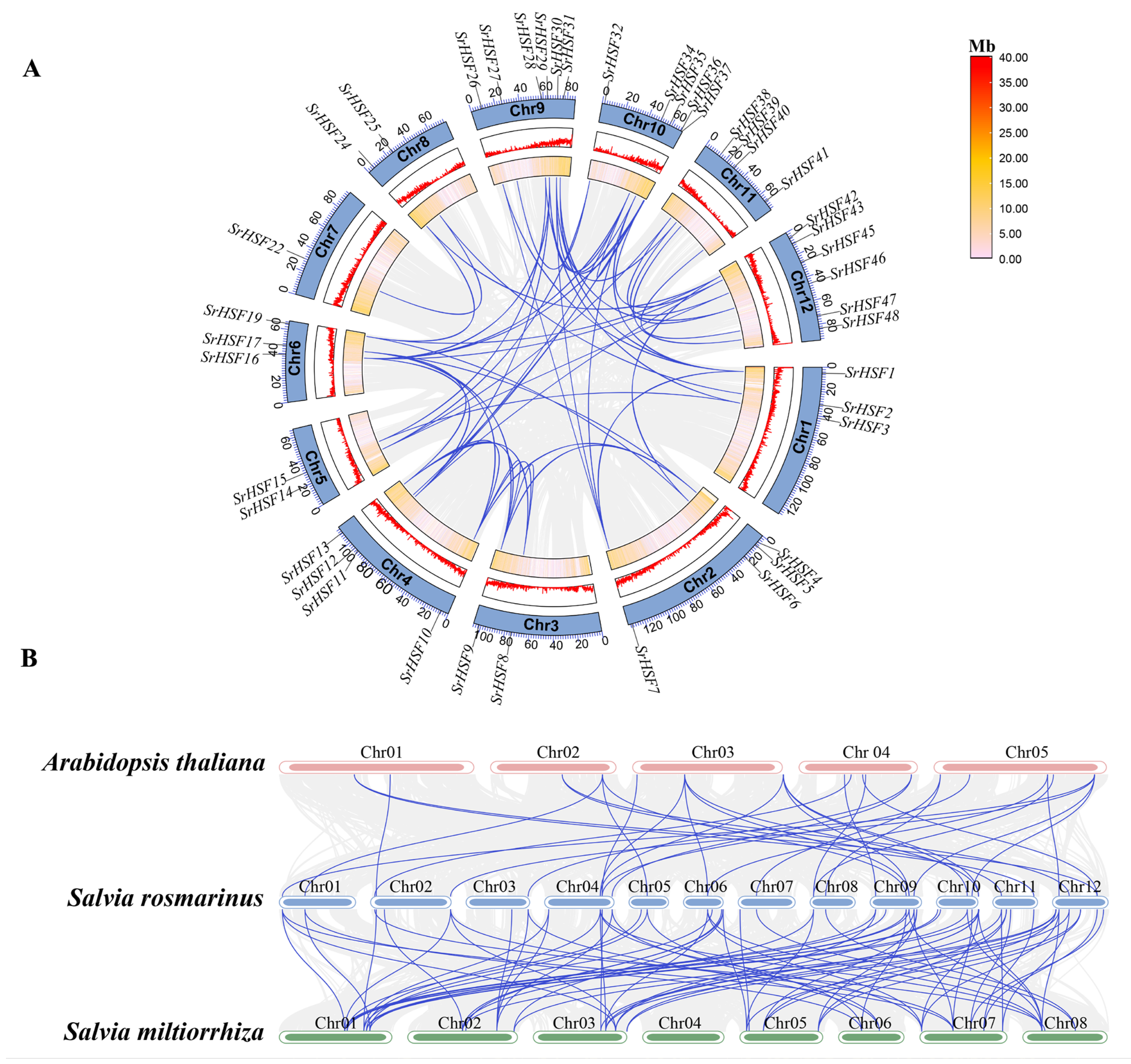
2.9. SrHSF Gene Duplication and Collinearity
2.10. Expression Pattern of SrHSF Genes
3. Results
3.1. Chromosomal Localization and Fundamental Information of SrHSF
3.2. Phylogenetic Characteristic of SrHSF Protein Sequences
3.3. SrHSF Gene Structure and Conserved Motifs
3.4. Promoter Function of SrHSF
3.5. Structure and Protein Interaction of SrHSF Proteins
3.6. Duplication Events and Collinearity of SrHSF Genes
3.7. Expression Pattern of SrHSF Genes in Different Tissues and Leaves Under MeJA Treatment
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Veenstra, J.P.; Johnson, J.J. Rosemary (Salvia rosmarinus): Health–promoting benefits and food preservative properties. Int. J. Nutr. 2021, 6, 1–10. [Google Scholar] [CrossRef]
- Faridzadeh, A.; Salimi, Y.; Deravi, N. Neuroprotective potential of aromatic herbs: Rosemary, sage, and lavender. Front. Neurosci. 2022, 16, 909833. [Google Scholar] [CrossRef] [PubMed]
- González-Minero, F.J.; Bravo–Díaz, L.; Ayala–Gómez, A. Rosmarinus officinalis L. (Rosemary): An ancient plant with uses in personal healthcare and cosmetics. Cosmetics 2020, 7, 77. [Google Scholar] [CrossRef]
- Hamann, E.; Denney, D.; DAY, S.; Lombardi, E.; Jameel, M.I.; MacTavish, R.; Anderson, J.T. Review: Plant eco-evolutionary responses to climate change: Emerging directions. Plant Sci. 2021, 304, 110737. [Google Scholar] [CrossRef]
- Kmiecik, S.W.; Mayer, M.P. Molecular mechanisms of heat shock factor 1 regulation. Trends Biochem. Sci. 2022, 47, 218–234. [Google Scholar] [CrossRef] [PubMed]
- Boopathy, L.R.A.; Jacob–Tomas, S.; Alecki, C.; Vera, M. Mechanisms tailoring the expression of heat shock proteins to proteostasis challenges. J. Biol. Chem. 2022, 298, 101796. [Google Scholar]
- Kmiecik, S.W.; Drzewicka, K.; Melchior, F.; Mayer, M.P. Heat shock transcription factor 1 is SUMOylated in the activated trimeric state. J. Biol. Chem. 2021, 296, 100324. [Google Scholar] [CrossRef]
- Jiang, D.; Xia, M.; Xing, H.; Gong, M.; Jiang, Y.; Liu, H.; Li, H.L. Exploring the heat shock transcription factor (HSF) gene family in Ginger: A genome-wide investigation on evolution, expression profiling, and response to developmental and abiotic stresses. Plants 2023, 12, 2999. [Google Scholar] [CrossRef]
- Kovács, D.; Kovács, M.; Ahmed, S.; Barna, J. Functional diversification of heat shock factors. Biol. Futur. 2022, 73, 427–439. [Google Scholar] [CrossRef]
- Haider, S.; Lqbal, J.; Naseer, S.; Shaukat, M.; Abbasi, B.A.; Yaseen, T.; Zahra, S.A.; Mahmood, T. Unfolding molecular switches in plant heat stress resistance: A comprehensive review. Plant Cell Rep. 2022, 41, 775–798. [Google Scholar] [CrossRef]
