Advances in Orchid Biology: Biotechnological Achievements, Translational Success, and Commercial Outcomes
Abstract
:1. Orchid Biology: Recent Aspects and Emerging Perspectives
2. Research Methodology: Literature Retrieval, Compilation, and Analysis
3. Conventional Versus Modern Approaches for Orchid Cultivation and Conservation
3.1. Classical Breeding Strategies
3.1.1. Crossbreeding
3.1.2. Selection Breeding
3.1.3. Mutation Breeding
3.1.4. Molecular Marker-Assisted Breeding (MMAB)
3.2. In Vitro Orchid Propagation in Plant Tissue Culture
3.3. Cryopreservation Techniques
3.3.1. Vitrification
3.3.2. Desiccation
3.3.3. Encapsulation–Dehydration
3.4. Genetic Engineering and Generation of Hybrids with ‘High-Value’ Traits
4. Biotechnological Interventions in Orchids: Existing and Upcoming Trends
4.1. Promoting Growth Vigor and Flowering
4.2. Diagnosis of Pathogens
4.3. CRISPR/Cas and Advanced Genome Editing in Orchids
5. Ethnomedicinal and Edible Importance of Orchids
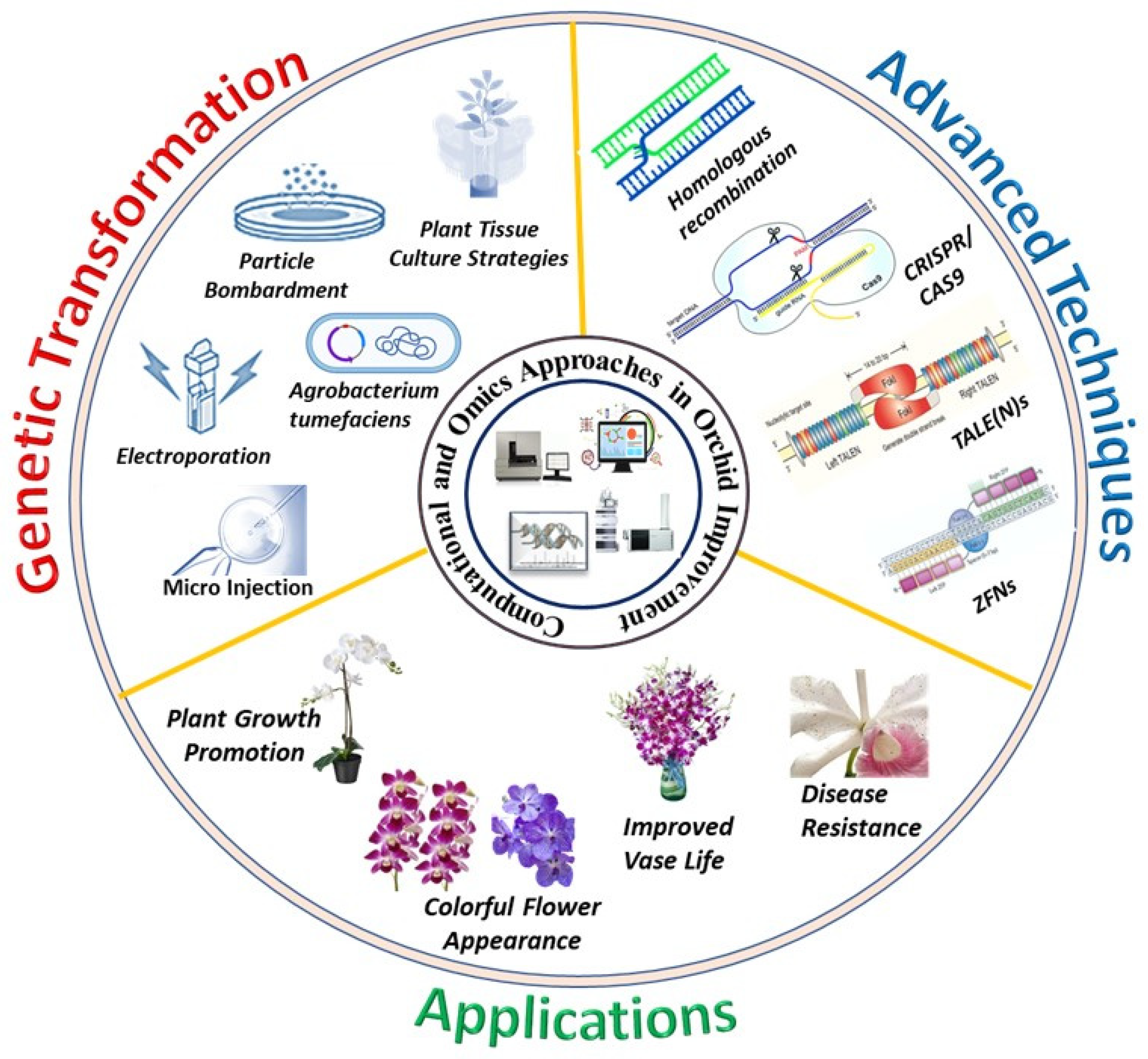
6. Computational and Omics Approaches in Orchid Biology
7. Strategies/Guidelines for Orchid Conservation and Utilization
International/National Guidelines for the Preservation of Orchid Biodiversity
8. Translational Success, Restoration Initiatives, and Future Research
9. Concluding Remarks
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Li, C.; Dong, N.; Zhao, Y.; Wu, S.; Liu, Z.; Zhai, J. A review for the breeding of orchids: Current achievements and prospects. Hortic. Plant J. 2021, 7, 380–392. [Google Scholar] [CrossRef]
- Vendrame, W.A.; Khoddamzadeh, A.A. Orchid biotechnology. In Horticulture Reviews; Janick, J., Ed.; World Scientific: London, UK, 2017; Volume 44, pp. 173–228. [Google Scholar]
- Hsu, C.C.; Chung, Y.L.; Chen, T.C.; Lee, Y.L.; Kuo, Y.T.; Tsai, W.C.; Hsiao, Y.Y.; Chen, Y.W.; Wu, W.L.; Chen, H.H. An overview of the Phalaenopsis orchid genome through BAC end sequence analysis. BMC Plant Biol. 2011, 11, 3. [Google Scholar] [CrossRef]
- Atwood, J.T. The size of the Orchidaceae and the systematic distribution of epiphytic orchids. Selbyana 1986, 9, 171–186. [Google Scholar]
- Hossain, M.M.; Kant, R.; Van, P.T.; Winarto, B.; Zeng, S.; Teixeira da Silva, J.A. The application of biotechnology to orchids. Crit. Rev. Plant Sci. 2013, 32, 69–139. [Google Scholar] [CrossRef]
- De, L.C.; Pathak, P.; Rao, A.N.; Rajeevan, P.K. Medicinal and Aromatic Orchids: In Commercial Orchids; De Gruyter Open: Warsaw, Poland, 2015. [Google Scholar] [CrossRef]
- Chen, F.-C.; Chin, S.-W. The Orchid Genome. In Compendium of Plant Genomes; Springer International Publishing: Berlin/Heidelberg, Germany, 2021. [Google Scholar] [CrossRef]
- Yuan, S.-C.; Lekawatana, S.; Amore, T.D.; Chen, F.-C.; Chin, S.-W.; Vega, D.M.; Wang, Y.-T. The global orchid markets. In The Orchid Genome, Compendium of Plant Genomes; Chen, F.-C., Chin, S.-W., Eds.; Springer Nature: Cham, Switzerland, 2021. [Google Scholar] [CrossRef]
- Thiruvengadam, M.; Chung, I.M.; Yang, C.H. Overexpression of Oncidium MADS-box (OMADS1) gene promotes early flowering in transgenic orchids (Oncidium gower Ramsey). Acta Physiol. Plant. 2012, 34, 1295–1302. [Google Scholar] [CrossRef]
- Liu, Y.C.; Yeh, C.W.; Chung, J.D.; Tsai, C.Y.; Chiou, C.Y.; Yeh, K.W. Petal-specific RNAi-mediated silencing of the phytoene synthase gene reduces xanthophyll levels to generate new Oncidium orchid varieties with white-colour blooms. Plant Biotechnol. J. 2019, 17, 2035–2037. [Google Scholar] [CrossRef] [PubMed]
- Krittiya, N.; Kisana, B.; Huehne, P. A direct gene transferring system for Oncidium orchids, a difficult crop for genetic transformation. Agric. Nat. Nat. Resour. 2018, 52, 424–442. [Google Scholar]
- Su, V.; Hsu, B.D. Cloning and expression of a putative cytochrome P450 gene that influences the colour of Phalaenopsis flowers. Biotechnol. Lett. 2003, 25, 1933–1939. [Google Scholar] [CrossRef] [PubMed]
- Semiarti, E.; Indrianto, A.; Purwantoro, A.; Suseno, N.; Isminingsih, S.; Yoshioka, Y.; Iwakawa, H.; Machida, Y.; Machida, C. Agrobacterium-mediated transformation of the wild orchid species Phalaenopsis amabilis. Plant Biotechnol. 2007, 24, 265–272. [Google Scholar] [CrossRef]
