Abstract
The circadian clock plays a vital role in facilitating plant adaptation to rhythmically changing environmental factors. Pseudo-response regulators (PRRs) are key components regulating the plant circadian clock and have been extensively characterized in model plants. However, the PRRs in the tea plant have not been comprehensively studied. In this study, seven CsPRRs were cloned from the tea plant. Domain, phylogenetic evolution, gene structure, motifs, and cis-acting element analysis revealed their sequence characters and suggested that the first subgroup members, CsPRR1a, 1b, 5a, 5b, 7, and 37, may be responsible for circadian rhythm regulation and abiotic stress responses, while the second subgroup member, CsPRR2, may be involved in development and chloroplast function regulation. Most CsPRRs showed relatively higher expression in flowers, implying their potential roles in photoperiod-regulated flower induction. Moreover, rhythmic expression of CsPRR7, 5b, 5a, 37, 1b, and 1a was observed under long-day conditions in a sequential manner. Additionally, CsPRRs were differently induced/inhibited by cold, heat, and drought stresses in tissue-specific and photoperiod-related manners. A stronger cold induction of CsPRRs was observed under long-day conditions than under short-day conditions. And, among the two tested tissues, changes in the expression of CsPRRs caused by various stresses were more obvious in young shoots. Studies using a floriferous cultivar (FDDB) and an oliganthous cultivar (PYTZ) implied that CsPRRs also played crucial roles in tea-plant flower induction. This study presents the first comprehensive analysis of CsPRRs in the tea plant, providing vital information for further elucidation of CsPRR functions. It also suggests that tissue type and photoperiod conditions should be taken into consideration when conducting gene function studies in the tea plant.
1. Introduction
Plants have evolved complex and sophisticated signal systems in order to adapt to daily and seasonal cycles. Circadian clocks are ubiquitous endogenous time-keeping machineries that respond to the rhythmically changing environmental factors caused by the Earth’s rotation [,,]. The molecular system of the plant’s circadian clock consists of input pathways, core oscillators, and output pathways []. Among these three parts, the core oscillators, which are composed of multiple transcriptional–translational feedback loops, have been extensively characterized [,]. CIRCADIAN CLOCK ASSOCIATED1 (CCA1), LATE ELONGATED HYPOCOTYL (LHY), and PSEUDO-RESPONSE REGULATORS (PRRs) have been proposed as key components of this network []. CCA1 and LHY proteins are MYB (myeloblastosis) transcription factors with a transcription peak at dawn []. In contrast, PRRs belong to the two-component system, containing the complete receiver domains but lacking the conserved aspartic acid residues for phosphorylation in the receiver domain compared with the authentic response regulators (ARRs) []. In Arabidopsis, PRRs, also called APRRs, include nine members, from APRR1 (also known as TIMING OF CAB EXPRESSION 1, TOC1) to APRR9 []. Moreover, ten BoPRRs have been identified in cabbage [], nine ZmPRRs in maize [] and four of the MtPRR family in Medicago truncatula []. APRRs can be further divided into two groups according to the type of domain at their C-terminus []. The first subgroup consists of typical PRR proteins with a pseudo-receiver (Pseudo-Rec) domain at the N-terminus and a CONSTANS/CONSTANS-LIKE (CCT) domain at the C-terminus [,]. This subgroup consists of five members, which are APRR1 (TOC1), 3, 5, 7, and 9, and it has been proposed that they are involved in circadian clock regulation. Most studies that refer to PRRs have focused on this group. The rice genome encodes five clock-associated PRR genes [], and four PpPRRs have been identified in Moss Physcomitrella patens []. The second subgroup contains APRR2, 4, and 6, with mostly unknown functions, and they possess a Pseudo-Rec domain at the N-terminus and an MYB-related DNA-binding domain at the C-terminus []. Although the distribution and function of APRR8 are unclear because only a Pseudo-Rec domain has been detected in its N-terminus, neither a CCT nor an MYB domain has been detected in its C-terminus []. Moreover, a sequence analysis revealed that APRR2 also possesses a conserved GCT box (Myb DNA-binding domain) that is only found in GLK (Golden2-Like) proteins, which provided ideas for studies on its function [].
The expression and function of PRRs have been well studied in Arabidopsis. APRR9, 7, 5, 3, and 1 are expressed in a sequential and overlapping order from early daytime to around midnight []. APRR9 showed the highest expression at dawn, APRR7 reached its peak in the morning, APRR5 reached its peak around noon, and APRR3 and 1 reached their peak in the evening []. The regulators of APRRs, CCA1 and LHY, which show the highest expression at dawn, can inhibit APRR9, 7, and 5 and APRR1 (TOC1) transcription []. In turn, APRRs (APRR9, 7, and 5) can also repress the expression of CCA1 and LHY by complexing with TOPLESS/TOPLESS-RELATED (TPL/TPR), members of the plant Groucho/TUP1 corepressor family []. Clock-associated PRRs have been proven to be involved in a wide array of plant adaptation processes, such as flowering induction, photosynthesis, and various biotic- and abiotic-stress responses [,]. Genetic studies found that the loss of function of a single PRR in the first subgroup only led to subtle phenotypes, suggesting a functional redundancy among PRRs []. However, the prr5prr7prr9 triple mutant (prr579) showed photoperiod insensitivity; a delayed-flowering-time phenotype; and stronger cold, drought, and high-salinity stresses under specific photoperiodic conditions [,]. In Arabidopsis, PRRs (TOC1 and APRR5) were found to coordinate with the evening complex (EC) to directly repress PIF4 and PIF5 (phytochrome-interacting factors 4 and 5) expression in order to regulate photoperiodic hypocotyl growth []. GmPRR37, a soybean pseudo-response regulator protein was identified through quantitative trait locus mapping. Its overexpression led to delayed flowering under long-photoperiod conditions, but no significant flowering time changes under short-day conditions []. Moreover, APRR9, 7, and 5 were proposed to directly repress the expression of genes in the clock output pathway, including the CYCLING DOF FACTOR (CDF) genes involved in flowering time control and the DREB1/CBF genes involved in cold-stress responses [,]. In Arabidopsis, APRR2 was found to act as a positive regulator of plant immunity by influencing salicylic acid (SA) biosynthesis and SA signaling responses []. SlPRR2, an ortholog of APRR2, was demonstrated to be associated with pigment accumulation in tomato and pepper []. Cucumber CsaPRR2 is a nuclear-localized protein, and it was proven to be involved in green immature-fruit-color regulation []. Even though the functions of APRR4, 6, and 8 and their homologs remain unknown, the abovementioned findings imply that the first subgroup of PRRs is mainly responsible for rhythm regulation, while the second subgroup of PRRs may play crucial roles in plant development and immunity. Collectively, PRRs play vital roles in plant development and fitness in changing environments.
The tea plant (Camellia sinensis (L.) O. Kuntze) is an important perennial evergreen and economically important crop in many countries, because of the remarkable composition of flavonoids, caffeine, and theanine in its leaves []. The tea plant is native to Southeast Asia and has been proposed to be susceptible to low temperatures during its growth and development []. Studies have proposed that temperature cues also act as important signals in entraining the circadian clock through a mechanism called temperature compensation. However, the knowledge of clock-specific thermosensors is limited. In Arabidopsis, the loss of function of APRR7 and APRR9 leads to seedlings unable to entrain the clock in response to warm–cold cycles, indicating that APRRs are also essential for the synergy of temperature and clock signals []. However, to date, most studies on PRRs have been carried out on the model plant Arabidopsis, and the expression and potential function of PRRs in other plants, especially in woody plants such as the tea plant, remain unclear. In the present study, we carried out a comprehensive identification of the CsPRR family in the tea plant; we conducted analyses of their phylogenetic evolution, sequence characteristics, and promoter cis-acting elements, and we examined their expression in particular tissues and under various photoperiod and abiotic-stress conditions. We found that the rhythmic expression of all CsPRRs, except for CsPRR2, was only exhibited under long-day conditions. Moreover, we also found that the cold, heat, and drought induction of CsPRRs was tissue-specific, with cold induction also being photoperiod-related. Overall, our study provides valuable information on PRR evolution and function elucidation in the tea plant. It is also suggested that photoperiod conditions and tissue type be taken into consideration when conducting gene function studies, especially on cold-related genes.