- Singh, G.; Sarkar, N.; Grover, A. Mapping of domains of heat stress transcription factor OsHsfA6a responsible for its transactivation activity. Plant Sci. 2018, 274, 80–90. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.L.; Wang, S.S.; Cai, J.; Zhang, T.; Yuan, D.D.; Li, Y. Genome–wide identification, phylogeny and expression analysis of Hsf gene family in Verbena bonariensis under low–temperature stress. BMC Genom. 2024, 25, 729. [Google Scholar] [CrossRef] [PubMed]
- Feng, N.; Feng, H.; Wang, S.; Puneka, A.S.; Ladenstein, R.; Wang, D.C.; Zhang, Q.; Ding, J.; Liu, W. Structures of heat shock factor trimers bound to DNA. IScience 2021, 24, 102951. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Wang, Z.; Jian, S. Functional characterization of heat shock factor (CrHSF) families provide comprehensive insight into the adaptive mechanisms of canavalia rosea (sw.) DC. to tropical coral islands. Int. J. Mol. Sci. 2022, 23, 12357. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Liu, Z.; Gichira, A.W.; Yang, M.; Mbichi, R.W.; Meng, L.; Wan, T. Deep evaluation of the evolutionary history of the Heat Shock Factor (HSF) gene family and its expansion pattern in seed plants. PeerJ 2022, 10, e13603. [Google Scholar] [CrossRef]
- Ma, L.; Gao, Z.; Wu, J.; Zhong, B.; Xie, Y.; Huang, W.; Lin, Y. Co–condensation between transcription factor and coactivator p300 modulates transcriptional bursting kinetics. Mol. Cell. 2021, 81, 1682–1689.e7. [Google Scholar] [CrossRef]
- Zhang, N.; Wang, Y.; Wang, Z.; Yue, Z.; Niu, Y. Heat shock transcription factor family in plants: A review. Chin. J. Biotechnol. 2021, 37, 1155–1167. [Google Scholar]
- Scharf, K.D.; Berberich, T.; Ebersberger, I.; Nover, L. The plant heat stress transcription factor (HSF) family: Structure, function and evolution. Biochim. Biophys. Acta 2012, 1819, 104–119. [Google Scholar] [CrossRef]
- Andrási, N.; Pettkó-Szandtner, A.; Szabados, L. Diversity of plant heat shock factors: Regulation, interactions, and functions. J. Exp. Bot. 2020, 72, 1558–1575. [Google Scholar] [CrossRef]
- Ul Haq, S.; Khan, A.; Ali, M.; Khattak, A.M.; Gai, W.X.; Zhang, H.X.; Wei, A.M.; Gong, Z.H. Heat shock proteins: Dynamic biomolecules to counter plant biotic and abiotic stresses. Int. J. Mol. Sci. 2019, 20, 5321. [Google Scholar] [CrossRef]