- Semiarti, E.; Indrianto, A.; Suyono, E.A.; Nurwulan, R.L.; Restiani, R.; Machida, Y.; Machida, C. Genetic transformation of the Indonesian black orchid (Coelogyne pandurata Lindley) through Agrobacterium tumefaciens for micropropagation. In Proceedings of the NIOC, Nagoya, Japan, 18–29 October 2010; pp. 16–20. [Google Scholar]
- Huang, W.T.; Wu, B.W.; Fang, Z.M. Temporal and spatial expression and functional analysis of FT homologous genes in Cymbidium sinense. J. Anhui Agric. Univ. 2017, 1, 141–147. (In Chinese) [Google Scholar]
- Shrestha, B.R.; Chin, D.P.; Tokuhara, K.; Mii, M. Efficient production of transgenic plants of Vanda through sonication-assisted Agrobacterium-mediated transformation of protocorm-like bodies. Plant Biotechnol. 2007, 24, 429–434. [Google Scholar] [CrossRef]
- Cao, Y.; Niimi, Y.; Hu, S.L. Meropenem as an alternative antibiotic agent for suppression of Agrobacterium in genetic transformation of orchid. Agric. Sci. China 2006, 5, 839–846. [Google Scholar] [CrossRef]
- Hsing, H.-X.; Lin, Y.-J.; Tong, C.-G.; Li, M.-J.; Chen, Y.-J.; Ko, S.-S. Efficient and heritable transformation of Phalaenopsis orchids. Bot. Stud. 2016, 57, 30. [Google Scholar] [CrossRef]
- Kantamaht, K.; Alisa, N.; Kamnoon, K.; Amornrat, P. Efficient biolistic transformation and regeneration capacity of an EgTCTP transgene in protocorm-like bodies of Phalaenopsis orchid. Notu-lae Botanicae Horti Agrobotanici Cluj-Napoca 2012, 40, 58–64. [Google Scholar]
- Chen, L.; Kawai, H.; Oku, T.; Takahashi, C.; Niimi, Y. Introduction of Odontoglossum ringspot virus coat protein gene into Cymbidium niveo-marginatum mediated by Agrobacterium tumefaciens to produce transgenic plants. J. Jpn. Soc. Hort. Sci. 2006, 75, 249–255. [Google Scholar] [CrossRef]
- Chen, Y.C.; Yang, Y.C.; Chen, Y.H.; Chao, Y.P.; Chang, L.Z.; Chen, Y.W. Modification of Phalaenopsis metabolism by genetic engineering. In International Orchid Symposium; ISHS: Leuven, Belgium, 2010; Volume 878, pp. 473–480. [Google Scholar]
- Lee, S.H.; Li, C.W.; Liau, C.H.; Chang, P.Y.; Liao, L.J.; Lin, C.S.; Chan, M.T. Establishment of an Agrobacterium-mediated genetic transformation procedure for the experimental model orchid Erycina pusilla. Plant Cell Tiss. Org. Cult. 2015, 120, 211–220. [Google Scholar] [CrossRef]
- Ratanasut, K.; Monmai, C.; Piluk, P. Transient hairpin RNAi-induced silencing in floral tissues of Dendrobium sonia ‘Earsakul’ by agroinfiltration for rapid assay of flower color modification. Plant Cell Tiss. Organ. Cult. 2015, 120, 643. [Google Scholar] [CrossRef]
- Zhang, L.; Chin, D.P.; Mii, M. Agrobacterium-mediated transformation of protocorm-like bodies in Cattleya. Plant Cell Tiss. Organ. Cult. 2010, 103, 41–47. [Google Scholar] [CrossRef]
- Qin, X.Y.; Liu, Y.; Mao, S.J.; Li, T.B.; Wu, H.K.; Chu, C.C.; Wang, Y.P. Genetic transformation of lipid transfer protein-encoding gene in Phalaenopsis amabilis to enhance cold resistance. Euphytica 2011, 177, 33–43. [Google Scholar] [CrossRef]
- Chang, L.; Chang, H.H.; Chiu, A.; Chang, J.C.; Hsu, D.W.; Tzean, Y.; Cheng, A.P.; Lu, H.C.; Yeh, H.H. Plant A20/AN1 proteins coordinate different immune responses including RNAi pathway for antiviral immunity. PLoS Pathog. 2019, 14, e1007288. [Google Scholar]
- Hsieh, R.M.; Huang, P.L. Studies on genetic transformation of Phalaenopsis via pollen tube pathway. J. Chin. Soc. Hort. Sci. 1995, 41, 309–324. [Google Scholar]
- Nan, G.L.; Kuehnle, A.R. Genetic transformation in Dendrobium (orchid). In Plant Protoplasts and Genetic Engineering VI. Biotechnology in Agriculture and Forestry; Bajaj, Y.P.S., Ed.; Springer: Berlin/Heidelberg, Germany, 1995; Volume 34, pp. 145–155. [Google Scholar]
- You, S.J.; Liau, C.H.; Huang, H.E.; Feng, T.Y.; Venkatesh, P.; Hsiao, H.H.; Lu, J.C.; Chan, M.T. Sweet pepper ferredoxin-like protein (pflp) gene as a novel selection marker for orchid transformation. Planta 2003, 217, 60–65. [Google Scholar] [CrossRef]
- Chia, T.F.; Lim, A.Y.H.; Luan, Y.; Ng, I. Transgenic Dendrobium (Orchid). In Biotechnology in Agriculture and Forestry 48, Transgenic Crops III; Bajaj, Y.P.S., Ed.; Springer: Berlin/Heidelberg, Germany, 2001; pp. 95–106. [Google Scholar]
- Anzai, H.; Tanaka, M. Transgenic Phalaenopsis (a moth orchid). In Biotechnology in Agriculture and Forestry 48, Trangenic Crops II; Bajaj, Y.P.S., Ed.; Springer: Berlun/Heidelberg, Germany, 2001; pp. 249–264. [Google Scholar]