2. Materials and Methods
2.1. Plant Materials and Treatments
Two tea-plant cultivars, ‘Longjing 43’ (LJ43) and ‘Zhongcha 108’ (ZC108), grown in the field or a growth chamber at the Tea Research Institute, Chinese Academy of Agricultural Sciences (TRI, CAAS, N 30°100, E120°50), Hangzhou, China, were used as materials. To detect the tissue-specific expression of target genes, the apical buds, young leaves, mature leaves, young stems, mature stems, flowers, seeds, and roots of the ZC108 plants were collected. The LJ43 plants were used as materials for various photoperiod and stress treatments. The parameters of the growth chamber were set to 25 °C/22 °C (light/dark) and 65% relative humidity. Four different photoperiod treatments, consisting of long-day (16 h/8 h) (light/dark), short-day (8 h/16 h) (light/dark), constant light, and constant dark conditions, were carried out. Mature leaves were collected every 4 h for 48 h, starting from 5:00 am. The stress treatments were based on our previous studies with slight modifications [,]. For cold stress, plants grown under long-day (16 h/8 h) (light/dark) or short-day (8 h/16 h) (light/dark) conditions were treated with 4 °C/2 °C (light/dark). For drought treatments, 5% (w/v) PEG 6000 (Sigma-Aldrich, Shanghai, China) solution and its control ddH2O was used under conditions of 14h light 25 °C/10 h dark 22 °C (light/dark) and 65% relative humidity. For high-temperature treatment, conditions of 14h light 38 °C/10 h dark 36 °C (light/dark) and 65% relative humidity were used. After the stress treatments, the young shoots (the apical buds and the first two leaves) and mature leaves (the fourth leaf) were collected at the indicated time points and stored at −80 °C until use. Axillary buds of a floriferous cultivar ‘Fudingdabai’ (FDDB) and an oliganthous cultivar ’Pingyangtezao’ (PYTZ) were collected and used to explore the potential relationship between CsPRRs expression and flower induction in tea plants.
2.2. Data Collection and Gene Identification
To obtain the CsPRRs in the tea plant, the genomes of four Camellia sinensis var. sinensis (CSS)-type cultivars, namely, ‘Longjing43’(LJ43), ‘Tieguanyin’ (TGY), ‘huangdan’ (HD), and ‘shuchazao’ (SCZ), were used as a database [,,], and the amino acid sequences of the known PRRs in Arabidopsis, soybean, and rice were used as the query. Blastp searching was carried out using the local blast platform (Blast + 2.13.0) with the parameter e-value cut-off > 1 × 10−5. Then, sequence alignments were conducted to remove duplicated sequences; after that, the unique sequences were submitted to the online tool InterPro (https://www.ebi.ac.uk/interpro/, accessed on 20 March 2023) for a domain analysis. Finally, sequences containing the Pseudo-Rec domain at the N-terminus and the CCT or MYB-like domain at the C-terminus were defined as CsPRRs. The identified CsPRRs were named according to their homologies with APRRs.
2.3. Phylogenetic Analysis
To determine the subgroup classification of the CsPRRs identified from ‘LJ43’ cultivar, a neighbor-joining tree (NJ) was constructed based on 45 PRR protein sequences using MEGA11 software (v. 11.0.13) with 1000 bootstrap replications. The evolutionary distances were computed using the p-distance method and are presented in the units of the number of amino acid differences per site []. Then, the result of the NJ tree was modified using Adobe illustrator software (v. 26.2.1).
2.4. Gene Structure, Motif, and Promoter Analyses
For an exon/intron distribution analysis, the genome sequences and coding sequences of the CsPRRs identified from ‘LJ43’ cultivar were applied in Gene Structure Display Server 2.0 (GSDS 2.0, http://gsds.cbi.pku.edu.cn/, accessed on 12 April 2023). The Multiple Em for Motif Elicitation (MEME, https://meme-suite.org/meme/, accessed on 15 April 2023) online tool was used to detect the top 10 motifs with default parameters. For a cis-acting element analysis, the 2000 bp upstream sequences from the translational start site (ATG) were retrieved from the LJ43 genome and submitted to the PLACE database (https://www.dna.affrc.go.jp/PLACE/, accessed on 18 April 2023). The subcellular location prediction of the CsPRR proteins was conducted using the online tool UniProt (https://www.uniprot.org, accessed on 15 April 2023).
2.5. RNA Isolation and RT-qPCR
The total RNA of the collected samples was extracted using a FastPure Plant Total RNA Isolation Kit (RC401-01, Vazyme, Nanjing, China), following the instructions. A PrimeScript™ RT reagent kit (RR037A, TaKaRa, Dalian, China) was used for first-strand cDNA synthesis with 1 µg of total RNA. RT-qPCR was conducted on a Light Cycler 480 (Roche, Mannheim, Germany) platform with SYBR Green I Master Mix (04913914001, Roche, Mannheim, Germany) in a 10 µL reaction volume. The amplification was performed at 95 °C for 10 s, 40 cycles at 95 °C for 10 s, then 58 °C for 45 s. The melt curve analysis was performed immediately after the completion of qRT-PCR to assess non-specific amplification. The relative gene expression was calculated using the comparative Ct value (2−∆ct) method, with normalization to CsEF1-α (elongation factor 1-alpha) [,]. Gene expression data without scale or transformation were applied to heatmap generating using TBtools (v. 1.6). GraphPad Prism 9 (v. 9.5.0) was used to plot the bar charts of gene expression and statistical analysis. Student’s t-tests were used for statistical significance analysis. Primers and their amplification efficiency are listed in Supplementary Table S1.
3. Results
3.1. Identification and Characterization of CsPRR Gene Family in Tea Plant
To systematically identify the CsPRR genes in the tea plant, comprehensive data retrieval was conducted using CSS-type genomes. A total of 14, 9, 16, and 18 putative targets were identified in the LJ43, HD, TGY, and SCZ genomes, respectively. Then, a pairwise comparison, domain detection, and a homologous analysis with Arabidopsis APRR genes were conducted to eliminate repeated sequences. Finally, seven CsPRRs with 363–580 amino acid residues were cloned from the tea plant variety ’LJ43’ (Table 1 and Supplementary Data S1). Molecular weight and theoretical isoelectric point (pI) predictions revealed that their protein size ranged from 61.59 kDa to 86.46 kDa and that their pI was 5.06–7.83, respectively. A chromosome (chr) location analysis showed that CsPRR1a was located on chr13, CsPRR1b was located on chr14, CsPRR2 was located on chr12, CsPRR5a was located on chr1, CsPRR5b and CsPRR37 were located on chr5, and CsPRR7 was located on chr9. The domain analysis revealed that all seven CsPRRs possessed the Pseudo-Rec domain at their N-terminus and, thus, were defined as PRRs. Further analysis showed that CsPRR1a, 1b, 5a, 5b, 7 and 37 also possessed CCT at their C-terminus, and CsPRR2 possessed a conserved GCT box only encountered in GLK (Golden2-Like) proteins. Subcellular localization prediction revealed that seven CsPRR proteins were mainly located in the nucleus, which provided a foundation for their transcriptional-regulation gene functions.

Table 1.
Basic bioinformatic information of CsPRR genes cloned from tea plant ‘Longjing43’ variety.
3.2. Phylogenetic Analysis of CsPRR Gene Family in Tea Plant and Other Species
An NJ tree was constructed to investigate the evolutionary relationships of the CsPRRs, and several GLK sequences were added because of their close relationship with the second subgroup of PRRs. As shown in Figure 1, 45 proteins from Arabidopsis (APRR or At), tea plant (Cs), rice (Os), grape (Vv), poplar (Pt), cucumber (Csa), tomato (Sl), pepper (Caa), soybean (Gm), and kiwi fruit (Ar) were divided into two main subgroups. Similar to previously reported studies, the first subgroup consisted of 29 members, and all of them were homologous to APRR1, 3, 5, 7, and 9, which function as core regulators of the circadian clock. The second subgroup included 16 proteins homologous to APRR2, 4, 6, and 8 and AtGLK. For the tea plant, the first subgroup consisted of six members, CsPRR1a, 1b, 5a, 5b, 7 and 37, while the second subgroup consisted of only CsPRR2. Compared with Arabidopsis, our study did not find homologs of APRR4, 6, 8, and 9 in the tea plant. Interestingly, CsPRR2 was clustered together with GLK proteins, except for VvGLK1; this was consistent with the domain analysis, which revealed that CsPRR2 and GLKs shared a conserved GCT box. Overall, the phylogenetic analysis, combined with the domain analysis, provides basic information for further function studies.