- Zhao, S.; Qing, J.; Yang, Z.; Tian, T.; Yan, Y.; Li, H.; Bai, Y. Genome-wide identification and expression analysis of the HSF hene family in Ammopiptanthus mongolicus. Curr. Issues Mol. Biol. 2024, 46, 11375–11393. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Zhang, R.; Zhou, J.; Du, J.; Li, X.; Zhang, Y.; Chen, Q.; Wang, Y.; Lin, Y.; Zhang, Y.; et al. Comprehensive analysis of HSF genes from celery (Apium graveolens L.) and functional characterization of AgHSFa6-1 in response to heat stress. Front. Plant Sci. 2023, 14, 1132307. [Google Scholar] [CrossRef] [PubMed]
- Bian, X.H.; Li, W.; Niu, C.F.; Wei, W.; Hu, Y.; Han, J.Q.; Lu, X.; Tao, J.J.; Jin, M.; Qin, H.; et al. A class B heat shock factor selected for during soybean domestication contributes to salt tolerance by promoting flavonoid biosynthesis. New Phytol. 2020, 225, 268–283. [Google Scholar] [CrossRef] [PubMed]
- Zou, X.W.; Yue, J.N.; Li, Z.Y.; Dai, L.Y.; Li, W. Functional analysis of heat shock transcription factor HSFA2b in rice regulating abiotic stress resistance. Biotechnol. Bull. 2024, 40, 90–98. [Google Scholar]
- Shah, Z.; Iqbal, A.; Khan, F.U.; Khan, H.U.; Durrani, F.; Ahmad, M.Z. Genetic manipulation of pea (Pisum sativum L.) with Arabidopsis’s heat shock factor HSFA1d improves ROS scavenging system to confront thermal stress. Genet. Resour. Crop Evol. 2020, 67, 2119–2127. [Google Scholar] [CrossRef]
- Sun, J.; Guo, C. Genome–wide identification and expression analysis of RR–TYPE MYB–Related transcription factors in tomato (Solanum lycopersicum L.). Horticulturae 2022, 8, 399. [Google Scholar] [CrossRef]
- Huang, Y.C.; Niu, C.Y.; Yang, C.R.; Jinn, T.L. The heat stress factor HSFa6b connects ABA signaling and ABA–Mediated heat responses. Plant Physiol. 2016, 172, 1182–1199. [Google Scholar] [CrossRef]
- Zhao, X.; Guo, Q.; Chen, Q.Q.; Li, B.l.; Zhang, D.Q. Isolation, expression and single nucleotide polymorphisms analysis of heat shock transcription factor Hsfa1d in populus. Sci. Silvae Sin. 2011, 47, 82–90. [Google Scholar]
- Majee, A.; Kumar, V.; Bano, N.; Kumari, A.; BagSane, S.K.; Sane, V.A. Elucidation of heat shock transcription factor family (HSFs) postulates significant insights for the identification of their putative roles in root development and hormonal regulation in tomato. J. Plant Growth Regul. 2023, 42, 2327–2344. [Google Scholar] [CrossRef]
- Yu, T.; Bai, Y.; Liu, Z.; Wang, Z.; Yang, Q.; Wu, T.; Feng, S.; Zhang, Y.; Shen, S.; Li, Q.; et al. Large-scale analyses of heat shock transcription factors and database construction based on whole-genome genes in horticultural and representative plants. Hortic. Res. 2022, 9, uhac035. [Google Scholar] [CrossRef]