- Griesbach, R.J. An improved method for transforming plants through Electrophoresis. Plant Sci. 1994, 102, 81–89. [Google Scholar] [CrossRef]
- Fay, M.; Rankou, H. Slipper orchids on the IUCN Red List. In The 2015 Annual Report to the Environment Agency-Abu Dhabi. Framework Support for Implementing the Strategic Plan of the IUCN Species Survival Commission; 2016; pp. 106–111. [Google Scholar]
- Vogt-Schilb, H.; Pradel, R.; Geniez, P.; Hugot, L.; Delage, A.; Richard, F.; Schatz, B. Responses of orchids to habitat change in Corsica over 27 years. Ann. Bot. 2016, 118, 115–123. [Google Scholar] [CrossRef] [PubMed]
- Fay, M.F. British and Irish orchids in a changing world. Curtis’s Bot. Mag. 2015, 32, 3–23. [Google Scholar] [CrossRef]
- Cribb, P.J.; Kell, S.P.; Dixon, K.W.; Barrett, R.L. Orchid conservation: A global perspective. In Orchid Conservation: A Global Perspective; Dixon, K.W., Kell, S.P., Barrett, R.L., Cribb, P.J., Eds.; Natural History Publications (Borneo): Kota Kinabalu, Malaysia, 2003. [Google Scholar]
- Ghorbani, A.; Gravendeel, B.; Zarre, S.; de Boer, H. Illegal wild collection and international trade of CITES-listed terrestrial orchid tubers in Iran. Traffic Bull. 2014, 26, 52–58. [Google Scholar]
- Hinsley, A.; Nuno, A.; Ridout, M.; St John, F.A.V.; Roberts, D.L. Estimating the extent of CITES noncompliance among traders and end-consumers, lessons from the global orchid trade. Cons. Lett. 2017, 10, 602–609. [Google Scholar] [CrossRef]
- Sharma, V. Agrotechniques in orchids. Adv. Tissue Eng. Regen. Med. 2017, 2, 105–108. [Google Scholar]
- Bose, S.K.; Tiwari, P. Orchid biology: A biotechnological perspective on the prospects and challenges in Orchid cultivation. Vitr. Cult. Commer. Flower. Plants Orchid. 2020, 2021, 162–210. [Google Scholar]
- Pansarin, E.; Pansarin, L.; Poleti, M.M.; Gobbo-Neto, L. Self-compatibility and specialization in a fly-pollinated Acianthera (Orchidaceae: Pleurothallidiinae). Aust. J. Bot. 2016, 64, 359–367. [Google Scholar] [CrossRef]
- Veitch, H.J. The hybridization of orchids. The report on the orchid conference held at South Kensignton. J. R. Hortic. Soc. 1886, 1, 22–49. [Google Scholar]
- de Chandra, L.; Pathak, P.; Rao, A.N.; Rajeevan, P.K. Breeding approaches for improved genotypes. In Commercial Orchids; De, L.C., Ed.; De Gruyter Open Poland: Warsaw, Polland, 2019; Volume 300. [Google Scholar]
- Su, J.S.; Jiang, J.F.; Zhang, F.; Liu, Y.; Ding, L.; Chen, S.M.; Chen, F.D. Current achievements and future prospects in the genetic breeding of Chrysanthemum: A review. Hort. Res. 2019, 6, 109. [Google Scholar] [CrossRef]
- Zhang, Z.S.; He, Q.Y.; Fu, X.L.; Ou, X.J.; Lin, W.Q.; Jiang, J.Y. Studies on the wide cross of Chinese orchids and the germination of their hybrid seeds. J. South China Agric. Univ. 2001, 2, 62–65. [Google Scholar]
- Luo, Y.H.; Huang, M.L.; Wu, J.S. Progress in Oncidium breeding study. Acta Agric. Jiangxi 2012, 24, 15–20. [Google Scholar]
- Lu, H.C.; Chen, H.H.; Tsai, W.C.; Chen, W.H.; Su, H.J.; Chang, C.N.; Yeh, H.H. Strategies for functional validation of genes involved in reproductive stages of orchids. Plant Physiol. 2007, 143, 558–569. [Google Scholar] [CrossRef]
- Pan, I.C.; Liao, D.C.; Wu, F.H.; Daniell, H.; Singh, N.D.; Chang, C.; Shih, M.C.; Chan, M.T.; Lin, C.S. Complete chloroplast genome sequence of an orchid model plant candidate: Erycina pusilla apply in tropical Oncidium breeding. PLoS ONE 2012, 7, e34738. [Google Scholar] [CrossRef]
- Wang, H.M.; Tong, C.G.; Jang, S. Current progress in orchid flowering/flower development research. Plant Signal. Behav. 2017, 12, e1322245. [Google Scholar] [CrossRef]
- Osadchuk, V.D.; Saranchuk, I.I.; Lesyk, O.B.; Olifirovych, V.O. Selective breeding in plant growing in Bukovina. Taurian Sci. Herald. 2020, 115, 16. [Google Scholar]
- Li, X.L.; An, D. Induction and identification of autotetraploids in Dendrobium. Hortic. J. 2009, 8, 1239–1242. [Google Scholar]
- Cui, G.R.; Zhang, Z.X.; Zhang, C.Y.; Hu, N.B.; Sui, Y.H.; Li, J.Q. Polyploid induction and identification of Oncidium. Acta Pratacult. Sinica 2010, 19, 184–190. [Google Scholar]
- Cui, G.R.; Zhang, Z.X.; Hu, N.B.; Zhang, C.Y.; Hou, X.L. Tetraploid of Phalaenopsis induction via colchicine treatment from protocorm-like bodies in liquid culture. J. Zhejiang Univ.—Agric. Life Sci. 2010, 1, 49–55. [Google Scholar]