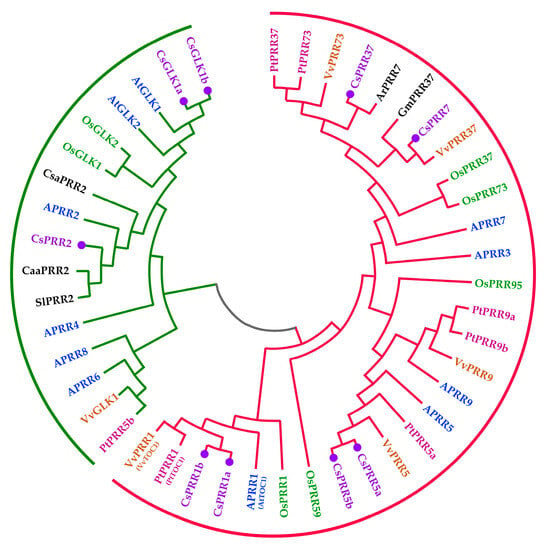
Figure 1.
Phylogenetic analysis of CsPRRs in the tea plant. The tree was constructed based on 45 PRR and GLK protein sequences using the neighbor-joining (NJ) method, with the no. of bootstrap replications set to 1000. Different color fronts represent PRR proteins from Arabidopsis (APRR), tea plant (Cs), rice (Os), grape (Vv), poplar (Pt), cucumber (Csa), tomato (Sl), pepper (Caa), soybean (Gm), and kiwi fruit (Ar).
3.3. Gene Structure and Motif Composition Analysis of CsPRR Genes in Tea Plant
To gain better insights into the sequence characteristics of the CsPRRs, the gene structures and top 10 motifs were analyzed using the GSDS2.0 and MEME online tools, respectively. As is shown in Figure 2a, the gene size of the CsPRRs ranged from 3927 bp to 25,352 bp, and the exon number ranged from 4 to 11. CsPRR1a possessed the minimum number of exons, while CsPRR2 possessed the maximum number of exons. Compared with Arabidopsis, the exon number of APRRs ranged from 5 to 10, and, similarly, APRR2 possessed the maximum number of exons (Figure 2a). The analysis of the top 10 motifs (sequences are listed in Supplementary Figure S1) revealed that CsPRR1a possessed only motifs 6 and 8, and CsPRR5a and 5b showed the same motif distribution patterns, with both of them possessing eight motifs each (Figure 2b). Further analysis revealed that motifs 1 and 6 were detected in all CsPRRs, except for CsPRR1a. Interestingly, we also analyzed the motif composition of CsGLKs (Supplementary Data S2) and found that CsPRR2 shared a similar motif distribution with CsGLKs, with both of them having a unique motif, motif 4.
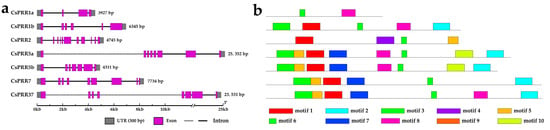
Figure 2.
Gene structure and motif analysis of CsPRR genes. (a) Gene structure of CsPRRs, showing 300 bp UTR (gray boxes), introns (lines), and exons (purple box). The black line below indicates the nucleic acid length. (b) Conserved motif distribution on CsPRR sequences (E values < 10−5). Various motifs are represented by different-colored boxes.
3.4. Cis-Acting Element Analysis of CsPRR Genes in Tea Plant
Cis-acting elements play key roles in gene expression regulation at the transcriptional level. Therefore, 2000 bp upstream sequences from the translation start site were applied in the PlantCARE online database to determine the compositions of the cis-acting elements in the CsPRR promoter regions. In total, 66 types of known and 8 types of unnamed cis-acting elements were detected (Figure 3). The identified cis-acting elements could be classified into five different categories: transcription-related, light-responsive, phytohormone-responsive, defense- and stress-responsive, and plant development and metabolism-regulating elements. Interestingly, in addition to gene transcription-related elements (TATA-box and CAAT-box) and various light-responsive elements (i.e., G-Box, Box 4, and TCT-motif), which were widely distributed in all CsPRR promoters, an anaerobic induction-related element, ARE, was also found. Six types of phytohormone-responsive elements were detected in the CsPRR promoters, including TGA-element for auxin-responsive, ABRE for abscisic acid-responsive, TATC-box for gibberellin-responsive, TCA-element for salicylic acid-responsive, and CGTCA-motif for MeJA-responsive elements. In addition, ABRE was the most widely distributed, TCA-element was only detected in CsPRR2 and CsPRR5a, and TGA-element was only detected in CsPRR1a and CsPRR5a (Figure 3). Defense- and stress-responsive-related elements, such as LTR (low-temperature responsiveness) and MBS (MYB binding site involved in drought inducibility), were found in CsPRR1b and CsPRR2, implying their involvement in cold and drought responses. As mentioned previously, the first subgroup of PRRs plays key roles in plant circadian clock regulation. However, the circadian control-related cis-acting regulatory element was only detected in CsPRR37, suggesting a much more complicated circadian rhythm-regulation network in the tea plant.
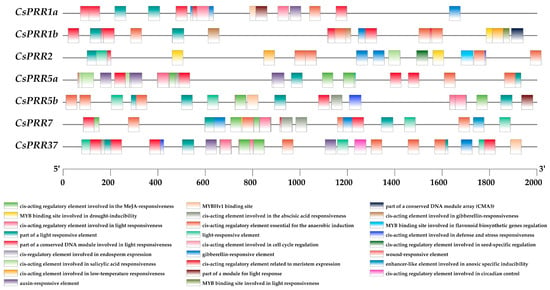
Figure 3.
Cis-acting element analysis of CsPRR genes. Colored boxes represent different cis-acting elements, and the black line below indicates the promoter length.
3.5. Tissue-Specific Expression of CsPRR Genes
qRT-PCR was conducted to investigate the tissue-specific expression of the CsPRRs. As shown in Figure 4, the lowest transcription of CsPRRs was mostly found in the roots, except for CsPRR5b, which was in the mature stems. In contrast, the highest expression of CsPRRs was observed mainly in the flowers, except for CsPRR2, which was in the mature stems. CsPRR5b, 7, and 37 expressed relatively higher than other members and with a relatively higher expression than other members. CsPRR1a, 1b and 5a clustered together, and all showed higher expression in flowers and mature leaves. Regarding the only member of the second subgroup, CsPRR2, higher expression was observed in the mature stems, buds, seeds, and flowers. Interestingly, the clustering patterns of these seven CsPRRs in various tissues were similar to the distribution pattern of the NJ tree generated via a phylogenetic analysis, suggesting that the genes in the same clusters have similar functions.
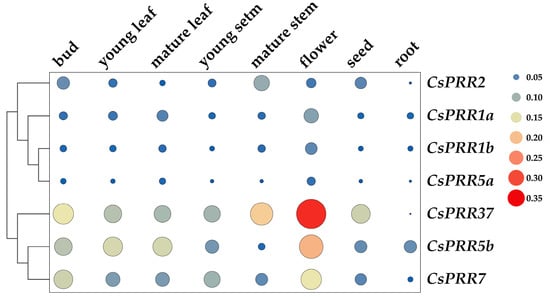
Figure 4.
Tissue-specific expression of CsPRR genes in bud, young leaf, mature leaf, young stem, mature stem, seed, and root. Size and color of the spots represent the levels of gene expression.