- Tian, F.X.; Hu, X.L.; Yao, T.; Yang, X.H.; Chen, J.G.; Lu, M.Z.; Zhang, J. Recent Advances in the Roles of HSFs and HSPs in Heat Stress Response in Woody Plants. Front Plant Sci. 2021, 12, 704905. [Google Scholar] [CrossRef]
- Xu, P.; Guo, Q.W.; Pang, X.; Zhang, P.; Kong, D.J.; Liu, J. New insights into evolution of plant heat shock factors (HSFs) and expression analysis of tea genes in response to abiotic stresses. Plants 2020, 9, 311. [Google Scholar] [CrossRef] [PubMed]
- Yurina, N. Heat shock proteins in plant protection from oxidative stress. Mol. Biol. 2023, 57, 951–964. [Google Scholar] [CrossRef]
- Han, D.L.; Li, W.L.; Hou, Z.W.; Lin, C.F.; Xie, Y.; Zhou, X.F.; Gao, Y.; Huang, J.W.; Lai, J.B.; Wang, L.; et al. The chromosome–scale assembly of the Salvia rosmarinus genome provides insight into carnosic acid biosynthesis. Plant J. 2023, 113, 819–832. [Google Scholar] [CrossRef] [PubMed]
- Lai, Y.; Ma, J.H.; Zhang, X.B.; Xuan, X.B.; Zhu, F.Y.; Ding, S.; Shang, F.D.; Chen, Y.Y.; Zhao, B.; Lan, C.; et al. High–quality chromosome–level genome assembly and multi–omics analysis of rosemary (Salvia rosmarinus) reveals new insights into the environmental and genome adaptation. Plant Biotechnol. J. 2024, 22, 1833–1847. [Google Scholar] [CrossRef]
- Zhao, M.X.; Hu, X.J. The research progress of heat shock transcription factors in plants. Bot. Res. 2018, 7, 158–163. [Google Scholar]
- Qu, R.J.; Wang, S.W.; Wang, X.X.; Peng, J.M.; Guo, J.; Cui, G.H.; Chen, M.L.; Mu, J.; Lai, C.J.S.; Huang, L.Q.; et al. Genome–wide characterization and expression of the HSF gene family in Salvia miltiorrhiza (Danshen) and the potential thermotolerance of SmHSF1 and SmHSF7 in yeast. Int. J. Mol. Sci. 2023, 24, 8461. [Google Scholar] [CrossRef]
- Dossa, K.; Diouf, D.; Cissé, N. Genome–wide investigation of HSF genes in Sesame reveals their segmental duplication expansion and their active role in drought stress response. Front. Plant Sci. 2016, 7, 1522. [Google Scholar] [CrossRef]
- Ling, C.C.; Liu, Y.Y.; Yang, Z.C.; Xu, J.L.; Ouyang, Z.Y.; Yang, J.; Wang, S.H. Genom–wide identification of HSF gene family in kiwifruit and the function of AeHSFa2b in salt tolerance. Int. J. Mol. Sci. 2023, 24, 15638. [Google Scholar] [CrossRef]
- Lohani, N.; Golicz, A.A.; Singh, M.B.; Bhalla, P.L. Genome–wide analysis of the HSF gene family in Brassica oleracea and a comparative analysis of the HSF gene family in B. oleracea, B. rapa and B. napus. Funct. Integr. Genom. 2019, 19, 515–531. [Google Scholar] [CrossRef]