- Zhang, J.X.; Wu, K.L.; Tian, L.N.; Zeng, S.J.; Duan, J. Cloning and characterization of a novel CONSTANS-like gene from Phalaenopsis hybrida. Acta Physiol. Plant. 2011, 33, 409–417. [Google Scholar] [CrossRef]
- Cheng, Q.Q. Studies on Leaf Culture and Induction of Octoploid of Phalaenopsis Cultivars. Master’s Dissertation, Shantou University, Shantou, China, 2011. [Google Scholar]
- Wang, S.Y.; Lee, P.F.; Lee, Y.I.; Hsiao, Y.Y.; Chen, Y.Y.; Pan, Z.J.; Liu, Z.J.; Tsai, W.C. Duplicated C-class MADS-box genes reveal distinct roles in gynostemium development in Cymbidium ensifolium (Orchidaceae). Plant Cell Physiol. 2011, 52, 563–577. [Google Scholar] [CrossRef] [PubMed]
- Jiang, G.L. Molecular marker-assisted breeding: A plant breeder’s review. In Advances in Plant Breeding Strategies: Breed, Biotechnology Molecular Tools; Al-Khayri Shri, J.M., Jain, S.M., Johnson, D.V., Eds.; Springer: New York, NY, USA, 2015; pp. 431–472. [Google Scholar]
- Li, X.; Jin, F.; Jin, L.; Jackson, A.; Huang, C.; Li, K.; Shu, X. Development of Cymbidium ensifolium genic SSR markers and their utility in genetic diversity and population structure analysis in Cymbidiums. BMC Genet. 2014, 15, 124. [Google Scholar] [CrossRef]
- Wu, W.L.; Chung, Y.L.; Kuo, Y.T. Development of SSR markers in Phalaenopsis orchids, their characterization, cross-transferability, and application for identification. In Orchid Biotechnology III; World Scientific: London, UK, 2017; pp. 91–107. [Google Scholar]
- Sudarsono, S.; Haristianita, M.D.; Handini, A.S.; Sukma, D. Molecular marker development based on diversity of genes associated with pigment biosynthetic pathways to support breeding for novel colors in Phalaenopsis. Acta Hortic. 2017, 1167, 305–312. [Google Scholar] [CrossRef]
- Lu, J.J.; Liu, Y.Y.; Xu, J.; Mei, Z.W.; Shi, Y.J.; Liu, P.L.; He, J.B.; Wang, X.T.; Meng, Y.J.; Feng, S.G.; et al. High-density genetic map construction and stem total polysaccharide content related QTL exploration for Chinese endemic Dendrobium (Orchidaceae). Front. Plant Sci. 2018, 9, 398. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.Y.; Wu, K.L.; Zhang, J.X.; Deng, R.F.; Duan, J.; Silva, J.A.T.; Huang, W.C.; Zeng, S.J. Embryo development in association with asymbiotic seed germination in vitro of Paphiopedilum armeniacum. Sci. Rep. 2015, 12, 16356. [Google Scholar] [CrossRef] [PubMed]
- Pakum, W.; Watthana, S.; Srimuang, K.O.; Kongbangkerd, A. Influence of the medium component on in vitro propagation of Thai’s endangered orchid: Bulbophyllum nipondhii Seidenf. Plant Tissue Cult. Biotechnol. 2016, 25, 37–46. [Google Scholar] [CrossRef]
- Diengdoh, R.V.; Kumaria, S.; Tandon, P.; Das, M.C. Asymbiotic germination and seed storage of Paphiopedilum insigne, an endangered lady’s slipper orchid. S. Afr. J. Bot. 2017, 112, 215–224. [Google Scholar] [CrossRef]
- Sherif, N.A.; Benjamin, J.H.F.; Kumar, T.S.; Rao, M.V. Somatic embryogenesis, acclimatization, and genetic homogeneity assessment of regenerated plantlets of Anoectochilus elatus Lindl., an endangered terrestrial jewel orchid. Plant Cell Tissue Organ Cult. 2018, 132, 303–316. [Google Scholar] [CrossRef]
- Pant, B. Medicinal orchids and their uses: Tissue culture a potential alternative for conservation. Afr. J. Plant Sci. 2013, 7, 448–467. [Google Scholar] [CrossRef]
- Shekarriz, P.; Kafi, M.; Deilamy, S.D.; Mirmasoumi, M. Coconut water and peptone improve seed germination and protocorm-like body formation of hybrid Phalaneopsis. Agric. Sci. Dev. 2014, 3, 317–322. [Google Scholar]
- Shukla, S.P.; Sharma, A. In vitro seed germination, proliferation, and ISSR marker-based clonal fidelity analysis of Shorea tumbuggaia Roxb.: An endangered and high trade medicinal tree of Eastern Ghats. Vitr. Cell. Dev. Biol. 2017, 53, 200–208. [Google Scholar] [CrossRef]
- Knudson, L. A New Nutrient Solution for Germination of Orchid Seed; American Orchid Society Bulletin: Cambridge, MA, USA, 1946; Volume 15, pp. 14–217. [Google Scholar]
- Long, B.; Niemiera, A.X.; Cheng, Z.; Long, C. In vitro propagation of four threatened Paphiopedilum species (Orchidaceae). Plant Cell Tissue Organ. Cult. 2010, 101, 151–162. [Google Scholar] [CrossRef]