3.6. Expression Analysis of CsPRR Genes Under Various Photoperiod Conditions
To explore their potential rhythmic-expression characteristics, we detected CsPRR expression changes under long-day, short-day, constant-light, and constant-dark conditions. As shown in Figure 5a, all CsPRR genes showed rhythmical expression, but with different patterns under long-day conditions (LD). CsPRR7 showed the highest expression from around morning (9:00) to noon (13:00), CsPRR5a and 5b showed the highest expression during noon (13:00) to dusk (17:00), CsPRR37 showed the highest expression near dusk (17:00), and CsPRR1a and 1b showed the highest expression from near dusk (17:00) to evening (21:00). For CsPRR2, it showed the highest expression at the beginning of the second 24 h cycle (Figure 5a). However, their rhythmic expression was disrupted under short-day conditions (Figure 5b). CsPRR7, 5b, and 37 showed higher expression at 5:00 and 9:00 during the first cycle, but showed higher expression during the night-time (21:00–5:00) in the second cycle. CsPRR5a, 2, and 1a showed relatively higher expression only during the night-time (21:00–5:00) of the second cycle (Figure 5b). Under constant light conditions, the heatmap was divided into two sub-clusters; CsPRR1a, 2,37,5b, and 7 were clustered together, and CsPRR1b and 5a were clustered together (Figure 5c). Similar expression patterns were observed under constant dark conditions (Figure 5d). However, no obvious rhythmic characteristics was found for any of the CsPRR genes under both conditions, indicating that light significantly affected CsPRR expression. Overall, these expression data reveal that the rhythmic expression of CsPRR genes is closely related to photoperiod conditions.
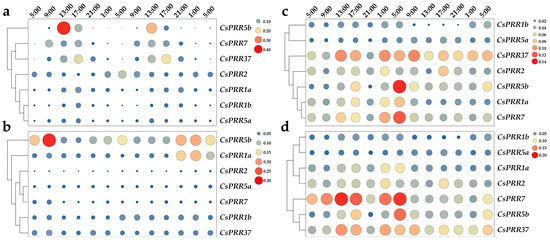
Figure 5.
Relative expression of CsPRR genes in mature leaves under long-day and short-day, constant-light and constant-dark conditions. (a) Heatmap of CsPRRs expression under long-day condition. (b) Heatmap of CsPRRs expression under short-day condition. (c) Heatmap of CsPRRs expression under constant-light condition. (d) Heatmap of CsPRRs expression under constant-dark condition. Size and color of the spots represent the levels of gene expression.
3.7. Cold Responses of CsPRR Genes Under Different Photoperiod Conditions
To investigate the involvement of the CsPRR genes in the tea plant cold responses, plants grown under long-day or short-day condition were treated with 4 °C/2 °C (light/dark), and then young shoots and mature leaves were collected for qRT-PCR assays. Considering their potential rhythmic-expression characters, samples of corresponding controls were also collected at each time point. As shown in Figure 6, the heatmap revealed that CsPRR genes expressed differently under the long-day and short-day conditions in both the young shoots and mature leaves. In young shoots under long-day condition, CsPRR1a, 1b, 37 and 7 were first down-regulated and then up-regulated after 48 h of treatment; CsPRR2 and 5a were gradually induced, and showed the highest expression after 48 h of treatment, while CsPRR5b was induced and peaked after 24 h treatment (Figure 6a). Under the short-day conditions, CsPRR37 and 7 were significantly inhibited at the beginning of cold stress; CsPRR5a, 1a, 1b and 2 initially showed inhibition, followed by significant induction after 48 h of cold treatment; and CsPRR5b was gradually induced and peaked at the end of cold stress (Figure 6b). For mature leaves, similar cluster patterns were observed under long-day and short-day conditions. CsPRR1a, 1b, and 37 were first downregulated, and then induced under both the long- and short-day conditions (Figure 6c,d). CsPRR2 was induced shortly after cold stress and then suppressed, and CsPRR7 showed dramatic down-regulation after 3–12 h of cold treatment under both photoperiod conditions. CsPRR5a was induced at the beginning of cold treatment under long-day condition, but it was inhibited under short-day conditions, while for CsPRR5b, it was firstly inhibited, then induced and peaked after 12 h and 24 h under long-day and short-day condition, respectively. Overall, those expression data reveal the photoperiod-related characteristics of the CsPRRs in response to cold stresses, and also suggest that the photoperiod conditions and tissue type, such as young or mature leaves, should be taken into consideration when conducting cold-related gene function studies.
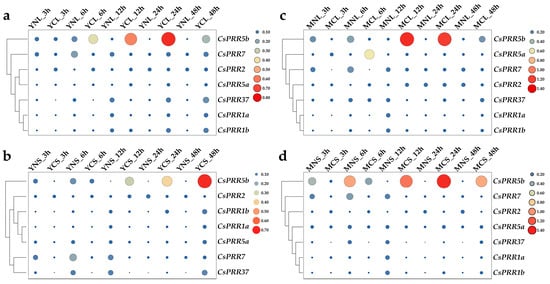
Figure 6.
Expression of CsPRR genes under cold, combined with long-day or short-day, conditions in young shoots and mature leaves. (a) Young shoots treated with normal (YNL) or cold (YCL) temperature under long-day conditions. (b) Young shoots treated with normal (YNS) or cold (YCS) temperature under short-day conditions. (c) Mature leaves treated with normal (MNL) or cold (MCL) temperature under long-day conditions. (d) Mature leaves treated with normal (MNS) or cold (MCS) temperature under short-day conditions. Size and color of the spots represent the levels of gene expression.
3.8. Response of CsPRR Genes to Heat and Drought Stresses
As mentioned above, various abiotic stress-related cis-acting elements were detected in the CsPRR promoter regions. To evaluate the potential function of the CsPRRs in response to heat and drought stresses, 38 °C and 5% (w/v) PEG 6000 treatments were carried out. As shown in Figure 7, the response of the CsPRRs to heat and drought stresses was dramatically different in the young shoots and mature leaves. CsPRR1a and 1b were significantly induced by heat and drought after 24 and 48 h of treatment, and a stronger induction was observed in the young shoots than in the mature leaves. CsPRR2 was significantly suppressed by heat after 12, 24, and 48 h of treatment, but it was significantly induced after 12 and 24 h of drought in the young shoots. However, in the mature leaves, CsPRR2 was downregulated to different degrees after the two stresses. CsPRR5a was significantly upregulated and peaked after 6 h of heat treatment in the young shoots, but it was first suppressed and then induced during drought stress. Drought significantly induced CsPRR5a, which was observed only after 24 h of treatment in the young shoots; however, in the mature leaves, CsPRR5a was significantly downregulated. CsPRR5b showed a similar expression in the young shoots and mature leaves, being suppressed at the beginning of the heat- and drought-stress treatment, but significantly upregulated after 48 h of the treatment. CsPRR7 was downregulated by heat and drought in both the young shoots and mature leaves, but a stronger inhibition was observed in the young shoots. Dramatically different patterns of CsPRR37 expression were observed in the young shoots and mature leaves in response to heat and drought stresses. CsPRR37 was first significantly inhibited after 12 h of heat treatment and then significantly induced after 48 h of treatment in the young shoots, while in the mature leaves, it was significantly upregulated after 12, 24, and 48 h of heat treatment. Regarding drought stress, CsPRR37 was induced to different levels, except for after 3 h of treatment, in the young shoots, while it was inhibited in the mature leaves. Overall, these expression profiles provide very important information for CsPRR function studies related to heat and drought stresses.
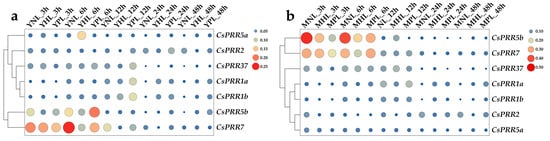
Figure 7.
Expression of CsPRR genes under heat and drought conditions in young shoots and mature leaves. (a) Young shoots treated under normal condition (YNL) or heat (YHL) or drought (YPL). (b) Mature leaves treated under normal condition (MNL) or heat (MHL) or drought (MPL). Size and color of the spots represent the levels of gene expression.