- Wang, L.L.; Liu, Y.H.; Chai, G.F.; Zhang, D.; Fang, Y.Y.; Deng, K.; Aslam, M.H.; Niu, X.P.; Zhang, W.B.; Qin, Y.; et al. Identification of passion fruit HSF gene family and the functional analysis of PeHSFC1a in response to heat and osmotic stress. Plant Physiol. Biochem. 2023, 200, 107800. [Google Scholar] [CrossRef] [PubMed]
- Liao, W.; Tang, X.; Li, J.; Zheng, Q.; Wang, T.; Cheng, S.; Chen, S.; Cao, S.; Cao, G. Genome wide investigation of HSF gene family in Phoebe bournei: Identification, evolution, and expression after abiotic stresses. J. For. Res. 2024, 35, 11. [Google Scholar] [CrossRef]
- Li, P.S.; Yu, T.F.; He, G.H.; Chen, M.; Zhou, Y.B.; Chai, S.C.; Xu, Z.S.; Ma, Y.Z. Genome–wide analysis of the HSF family in soybean and functional identification of GmHSF34 involvement in drought and heat stresses. BMC Genom. 2014, 15, 1009. [Google Scholar] [CrossRef] [PubMed]
- Xiong, H.; Wang, D.; Shao, C.; Yang, X.; Yang, J.; Ma, T.; Davis, C.C.; Liu, L.; Xi, Z. Species tree estimation and the impact of gene loss following whole–genome duplication. Syst. Biol. 2022, 71, 1348–1361. [Google Scholar] [CrossRef]
- Huang, B.; Huang, Z.; Ma, R.; Chen, J.; Zhang, Z.; Yrjälä, K. Genome–wide identification and analysis of the heat shock transcription factor family in moso bamboo (Phyllostachys edulis). Sci. Rep. 2021, 11, 16492. [Google Scholar] [CrossRef]
- Wang, J.; Cui, Y.P.; Li, S.Y.; Gao, X.Q.; Zhang, K.P.; Shen, X.L. Transcriptome analysis of Artemisia argyi following methyl jasmonate (MeJA) treatment and the mining of genes related to the stress resistance pathway. Front. Genet. 2023, 14, 1279850. [Google Scholar] [CrossRef]
- Gong, C.; Pang, Q.; Li, Z.; Li, Z.; Chen, R.; Sun, G.; Sun, B. Genome–wide identification and characterization of HSF and Hsp gene families and gene expression analysis under heat stress in eggplant (Solanum melongema L.). Horticulturae 2021, 7, 149. [Google Scholar] [CrossRef]
- Zhang, J.; Liu, B.B.; Li, J.B.; Zhang, L.; Wang, Y.; Zheng, H.Q.; Lu, M.Z.; Chen, J. HSF and Hsp gene families in Populus: Genome–wide identification, organization and correlated expression during development and in stress responses. BMC Genom. 2015, 16, 181. [Google Scholar] [CrossRef]
- Begum, T.; Reuter, R.; Schöffl, F. Overexpression of AtHSFB4 induces specific effects on root development of Arabidopsis. Mech. Dev. 2013, 130, 54–60. [Google Scholar] [CrossRef]
- Garrido–Bigotes, A.; Figueroa, P.M.; Figueroa, C.R. Jasmonate metabolism and its relationship with abscisic acid during strawberry fruit development and ripening. J. Plant Growth Regul. 2018, 37, 101–113. [Google Scholar] [CrossRef]