- Parthibhan, S.; Benjamin, J.H.; Muthukumar, M.; Sherif, N.A.; Kumar, T.S.; Rao, M.V. Influence of nutritional media and photoperiods on in vitro asymbiotic seed germination and seedling development of Dendrobium aqueum Lindley. Afr. J. Plant Sci. 2012, 6, 383–393. [Google Scholar] [CrossRef]
- Zeng, S.; Huang, W.; Wu, K.; Zhang, J.; Silva, J.A.T.; Duan, J. In vitro propagation of Paphiopedilum orchids. Crit. Rev. Biotechnol. 2016, 36, 521–534. [Google Scholar] [PubMed]
- Gow, W.P.; Chen, J.T.; Chang, W.C. Influence of growth regulators on direct embryo formation from leaf explants of Phalaenopsis orchids. Acta Physiol. Plant 2008, 30, 507. [Google Scholar] [CrossRef]
- Lindley, C.P.; Arniputri, R.B.; Soliah, L.A.; Cahyono, O. Effects of organic additives and naphthalene acetic acid (NAA) application on the in vitro growth of black orchid hybrid Coelogyne pandurata lindley. Bulg. J. Agric. Sci. 2017, 23, 951–957. [Google Scholar]
- Zhang, Y.; Lee, Y.I.; Deng, L.; Zhao, S. Asymbiotic germination of immature seeds and the seedling development of Cypripedium macranthos Sw., an endangered lady’s slipper orchid. Sci. Hortic. 2013, 164, 130–136. [Google Scholar] [CrossRef]
- Mii, M.; Chin, D.P. Genetic transformation of orchid species: An overview of approaches and methodologies. In Orchid Propagation: From Laboratories to Greenhouses-Methods and Protocols; ISHS: Leuven, Belgium, 2018; pp. 347–365. [Google Scholar]
- Teixeira da Silva, J.A.; Tanaka, M. Multiple regeneration pathways via thin cell layers in hybrid Cymbidium (Orchidaceae). J. Plant Growth Regul. 2006, 25, 203–210. [Google Scholar] [CrossRef]
- Sivasubramaniam, S.; Loh, C.S.; Lee, Y.K.; Hew, C.S. Low-temperature storage of Dendrobium multico white callus tissue. Malay. Orchid. Rev. 1987, 21, 22–26. [Google Scholar]
- Tiwari, P.; Jen Tsung, C. Advances in Orchid Biology, Biotechnology, and Omics; Springer Publication: Berlin/Heidelberg, Germany, 2023; ISBN 13-978-9819910786. [Google Scholar]
- Chang, C.; Sung, P.G.; Chang, C.H.; Chen, Y.C.; Lin, Y.H. Seed development and storage of Bletilla formosana (Hayata) Schltr. Seed Nurs. 2006, 8, 29–38. [Google Scholar]
- Hirano, T.; Godo, T.; Miyoshi, K.; Ishikawa, K.; Ishikawa, M.; Mii, M. Cryopreservation and low-temperature storage of seeds of Phaius tankervilleae. Plant Biotechnol. Rep. 2009, 3, 103–109. [Google Scholar] [CrossRef]
- Wu, R.-Y.; Chang, S.-Y.; Hsieh, T.-F.; Chuang, K.-C.; Ting, I.; Lai, Y.-H.; Chang, Y.-S. Cryopreservation of Orchid Genetic Resources by Desiccation: A Case Study of Bletilla formosana; IntechOpen: London, UK, 2016. [Google Scholar] [CrossRef]
- Khoddamzadeh, A.A.; Sinniah, U.R.; Lynch, P.; Kadir, M.A.; Kadzimin, S.B.; Mahmood, M. Cryopreservation of protocorm-like bodies (PLBs) of Phalaenopsis bellina (Rchb.f.) Christenson by encapsulation-dehydration. Plant Cell Tissue Organ. Cult. 2011, 107, 4. [Google Scholar] [CrossRef]
- Popova, E.; Kim, H.H.; Saxena, P.K.; Engelmann, F. Frozen beauty: The cryo-biotechnology of orchid diversity. Biotechnol. Adv. 2016, 34, 380–403. [Google Scholar] [CrossRef] [PubMed]
- Sakai, A. Development of cryopreservation techniques. In Cryopreservation of Tropical Plant Germplasm; Engellmann, F., Takagi, H., Eds.; IPGRI: Rome, Italy, 2000; pp. 1–7. [Google Scholar]
- Volk, G.M.; Walters, C. Plant vitrification solution lowers water content and alters freezing behavior in shoot tips during cryoprotection. Cryobiology 2006, 52, 48–61. [Google Scholar] [CrossRef]
- Hirano, T.; Ishikawa, K.; Mii, M. Advances on orchid cryopreservation. In Floriculture Ornamental and Plant Biotechnology: Advances and Topical Issues, 1st ed.; Global Science Book; Teiceira da Silva, J.A., Ed.; Ikenobe: London, UK, 2006; Volume 2, pp. 410–414. [Google Scholar]
- Fabre, J.; Dereuddre, J. Encapsulation-dehydration: A new approach to cryopreservation of Solanum shoot tips. Cryo-Lett. 1990, 11, 413–426. [Google Scholar]
- Flachsland, E.; Terada, G.; Scocchi, A.; Rey, H.; Mroginski, L.; Engelmann, F. Cryopreservation of seeds and in vitro-cultured protocorms of Oncidium bifolium sims. (Orchidaceae) by encapsulation-dehydration. Cryo-Lett. 2006, 27, 235–242. [Google Scholar]