3.9. Expression Analysis of CsPRRs in Floriferous and Oliganthous Tea Plant Cultivars
Studies have suggested that the period from 16 May to 16 June is critical for floral induction in the axillary bud []. Therefore, we collected axillary buds from a floriferous cultivar (FDDB) and an oliganthous cultivar (PYTZ) to explore the potential relationship between CsPRR expression and flower induction in tea plants. As shown in Figure 8, CsPRR members, except CsPRR7, exhibited relatively higher transcription levels in FDDB compared to PYTZ at the onset of floral induction on 16 May. As the floral induction process progressed, the fold change of CsPRR1a, 1b, and 37 gradually decreased in both cultivars, while CsPRR5a gradually increased. CsPRR5b and CsPRR7 initially increased, peaked on 31 May, and then declined by June 16th. Unlike the first sub-family members, the expression of CsPRR2 first decreased, and was then upregulated. On 27 June, the floral meristem in FDDB had formed and was beginning to differentiate into sepal primordia. During this stage, CsPRR1a and CsPRR1b exhibited lower expression in FDDB compared to PYTZ, while CsPRR2, 5a, and CsPRR7 were expressed at higher levels in FDDB. No differences were observed for CsPRR37 and CsPRR5b. Overall, these data suggested that CsPRRs may also played vital roles in tea-plant flower induction. However, to identify the key CsPRR member, much more work is needed following the successful breakthrough of the transgenic technology system in tea plants.
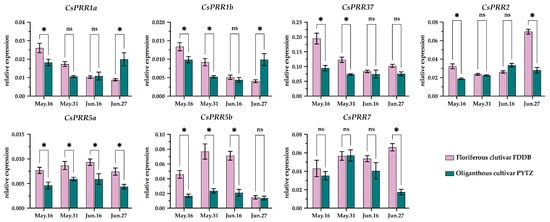
Figure 8.
Expression of CsPRR genes in axillary buds of floriferous cultivar (FDDB) and an oliganthous cultivar (PYTZ). The data were calculated using the 2−ΔCt method, with normalization to the reference gene CsEF1-α. The bar of each time point represents individual data points from replicates (mean ± SE, n = 4). Statistical significance was determined using Student’s t tests: *, p < 0.05. ns, non-significant.
4. Discussion
The circadian rhythms of biological activity and behavior are adaptively regulated in changing environments, enabling plants to predict daily and seasonal fluctuations []. Pseudo-response regulators (PRRs), variants of the two-component regulatory systems, act as dominant regulators of circadian rhythms in higher plants []. PRRs were highly conserved in the angiosperm evolutionary lineage, and PRRs have been reported to be involved in the regulation of photoperiod, flowering time, transition from the vegetative to reproductive phase, and various stress responses []. Most previously studied species possess 5–9 PRR genes. The model plant Arabidopsis possesses nine APRRs, and five of them are clock-associated; Vitis vinifera and Carica papaya have five PRRs, while Populus trichocarpa has seven [,,]. In this study, seven CsPRR genes, located on 6 out of 15 chromosomes, encoding proteins with 363–580 amino acid residues, were identified in the tea plant ‘LJ43’ genome (Table 1). A phylogenetic analysis revealed a two-subgroup clustering tree, and the first subgroup consisted of six members, CsPRR1a, 1b, 5a, 5b, 7, and 37, while the second subgroup consisted of only CsPRR2 (Figure 1). Like other known clock-associated PRRs [], all members of the first subgroup contained Pseudo-Rec and CCT domains, implying their potential roles in the tea plant. CsPRR2 clustered together with GLK proteins, suggesting a close relationship between these two proteins (Figure 1). A previous study found that SlGlk16 positively regulated cold tolerance in tomato plants []. Additionally, the silencing of cotton GhGLK1 induced sensitivity to drought and low-temperature stress, while GhGLK1 overexpression could enhance plant tolerance to identical stresses, correspondingly []. CsPRR2 possessed a Pseudo-Rec domain, indicating that it belongs to the PRR family. Moreover, the sharing of a conserved GCT box domain with GLK proteins suggests that CsPRR2 may play vital roles in the stress response involving chloroplast function regulation, similar to GLKs. Gene structure and motif diversity is important for gene family evolution and function prediction. The CsPRRs possessed 4–11 exons and 2–8 motifs (Figure 2), which were different from their corresponding homologs in Arabidopsis, implying some specialties in CsPRRs’ evolution. As specific targets identified and combined by transcription factors, various cis-acting elements, including those related to light, hormones, stress, and flavonoid biosynthesis, were detected in the CsPRR promoters, suggesting their potential roles in the regulation of the physiological processes of tea plant. The abovementioned analyses revealed the diversity and complexity of CsPRRs in evolution and function, which require further study in the tea plant.
The first report on the tissue-specific regulation of the circadian clock examined bean plants [], and clock-associated gene expression profiles were also found to be distinct in different tissues. The transcription rhythms of PRR9, 7, 5, and 1 were different in roots and shoots under two temperature conditions of 22 °C and 28 °C, indicating the organ-specific characteristics of PRR expression []. In the tea plant, the first subgroup of CsPRRs showed a higher expression in flowers than in other tissues (Figure 4). Various studies have proved that PRRs played vital roles in plant flower induction. In Arabidopsis, aberrant expression of APRR5 leads to early flowering and hypersensitiveness to light in early photomorphogenesis []. In Rosa chinensis, silencing of RcPRR1b and RcPRR5 led to earlier flowering, implying that they were negative flowering regulators []. In soybean knockout of GmPRR3b, a PRR family gene identified through a genome-wide association study led to delayed growth and floral transition []. Other reported genes, such as Ppd-H1, Ppd-1, PRR37, and SbPRR37 encode PRR proteins in barley, wheat, rice, and sorghum, respectively, and allelic variation at these genes confers natural diversity in flowering time [,,,]. In the tea plant, all CsPRR members, except CsPRR7, showed relatively higher expression in floriferous cultivar (FDDB) compared to oliganthous cultivar (PYTZ), at least at one time point during the floral induction procedure (Figure 8), which implied their crucial roles in tea-plant flowering control.
Maintaining correct circadian rhythms and behaviors is undoubtedly essential for plants to achieve optimal growth and performance in changing environments []. A key physiological property of circadian rhythms is maintaining a period of about 24 h when the plant is removed from natural fluctuating conditions and placed under constant light or dark conditions []. PRRs are important for maintaining an approximate 24 h circadian clock rhythm. A previous study found that a mutation in APRR1 or APRR5 resulted in a short period, while the loss of function of APRR9 or APRR7 led to an extended period []. Additionally, the overexpression of B-Box proteins, BBX28 or BBX29, significantly lengthened the circadian period through interactions with PRR5, 7, and 9 []. In rice, OsPRR37 could downregulate Hd3a (Heading date3a) and affect plant photoperiod sensitivity under long-day conditions. APRR9, 7, 5, 3, and 1 were expressed sequentially and reached their acrophase in a consecutive manner from morning to evening, with each of them functioning in a distinct period []. In addition, studies on various species have proposed that the rhythmic expression of PRRs is usually not significantly affected by photoperiod under normal conditions. In the tea plant, CsPRRs were expressed differently under long-day, short-day, continuous-light, and continuous-dark conditions (Figure 5). The rhythmic expression of CsPRR1a, 1b, 5a, 5b, 7, and 37 was observed only under long-day (LD) conditions and was disrupted under other conditions, suggesting that, unlike in Arabidopsis and rice, the rhythmic expression of CsPRRs is photoperiod-related. In addition, the rhythmic expression of CsPRR7, 5b, 5a, 37, 1b, and 1a occurred in a sequential manner from morning to evening under long-day conditions, similar to the APRRs mentioned above, implying some type of functional conservation of CsPRRs.