- Munemasa, S.; Mori, I.C.; Murata, Y. Methyl jasmonate signaling and signal crosstalk between methyl jasmonate and abscisic acid in guard cells. Plant Signal. Behav. 2011, 6, 939–941. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Zhang, X. Genome–wide dynamic network analysis reveals the potential genes for MeJA-induced growth-to-defense transition. BMC Plant Biol. 2021, 21, 450. [Google Scholar] [CrossRef] [PubMed]
- Huan, C.; Yang, X.; Wang, L.; Kebbeh, M.; Wang, Y.; Dai, B.; Zhou, H. Methyl jasmonate treatment regulates α–linolenic acid metabolism and jasmonate acid signaling pathway to improve chilling tolerance in both stony hard and melting flesh peaches. Postharvest Biol. Technol. 2022, 190, 111960. [Google Scholar] [CrossRef]
- Ding, C.K.; Wang, C.Y.; Gross, K.C.; Smith, D.L. Reduction of chilling injury and transcript accumulation of heat shock proteins in tomato fruit by methyl jasmonate and methyl salicylate. Plant Sci. 2001, 161, 1153–1159. [Google Scholar] [CrossRef]
- Nie, G.; Zhou, J.; Jiang, Y.; He, J.; Wang, Y.; Liao, Z.; Appiah, C.; Li, D.; Feng, G.; Huang, L.; et al. Transcriptome characterization of candidate genes for heat tolerance in perennial ryegrass after exogenous Methyl jasmonate application. BMC Plant Biol. 2022, 22, 68. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cui, W.; Xu, Z.; Kong, Y.; Yang, L.; Dou, H.; Zhang, D.; Li, M.; Chen, Y.; Ding, S.; Yang, C.; et al. Genome-Wide Identification of the Heat Shock Transcription Factor Gene Family in Rosemary (Salvia rosmarinus). Horticulturae 2024, 10, 1250. https://doi.org/10.3390/horticulturae10121250
Cui W, Xu Z, Kong Y, Yang L, Dou H, Zhang D, Li M, Chen Y, Ding S, Yang C, et al. Genome-Wide Identification of the Heat Shock Transcription Factor Gene Family in Rosemary (Salvia rosmarinus). Horticulturae. 2024; 10(12):1250. https://doi.org/10.3390/horticulturae10121250
Chicago/Turabian StyleCui, Weitong, Zongle Xu, Yuhua Kong, Lin Yang, Hao Dou, Dangquan Zhang, Mingwan Li, Yuanyuan Chen, Shen Ding, Chaochen Yang, and et al. 2024. "Genome-Wide Identification of the Heat Shock Transcription Factor Gene Family in Rosemary (Salvia rosmarinus)" Horticulturae 10, no. 12: 1250. https://doi.org/10.3390/horticulturae10121250
APA StyleCui, W., Xu, Z., Kong, Y., Yang, L., Dou, H., Zhang, D., Li, M., Chen, Y., Ding, S., Yang, C., & Lai, Y. (2024). Genome-Wide Identification of the Heat Shock Transcription Factor Gene Family in Rosemary (Salvia rosmarinus). Horticulturae, 10(12), 1250. https://doi.org/10.3390/horticulturae10121250