- Surenciski, M.R.; Flachsland, E.A.; Terada, G.; Mroginski, L.A.; Rey, H.Y. Cryopreservation of Cyrtopodium hatschbachii Pabst (Orchidaceae) immature seeds by encapsulation dehydration. Biocell 2012, 36, 31–36. [Google Scholar] [CrossRef]
- Mishiba, K.; Chin, D.P.; Mii, M. Agrobacterium-mediated transformation of Phalaenopsis by targeting protocorms at an early stage after germination. Plant Cell Rep. 2005, 24, 297–303. [Google Scholar] [CrossRef] [PubMed]
- Knapp, J.E.; Kausch, A.P.; Chandlee, J.M. Transformation of three genera of orchid using the bar gene as a selectable marker. Plant Cell Rep. 2000, 19, 893–898. [Google Scholar] [CrossRef]
- Tee, C.S.; Maziah, M.; Tan, C.S.; Abdullah, M.P. Evaluation of different promoters Belarmino, driving the GFP reporter gene and selected target tissues for particle bombardment of Dendrobium Sonia 17. Plant Cell Rep. 2003, 21, 452–458. [Google Scholar] [CrossRef] [PubMed]
- Belarmino, M.M.; Mii, M. Agrobacterium-mediated genetic transformation of a Phalaenopsis orchid. Plant Cell Rep. 2000, 19, 435–442. [Google Scholar] [CrossRef]
- Hiei, Y.; Komari, T.; Ishida, Y.; Saito, H. Development of Agrobacterium-mediated transformation method for monocotyledonous plants. Breed. Res. 2000, 2, 205–213. [Google Scholar] [CrossRef]
- Chin, D.P.; Mishiba, K.; Mii, M. Agrobacterium-mediated transformation of protocorm-like bodies in Cymbidium. Plant Cell Rep. 2007, 26, 735–743. [Google Scholar] [CrossRef]
- Ovcharenko, O.O.; Rudas, V.A. Modern approaches to genetic engineering in the Orchidaceae family. Cytol. Genet. 2023, 57, 142–156. [Google Scholar] [CrossRef]
- Phlaetita, W.; Chin, D.P.; Otang, N.V.; Nakamura, I.; Mii, M. High-efficiency Agrobacterium-mediated transformation of Dendrobium orchid using protocorms as a target material. Plant Biotechnol. 2015, 32, 323–327. [Google Scholar] [CrossRef]
- Teixeira da Silva, J.A.; Dobranszki, J.; Cardoso, J.C.; Chandler, S.F.; Zeng, S. Methods for genetic transformation in Dendrobium. Plant Cell Rep. 2016, 35, 483–504. [Google Scholar] [CrossRef]
- Liu, J.; Chiou, C.; Shen, C.; Chen, P.J.; Liu, Y.C.; Jian, C.D.; Shen, X.L.; Shen, F.Q.; Yeh, K.W. RNA Interference-Based Gene Silencing of Phytoene Synthase Impairs Growth, Carotenoids, and Plastid Phenotype in Oncidium Hybrid Orchid; Springer Plus: Berlin/Heidelberg, Germany, 2014; Volume 478. [Google Scholar]
- Iiyama, C.M.; Vilcherrez-Atoche, J.A.; Germanà, M.A.; Vendrame, W.A.; Cardoso, J.C. Breeding of ornamental orchids with focus on Phalaenopsis: Current approaches, tools, and challenges for this century. Heredity 2024. [Google Scholar] [CrossRef]
- Tiwari, P.; Bose, S.K.; Gautam, A.; Chen, J.-T. Emerging insights into the cultivation strategies, ethnomedicinal uses, and socioeconomic attributes of Orchids. J. Hortic. Sci. Biotechnol. 2023, 20, 420–438. [Google Scholar]
- Hsiao, Y.Y.; Pan, Z.J.; Hsu, C.C.; Yang, Y.P.; Hsu, Y.C.; Chuang, Y.C.; Shih, H.-H.; Chen, W.-h.; Tsai, W.-C.; Chen, H.-H. Research on orchid biology and biotechnology. Plant Cell Physiol. 2011, 52, 1467–1486. [Google Scholar] [CrossRef]
- Yu, H.; Goh, C.J. Identification and characterization of three orchid MADS-box genes of the AP1/AGL9 subfamily during floral transition. Plant Physiol. 2000, 123, 1325–1336. [Google Scholar] [CrossRef]
- Ding, L.; Wang, Y.; Yu, H. Overexpression of DOSOC1, an ortholog of Arabidopsis SOC1, promotes flowering in the orchid Dendrobium Chao Parya Smile. Plant Cell Physiol. 2013, 54, 595–608. [Google Scholar] [CrossRef]
- Liang, S.; Ye, Q.S.; Li, R.H.; Leng, J.Y.; Li, M.R.; Wang, X.J.; Li, H.Q. Transcriptional regulations on the low-temperature-induced floral transition in an Orchidaceae species, Dendrobium nobile: An expressed sequence tags analysis. Int. J. Genom. 2012, 2012, 757801. [Google Scholar]
- Liu, X.R.; Pan, T.; Liang, W.Q.; Gao, L.; Wang, X.J.; Li, H.Q.; Liang, S. Overexpression of an orchid (Dendrobium nobile) SOC1/TM3-Like Ortholog, DnAGL19, in Arabidopsis regulates HOS1-FT expression. Front. Plant Sci. 2016, 7, 99. [Google Scholar] [CrossRef]
- Wang, Y.W.; Liu, L.; Song, S.Y.; Li, Y.; Shen, L.S.; Yu, H. DOFT and DOFTIP1 affect reproductive development in the orchid Dendrobium Chao Praya Smile. J. Exp. Bot. 2017, 68, 5759–5772. [Google Scholar] [CrossRef]