An increasing number of studies have proven that PRRs are essential for tolerance to various stresses. APRR1 (TOC1) acts as a molecular switch between the drought-stress signaling pathway and the biological clock, through interactions with ABA-related genes []. While in the tea plant, both CsPRR1a and 1b were also differently induced by cold and drought stress in young and mature leaves (Figure 6 and Figure 7), which implied their potential roles. The increased degradation of PRR5 by LOV KELCH PROTEIN 2 (LKP2) overexpression led to stronger drought-stress tolerance in Arabidopsis []. In rice, OsPRR37, 59, and 95 could be induced and activated by cold and drought stresses, and OsPRR73 was found to confer salt tolerance []. Similarly, CsPRR5b was also strongly changed under cold and drought conditions in the tea plant (Figure 6 and Figure 7). GmPRR3b was found to play a pivotal role in the negative regulation of the drought response in soybeans by suppressing the expression of abscisic acid-responsive element-binding factor 3 (GmABF3) []. High temperature regulated PRR7 expression by adjusting the binding of heat shock HsfB2b (HEAT SHOCK FACTOR B2B) to its promoters []. Moreover, the Arabidopsis prr9prr7prr5 triple mutant showed stronger tolerance to cold, drought, and high-salinity stresses []. Both CsPRR7 and CsPRR37 were greatly affect by cold, but only slightly affected by drought and heat stress in the tea plant (Figure 6 and Figure 7), which suggested their major roles in cold response. For the second sub-group member, pepper CaPRR2 was proven to be involved in the regulation of drought and high salt tolerance; it could be gradually induced by cold, and peaked after 12 h of treatment []. Similarly, in the tea plant, CsPRR2 could be slightly induced by cold and drought, but was inhibited by heat (Figure 6 and Figure 7). Moreover, those qRT-PCR assays revealed that the cold induction and inhibition of the CsPRRs were obviously photoperiod-related and tissue-specific (Figure 6). A stronger cold effect was observed under long-day conditions than under short-day conditions, and in the young shoots when compared to the mature leaves. Regarding heat and drought stresses, a stronger induction of CsPRR1a, 1b and inhibition of CsPRR7 were observed in young shoots than in mature leaves (Figure 7). In general, CsPRRs were differently expressed under cold, heat, and drought stresses, suggesting that these genes play an essential role in the development and growth of tea plant under stress conditions.
5. Conclusions
In this study, we focused on a core circadian clock-regulator gene family, pseudo-response regulators (PRRs), in the tea plant, conducted comprehensive genome identification, determined sequence characteristics, and examined gene expression. Seven CsPRRs with a Pseudo-Rec domain in their N-terminus were identified. Phylogenetic, gene structure, and motif analyses revealed common and special characteristics of the CsPRR family. A cis-acting element analysis and qRT-PCR assays showed that CsPRR expression was tissue-specific and photoperiod-related, and rhythmic expression was observed only under long-day conditions. These findings suggest that CsPRRs play a crucial role in flower induction, photoperiod responses, and abiotic-stress adaptation in tea plant. Our results provide a strong basis for CsPRR function studies, and it is proposed that photoperiod and tissue type should be taken into consideration when conducting gene function studies.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/horticulturae10121294/s1, Table S1: Primers and their amplification efficiency; Figure S1: Sequences of the top 10 conserved motifs of CsPRRs. Letter size indicates the occurrence probability of the amino acid at a given position; Data S1: Coding sequence of CsPRRs cloned from tea plant variety ‘Longjing43’; Data S2: Protein sequences used for phylogenetic tree construction.
Author Contributions
C.D., J.W. and X.W. conceived and designed the research. C.D., M.Y. and L.Y. wrote the manuscript. L.Y., H.X., N.L. and J.H. performed the experiments and analyzed the data. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Zhejiang Provincial Natural Science Foundation of China (LY21C160006), the National Natural Science Foundation of China (32102440), and the Central Public-interest Scientific Institution Basal Research Fund (Y2023PT03 and 1610212022001).
Data Availability Statement
Data is contained within the article or Supplementary Materials.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Yuan, L.; Yu, Y.J.; Liu, M.M.; Song, Y.; Li, H.M.; Sun, J.Q.; Wang, Q.; Xie, Q.G.; Wang, L.; Xu, X.D. BBX19 fine-tunes the circadian rhythm by interacting with PSEUDO-RESPONSE REGULATOR proteins to facilitate their repressive effect on morning-phased clock genes. Plant Cell 2021, 33, 2602–2617. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.L.; Shao, Y.J.; Ding, L.; Wang, M.J.; Davis, S.J.; Liu, J.X. XBAT31 regulates thermoresponsive hypocotyl growth through mediating degradation of the thermosensor ELF3 in Arabidopsis. Sci. Adv. 2021, 7, eabf4427. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.Y.; Bo, C.P.; Wang, L. Novel crosstalks between circadian clock and jasmonic acid pathway finely coordinate the tradeoff among plant growth, senescence and defense. Int. J. Mol. Sci. 2019, 20, 5254. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Yuan, L.; Yang, X.; Zhang, X.; Wang, L.; Xie, Q. Circadian clock in plants: Linking timing to fitness. J. Integr. Plant Biol. 2022, 64, 792–811. [Google Scholar] [CrossRef]
- Greenham, K.; McClung, C.R. Integrating circadian dynamics with physiological processes in plants. Nat. Rev. Genet. 2015, 16, 598–610. [Google Scholar] [CrossRef]
- Farré, E.M.; Liu, T. The PRR family of transcriptional regulators reflects the complexity and evolution of plant circadian clocks. Curr. Opin. Plant Biol. 2013, 16, 621–629. [Google Scholar] [CrossRef]
- Yu, Y.; Su, C.; He, Y.; Wang, L. B-Box proteins BBX28 and BBX29 interplay with PSEUDO-RESPONSE REGULATORS to fine-tune circadian clock in Arabidopsis. Plant Cell Environ. 2023, 46, 2810–2826. [Google Scholar] [CrossRef]
- Schaller, G.E.; Kieber, J.J.; Shiu, S.H. Two-component signaling elements and histidyl-aspartyl phosphorelays. Arab. Book 2008, 6, e0112. [Google Scholar] [CrossRef]
- Xing, Y.; Jiang, Y.; Muhammad, A.R.; Song, J. Genome-wide identification of pseudo-response regulator (PRR) family members in cabbage (Brassica oleracea var. capitata L.) and their expression in response to abiotic stress. J. Hortic. Sci. Biotech. 2023, 99, 168–178. [Google Scholar]
- Wang, C.L.; Wang, L.L.; Liu, Q.Q.; Zhang, Y.L.; Dong, K.Q. Genome-wide identification and characterization of PRR gene family and their diurnal rhythmic expression profile in maize. Int. J. Genomics 2022, 2022, 6941607. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, J.; Liu, X.; Kong, Y.; Han, L. The Roles of the PSEUDO-RESPONSE REGULATORs in Circadian Clock and Flowering Time in Medicago truncatula. Int. J. Mol. Sci. 2023, 24, 16834. [Google Scholar] [CrossRef] [PubMed]
- Mizuno, T. Plant response regulators implicated in signal transduction and circadian rhythm. Curr. Opin. Plant Biol. 2004, 7, 499–505. [Google Scholar] [CrossRef] [PubMed]
- Nakamichi, N.; Kiba, T.; Henriques, R.; Mizuno, T.; Chua, N.H.; Sakakibara, H. PSEUDO-RESPONSE REGULATORS 9, 7, and 5 are transcriptional repressors in the Arabidopsis circadian clock. Plant Cell 2010, 22, 594–605. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Perez-Garcia, P.; Pokhilko, A.; Millar, A.J.; Antoshechkin, I.; Riechmann, J.L.; Mas, P. Mapping the core of the Arabidopsis circadian clock defines the network structure of the oscillator. Science 2012, 336, 75–79. [Google Scholar] [CrossRef] [PubMed]
- Murakami, M.; Ashikari, M.; Miura, K.; Yamashino, T.; Mizuno, T. The evolutionarily conserved OsPRR quintet: Rice pseudo-response regulators implicated in circadian rhythm. Plant Cell Physiol. 2003, 44, 1229–1236. [Google Scholar] [CrossRef] [PubMed]
- Satbhai, S.B.; Yamashino, T.; Okada, R.; Nomoto, Y.; Mizuno, T.; Tezuka, Y.; Itoh, T.; Tomita, M.; Otsuki, S.; Aoki, S. Pseudo-response regulator (PRR) homologues of the moss Physcomitrella patens: Insights into the evolution of the PRR family in land plants. DNA Res. 2011, 18, 39–52. [Google Scholar] [CrossRef]
- Chen, M.; Ji, M.; Wen, B.; Liu, L.; Li, S.; Chen, X.; Gao, D.; Li, L. Golden 2-like transcription factors of plants. Front. Plant Sci. 2016, 7, 1509. [Google Scholar] [CrossRef]
- Fujiwara, S.; Wang, L.; Han, L.; Suh, S.S.; Salome, P.A.; McClung, C.R.; Somers, D.E. Post-translational regulation of the Arabidopsis circadian clock through selective proteolysis and phosphorylation of pseudo-response regulator proteins. J. Biol. Chem. 2008, 283, 23073–23083. [Google Scholar] [CrossRef]
- Matsushika, A.; Makino, S.; Kojima, M.; Mizuno, T. Circadian waves of expression of the APRR1/TOC1 family of pseudo-response regulators in Arabidopsis thaliana: Insight into the plant circadian clock. Plant Cell Physiol. 2000, 41, 1002–1012. [Google Scholar] [CrossRef]
- Gil, K.E.; Park, C.M. Thermal adaptation and plasticity of the plant circadian clock. New Phytol. 2019, 221, 1215–1229. [Google Scholar] [CrossRef]
- Wang, L.; Kim, J.; Somers, D.E. Transcriptional corepressor TOPLESS complexes with pseudoresponse regulator proteins and histone deacetylases to regulate circadian transcription. Proc. Natl. Acad. Sci. USA 2013, 110, 761–766. [Google Scholar] [CrossRef] [PubMed]
- Dong, M.A.; Farre, E.M.; Thomashow, M.F. Circadian clock-associated 1 and late elongated hypocotyl regulate expression of the C-repeat binding factor (CBF) pathway in Arabidopsis. Proc. Natl. Acad. Sci. USA 2011, 108, 7241–7246. [Google Scholar] [CrossRef] [PubMed]
- Dodd, A.N.; Salathia, N.; Hall, A.; Kevei, E.; Toth, R.; Nagy, F.; Hibberd, J.M.; Millar, A.J.; Webb, A.A. Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science 2005, 309, 630–633. [Google Scholar] [CrossRef] [PubMed]
- Nakamichi, N.; Kita, M.; Niinuma, K.; Ito, S.; Yamashino, T.; Mizoguchi, T.; Mizuno, T. Arabidopsis clock-associated pseudo-response regulators PRR9, PRR7 and PRR5 coordinately and positively regulate flowering time through the canonical CONSTANS-dependent photoperiodic pathway. Plant Cell Physiol. 2007, 48, 822–832. [Google Scholar] [CrossRef]
- Nakamichi, N.; Kusano, M.; Fukushima, A.; Kita, M.; Ito, S.; Yamashino, T.; Saito, K.; Sakakibara, H.; Mizuno, T. Transcript profiling of an Arabidopsis PSEUDO RESPONSE REGULATOR arrhythmic triple mutant reveals a role for the circadian clock in cold stress response. Plant Cell Physiol. 2009, 50, 447–462. [Google Scholar] [CrossRef]
- Li, N.; Zhang, Y.; He, Y.; Wang, Y.; Wang, L. Pseudo response regulators regulate photoperiodic hypocotyl growth by repressing PIF4/5 transcription. Plant Physiol. 2020, 183, 686–699. [Google Scholar] [CrossRef]
- Wang, L.; Sun, S.; Wu, T.; Liu, L.; Sun, X.; Cai, Y.; Li, J.; Jia, H.; Yuan, S.; Chen, L.; et al. Natural variation and CRISPR/Cas9-mediated mutation in GmPRR37 affect photoperiodic flowering and contribute to regional adaptation of soybean. Plant Biotechnol. J. 2020, 18, 1869–1881. [Google Scholar] [CrossRef]
- Nakamichi, N. Molecular mechanisms underlying the Arabidopsis circadian clock. Plant Cell Physiol. 2011, 52, 1709–1718. [Google Scholar] [CrossRef]
- Liu, T.; Carlsson, J.; Takeuchi, T.; Newton, L.; Farre, E.M. Direct regulation of abiotic responses by the Arabidopsis circadian clock component PRR7. Plant J. 2013, 76, 101–114. [Google Scholar] [CrossRef]
- Cheval, C.; Perez, M.; Leba, L.J.; Ranty, B.; Perochon, A.; Reichelt, M.; Mithofer, A.; Robe, E.; Mazars, C.; Galaud, J.P.; et al. PRR2, a pseudo-response regulator, promotes salicylic acid and camalexin accumulation during plant immunity. Sci. Rep. 2017, 7, 6979. [Google Scholar] [CrossRef]
- Pan, Y.; Bradley, G.; Pyke, K.; Ball, G.; Lu, C.G.; Fray, R.; Marshall, A.; Jayasuta, S.; Baxter, C.; van Wijk, R.; et al. Network inference analysis identifies an APRR2-like gene linked to pigment accumulation in tomato and pepper fruits. Plant Physiol. 2013, 161, 1476–1485. [Google Scholar] [CrossRef] [PubMed]
- Jiao, J.; Liu, H.; Liu, J.; Cui, M.; Xu, J.; Meng, H.; Li, Y.; Chen, S.; Cheng, Z. Identification and functional characterization of APRR2 controlling green immature fruit color in cucumber (Cucumis sativus L.). Plant Growth Regul. 2017, 83, 233–243. [Google Scholar] [CrossRef]
- Ye, M.; Liu, C.; Li, N.; Yuan, C.; Liu, M.; Xin, Z.; Lei, S.; Sun, X. A constitutive serine protease inhibitor suppresses herbivore performance in tea (Camellia sinensis). Hortic. Res. 2023, 10, uhad178. [Google Scholar] [CrossRef] [PubMed]
- Kingdom-Ward, F. Does wild tea exist? Nature 1950, 165, 297–299. [Google Scholar] [CrossRef]
- Ding, C.Q.; Ng, S.; Wang, L.; Wang, Y.C.; Li, N.N.; Hao, X.Y.; Zeng, J.M.; Wang, X.C.; Yang, Y.J. Genome-wide identification and characterization of ALTERNATIVE OXIDASE genes and their response under abiotic stresses in Camellia sinensis (L.) O. Kuntze. Planta 2018, 248, 1231–1247. [Google Scholar] [CrossRef]
- Wang, L.; Feng, X.; Yao, L.; Ding, C.; Lei, L.; Hao, X.; Li, N.; Zeng, J.; Yang, Y.; Wang, X. Characterization of CBL-CIPK signaling complexes and their involvement in cold response in tea plant. Plant Physiol. Biochem. 2020, 154, 195–203. [Google Scholar] [CrossRef]
- Zhang, X.; Chen, S.; Shi, L.; Gong, D.; Zhang, S.; Zhao, Q.; Zhan, D.; Vasseur, L.; Wang, Y.; Yu, J.; et al. Haplotype-resolved genome assembly provides insights into evolutionary history of the tea plant Camellia sinensis. Nat. Genet. 2021, 53, 1250–1259. [Google Scholar] [CrossRef]
- Wang, X.; Feng, H.; Chang, Y.; Ma, C.; Wang, L.; Hao, X.; Li, A.; Cheng, H.; Wang, L.; Cui, P.; et al. Population sequencing enhances understanding of tea plant evolution. Nat. Commun. 2020, 11, 4447. [Google Scholar] [CrossRef]