- Sawettalake, N.; Bunnag, S.; Wang, Y.; Shen, L.; Yu, H. DOAP1 promotes flowering in the orchid Dendrobium Chao Praya Smile. Front. Plant Sci. 2017, 8, 400. [Google Scholar] [CrossRef]
- Chen, W.; Qin, Q.; Zheng, Y.; Wang, C.; Wang, S.; Zhou, M.; Zhang, C. Overexpression of Doritaenopsis hybrid EARLY FLOWERING 4-like4 Gene, DhEFL4, postpones flowering in transgenic Arabidopsis. Plant Mol. Biol. Rep. 2016, 34, 103–117. [Google Scholar] [CrossRef]
- Hsu, H.F.; Huang, C.H.; Chou, L.T.; Yang, C.H. Ectopic expression of an orchid (Oncidium Gower Ramsey) AGL6-like gene promotes flowering by activating flowering time genes in Arabidopsis thaliana. Plant Cell Physiol. 2003, 44, 783–794. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Li, Y.; Zhang, M.-M.; He, X.; Ahmad, S.; Lan, S.; Liu, Z.-J. Research advances on the gene regulation of floral development and color in orchids. Gene 2023, 888, 147751. [Google Scholar] [CrossRef] [PubMed]
- Tsai, W.C.; Lee, P.F.; Chen, H.I.; Hsiao, Y.Y.; Wei, W.J.; Pan, Z.J.; Chuang, M.H.; Kuoh, C.S.; Chen, W.H.; Chen, H.H. PeMADS6, a GLOBOSA/PISTILLATA-like gene in Phalaenopsis equestris involved in petaloid formation, and correlated with flower longevity and ovary development. Plant Cell Physiol. 2005, 46, 1125–1139. [Google Scholar] [CrossRef]
- Pan, Z.J.; Chen, Y.Y.; Du, J.S.; Chen, Y.Y.; Chung, M.C.; Tsai, W.C.; Wang, C.N.; Chen, H.H. Flower development of Phalaenopsis orchid involves functionally divergent SEPALLATA-like genes. New Phytol. 2014, 202, 1024–1042. [Google Scholar] [CrossRef]
- Jang, S.; Choi, S.C.; Li, H.Y.; An, G.; Schmelzer, E. Functional characterization of Phalaenopsis aphrodite flowering genes PaFT1 and PaFD. PLoS ONE 2015, 10, e0134987. [Google Scholar] [CrossRef]
- Wang, S.-L.; Viswanath, K.K.; Tong, C.-G.; An, H.R.; Jang, S.; Chen, F.-C. Floral induction and flower development of Orchids. Front. Plant Sci. 2019, 10, 1258. [Google Scholar] [CrossRef]
- Chen, D.; Guo, B.; Hexige, S.; Zhang, T.; Shen, D.; Ming, F. SQUA-like genes in the orchid Phalaenopsis are expressed in both vegetative and reproductive tissues. Planta 2007, 226, 369–380. [Google Scholar] [CrossRef]
- Su, C.-l.; Chen, W.-C.; Lee, A.-Y.; Chen, C.-Y.; Chang, Y.-C.A.; Chao, Y.-T. A modified ABCDE model of flowering in orchids based on gene expression profiling studies of the moth orchid Phalaenopsis aphrodite. PLoS ONE 2013, 8, e80462. [Google Scholar] [CrossRef]
- Teo, Z.W.N.; Zhou, W.; Shen, L. Dissecting the function of MADS-box transcription factors in Orchid reproductive development. Front. Plant Sci. 2019, 10, 1474. [Google Scholar] [CrossRef]
- Raffeiner, B.; Serek, M.; Winkelmann, T. Agrobacterium tumefaciens mediated transformation of Oncidium and Odontoglossum orchid species with the ethylene receptor mutant gene etr1-1. Plant Cell Tissue Organ Cult. 2009, 98, 125–134. [Google Scholar] [CrossRef]
- Chen, W.H.; Hsu, C.Y.; Cheng, H.Y.; Chang, H.; Chen, H.H.; Ger, M.J. Down-regulation of putative UDP-glucose: Flavonoid 3-O-glucosyltransferase gene alters flower coloring in Phalaenopsis. Plant Cell Rep. 2011, 30, 1007–1017. [Google Scholar] [CrossRef]
- Yu, H.; Yang, S.H.; Goh, C.J. Agrobacterium-mediated transformation of a Dendrobium orchid with the 1 knox gene DOH1. Plant Cell Rep. 2001, 20, 301–305. [Google Scholar] [CrossRef]
- Yang, S.H.; Yu, H.; Goh, C.J. Functional characterization of a cytokinin oxidase gene DSCKX1 in Dendrobium orchid. Plant Mol. Biol. 2003, 51, 237–248. [Google Scholar] [CrossRef]
- Yu, H.; Xu, Y. Orchids. In Biotechnology in Agriculture and Forestry, Transgenic Crops IV; Pua, E.C., Davey, M.R., Eds.; Springer: Berlin/Heidelberg, Germany, 2007; pp. 273–286. [Google Scholar]
- Srivastava, S.; Kadooka, C.; Uchida, J.Y. Fusarium species as pathogen on orchids. Microbiol. Res. 2018, 207, 188–195. [Google Scholar] [CrossRef]
- Adil, M.; Tiwari, P.; Chen, J.-T.; Kanwal, S. Major bioactive metabolites and antimicrobial potential of Orchidaceae Fungal endophytes. In Advances in Orchid Biology, Biotechnology, and Omics; Tiwari, P., Chen, J.-T., Eds.; Springer Publishers: Berlin/Heidelberg, Germany, 2023; ISBN 978-9819910786. [Google Scholar]
- Taylor, S.C.; Laperriere, G.; Germain, H. Droplet digital PCR versus q-PCR for gene expression analysis with low abundant targets: From variable nonsense to publication quality data. Sci. Rep. 2017, 7, 2409. [Google Scholar] [CrossRef] [PubMed]