- Wang, P.; Yu, J.; Jin, S.; Chen, S.; Yue, C.; Wang, W.; Gao, S.; Cao, H.; Zheng, Y.; Gu, M.; et al. Genetic basis of high aroma and stress tolerance in the oolong tea cultivar genome. Hortic. Res. 2021, 8, 107. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Ma, Q.P.; Hao, S.; Chen, X.; Li, X.H. Validation of reliability for reference genes under various abiotic stresses in tea plant. Russ. J. Plant Physiol. 2016, 63, 423–432. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Hao, X.; Lu, Q.; Zhang, W.; Zhang, H.; Wang, L.; Yang, Y.; Xiao, B.; Wang, X. Genome-wide identification and expression analysis of flowering-related genes reveal putative floral induction and differentiation mechanisms in tea plant (Camellia sinensis). Genomics 2020, 112, 2318–2326. [Google Scholar] [CrossRef] [PubMed]
- Troein, C.; Locke, J.C.; Turner, M.S.; Millar, A.J. Weather and seasons together demand complex biological clocks. Curr. Biol. 2009, 19, 1961–1964. [Google Scholar] [CrossRef] [PubMed]
- Murakami, M.; Tago, Y.; Yamashino, T.; Mizuno, T. Comparative overviews of clock-associated genes of Arabidopsis thaliana and Oryza sativa. Plant Cell Physiol. 2007, 48, 110–121. [Google Scholar] [CrossRef]
- Kim, J.A.; Kim, J.S.; Hong, J.K.; Lee, Y.H.; Choi, B.S.; Seol, Y.J.; Jeon, C.H. Comparative mapping, genomic structure, and expression analysis of eight pseudo-response regulator genes in Brassica rapa. Mol. Genet. Genom. 2012, 287, 373–388. [Google Scholar] [CrossRef]
- Ming, R.; Hou, S.; Feng, Y.; Yu, Q.; Dionne-Laporte, A.; Saw, J.H.; Senin, P.; Wang, W.; Ly, B.V.; Lewis, K.L.; et al. The draft genome of the transgenic tropical fruit tree papaya (Carica papaya Linnaeus). Nature 2008, 452, 991–996. [Google Scholar] [CrossRef]
- Takata, N.; Saito, S.; Saito, C.T.; Uemura, M. Phylogenetic footprint of the plant clock system in angiosperms: Evolutionary processes of pseudo-response regulators. BMC Evol. Biol. 2010, 10, 126. [Google Scholar] [CrossRef]
- Wang, Z.Y.; Zhao, S.; Liu, J.F.; Zhao, H.Y.; Sun, X.Y.; Wu, T.R.; Pei, T.; Wang, Y.; Liu, Q.F.; Yang, H.H.; et al. Genome-wide identification of tomato golden 2-like transcription factors and abiotic stress related members screening. BMC Plant Biol. 2022, 22, 82. [Google Scholar] [CrossRef]
- Liu, J.; Mehari, T.G.; Xu, Y.; Umer, M.J.; Hou, Y.; Wang, Y.; Peng, R.; Wang, K.; Cai, X.; Zhou, Z.; et al. GhGLK1 a key candidate gene from GARP family enhances cold and drought stress tolerance in cotton. Front. Plant Sci. 2021, 12, 759312. [Google Scholar] [CrossRef]
- Hennessey, T.L.; Field, C.B. Evidence of multiple circadian oscillators in bean plants. J. Biol. Rhythm. 1992, 7, 105–113. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wang, L.; Yuan, L.; Song, Y.; Sun, J.; Jia, Q.; Xie, Q.; Xu, X. Molecular investigation of organ-autonomous expression of Arabidopsis circadian oscillators. Plant Cell Environ. 2020, 43, 1501–1512. [Google Scholar] [CrossRef] [PubMed]
- Sato, E.; Nakamichi, N.; Yamashino, T.; Mizuno, T. Aberrant expression of the circadian-regulated gene belonging to the APRR1/TOC1 quintet results in early flowering and hypersensitiveness to light in early photomorphogenesis. Plant Cell Physiol. 2002, 43, 1374–1385. [Google Scholar] [CrossRef] [PubMed]
- Jalal, A.; Sun, J.; Chen, Y.; Fan, C.; Liu, J.; Wang, C. Evolutionary analysis and functional identification of clock-associated PSEUDO-RESPONSE REGULATOR (PRRs) genes in the flowering regulation of roses. Int. J. Mol. Sci. 2022, 23, 7335. [Google Scholar] [CrossRef]
- Li, C.; Li, Y.H.; Li, Y.; Lu, H.; Hong, H.; Tian, Y.; Li, H.; Zhao, T.; Zhou, X.; Liu, J.; et al. A domestication-associated gene gmPRR3b regulates the circadian clock and flowering time in soybean. Mol. Plant 2020, 13, 745–759. [Google Scholar] [CrossRef]
- Koo, B.H.; Yoo, S.C.; Park, J.W.; Kwon, C.T.; Lee, B.D.; An, G.; Zhang, Z.Y.; Li, J.J.; Li, Z.C.; Paek, N.C. Natural variation in OsPRR37 regulates heading date and contributes to rice cultivation at a wide range of latitudes. Mol. Plant 2013, 6, 1877–1888. [Google Scholar] [CrossRef]
- Murphy, R.L.; Klein, R.R.; Morishige, D.T.; Brady, J.A.; Rooney, W.L.; Miller, F.R.; Dugas, D.V.; Klein, P.E.; Mullet, J.E. Coincident light and clock regulation of pseudoresponse regulator protein 37 (PRR37) controls photoperiodic flowering in sorghum. Proc. Natl. Acad. Sci. USA 2011, 108, 16469–16474. [Google Scholar] [CrossRef]
- Shaw, L.M.; Turner, A.S.; Laurie, D.A. The impact of photoperiod insensitive Ppd-1a mutations on the photoperiod pathway across the three genomes of hexaploid wheat (Triticum aestivum). Plant J. 2012, 71, 71–84. [Google Scholar] [CrossRef]
- Turner, A.; Beales, J.; Faure, S.; Dunford, R.P.; Laurie, D.A. The pseudo-response regulator Ppd-H1 provides adaptation to photoperiod in barley. Science 2005, 310, 1031–1034. [Google Scholar] [CrossRef]
- Petersen, J.; Rredhi, A.; Szyttenholm, J.; Mittag, M. Evolution of circadian clocks along the green lineage. Plant Physiol. 2022, 190, 924–937. [Google Scholar] [CrossRef]
- Li, C.; Chen, Y.H.; Hu, Q.; Yang, X.L.; Zhao, Y.F.; Lin, Y.; Yuan, J.B.; Gu, J.B.; Li, Y.; He, J.; et al. PSEUDO-RESPONSE REGULATOR 3b and transcription factor ABF3 modulate abscisic acid-dependent drought stress response in soybean. Plant Physiol. 2024, 195, 3053–3071. [Google Scholar] [CrossRef] [PubMed]
- Lim, J.; Lim, C.W.; Lee, S.C. Pepper novel pseudo response regulator protein CaPRR2 modulates drought and high salt tolerance. Front. Plant Sci. 2021, 12, 736421. [Google Scholar] [CrossRef] [PubMed]
- Legnaioli, T.; Cuevas, J.; Mas, P. TOC1 functions as a molecular switch connecting the circadian clock with plant responses to drought. Embo J. 2009, 28, 3745–3757. [Google Scholar] [CrossRef] [PubMed]
- Wei, H.; Wang, X.; He, Y.; Xu, H.; Wang, L. Clock component OsPRR73 positively regulates rice salt tolerance by modulating OsHKT2;1-mediated sodium homeostasis. EMBO J. 2020, 40, e105086. [Google Scholar] [CrossRef]
- Yamamoto, Y.; Sato, E.; Shimizu, T.; Nakamich, N.; Sato, S.; Kato, T.; Tabata, S.; Nagatani, A.; Yamashino, T.; Mizuno, T. Comparative genetic studies on the and genes belonging to the APRR1/TOC1 quintet implicated in circadian rhythm, control of flowering time, and early photomorphogenesis. Plant Cell Physiol. 2003, 44, 1119–1130. [Google Scholar] [CrossRef]
- Nakamichi, N.; Kiba, T.; Kamioka, M.; Suzuki, T.; Yamashino, T.; Higashiyama, T.; Sakakibara, H.; Mizuno, T. Transcriptional repressor PRR5 directly regulates clock-output pathways. Proc. Natl. Acad. Sci. USA 2012, 109, 17123–17128. [Google Scholar] [CrossRef]
- Kolmos, E.; Chow, B.Y.; Pruneda-Paz, J.L.; Kay, S.A. HsfB2b-mediated repression of PRR7 directs abiotic stress responses of the circadian clock. Proc. Natl. Acad. Sci. USA 2014, 111, 16172–16177. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).