- Chocarro-Ruiz, B.; Fernández-Gavela, A.; Herranz, S.; Lechuga, L.M. Nanophotonic label-free biosensors for environmental monitoring. Curr. Opin. Biotechnol. 2017, 45, 175–183. [Google Scholar] [CrossRef]
- Jain, A.; Sarsaiya, S.; Chen, J.; Wu, Q.; Lu, Y.; Shi, J. Changes in global Orchidaceae disease geographical research trends: Recent incidences, distributions, treatment, and challenges. Bioengineered 2021, 12, 13–29. [Google Scholar] [CrossRef]
- Chang, W.H.; Yang, S.Y.; Lin, C.L.; Wang, C.-H.; Li, P.-C.; Chen, T.-Y.; Jan, F.-J.; Lee, G.-B. Detection of viruses directly from the fresh leaves of a Phalaenopsis orchid using a microfluidic system. Nanomedicine 2013, 9, 1274–1282. [Google Scholar] [CrossRef]
- Tong, C.G.; Wu, F.H.; Yuan, Y.H.; Chen, Y.R.; Lin, C.S. High-efficiency CRISPR/Cas-based editing of Phalaenopsis orchid MADS-genes. Plant Biotechnol. J. 2019, 18, 889–891. [Google Scholar] [CrossRef]
- Ma, X.; Zhang, Q.; Zhu, Q.; Liu, W.; Chen, Y.; Qiu, R.; Wang, B.; Yang, Z.; Li, H.; Lin, Y.; et al. A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol. Plant 2015, 8, 1274–1284. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.S.; Hsu, C.T.; Yang, L.H.; Lee, L.Y.; Fu, J.Y.; Cheng, Q.W.; Wu, F.H.; Hisao, H.C.W.; Zhang, Y.S.; Zhang, R.; et al. Application of protoplast technology to CRISPR/Cas9 mutagenesis: From single-cell mutation detection to mutant plant regeneration. Plant Biotechnol. J. 2018, 16, 1295–1310. [Google Scholar] [CrossRef] [PubMed]
- Semiarti, E.; Nopitasari, S.; Setiawati, Y.; Lawrie, M.D.; Purwantoro, A.; Widada, J.; Ninomiya, K.; Asano, Y.; Matsumoto, S.; Yoshioka, Y. Application of CRISPR/Cas9 genome editing system for molecular breeding of orchids. Indones. J. Biotechnol. 2020, 25, 61–68. [Google Scholar] [CrossRef]
- Xia, K.; Zhang, D.; Xu, X.; Liu, G.; Yang, Y.; Chen, Z.; Wang, X.; Zhang, G.-Q.; Sun, H.-X.; Gu, Y. Protoplast technology enables the identification of efficient multiplex genome editing tools in Phalaenopsis. Plant Sci. 2022, 322, 111368. [Google Scholar] [CrossRef]
- Kui, L.; Chen, H.; Zhang, W.; He, S.; Xiong, Z.; Zhang, Y.; Yan, L.; Zhong, C.; He, F.; Chen, J.; et al. Building a genetic manipulation toolbox for orchid biology: Identification of constitutive promoters and application of CRISPR/Cas9 in the orchid, Dendrobium officinale. Front. Plant Sci. 2017, 7, 2036. [Google Scholar] [CrossRef]
- Jalal, J.S.; Kumar, P.; Pangtey, Y. Ethnomedicinal orchids of Uttarakhand, Western Himalaya. Ethnobot. Leafl. 2008, 12, 1227–1230. [Google Scholar]
- Reinikka, M.A. A History of the Orchid; Portland Timber Press: Oregon, Poland, 1995. [Google Scholar]
- Bulpitt, C. The uses and misuses of orchids in medicine. QJM Int. J. Med. 2005, 98, 625–631. [Google Scholar] [CrossRef]
- Li, P.-Y.; Li, L.; Wang, Y.-Z. Traditional uses, chemical compositions and pharmacological activities of Dendrobium: A review. J. Ethnopharmacol. 2023, 310, 116382. [Google Scholar] [CrossRef]
- Gutierrez, R.M.P. Orchids: A review of uses in traditional medicine, its phytochemistry, and pharmacology. J. Med. Plant Res. 2010, 4, 592–638. [Google Scholar]
- Pant, B.; Shrestha, S.; Pradhan, S. In vitro seed germination and seedling development of Phaius tancarvilleae (L’Her.) Blume. Sci. World 2011, 9, 50–52. [Google Scholar] [CrossRef]
- Arditti, J.; Emst, R. Micropropagation of Orchid; Wiley: New York, NY, USA, 1993. [Google Scholar]
- Teoh, E.S. Genus: Calanthe to Cyrtosia. In Medicinal orchids of Asia; Springer: Cham, Switzerland, 2015; pp. 171–250. [Google Scholar]
- Singh, S.; Singh, A.K.; Kumar, S.; Kumar, M.; Pandey, P.K.; Singh, M.C.K. Medicinal properties and uses of orchids: A concise review. Elixir Appl. Bot. 2012, 52, 11627–11634. [Google Scholar]
- Neelam, S.; Chandel, K.P.S. In Vitro conservation of Orchis latifolia: A threatened, medicinal terrestrial orchid. Ind. J. Plant Genet. Resour. 1996, 9, 109–113. [Google Scholar]
- Radice, M.; Scalvenzi, L.; Gutierrez, D. Ethnopharmacology, bioactivity and phytochemistry of Maxillaria densa Lindl. Scientific review and biotrading in the neotropics. Rev. Colomb. For. 2020, 23, 20–33. [Google Scholar]
- Pradhan, S.; Tiruwa, B.; Subedee, B.R.; Pant, B. In vitro germination and propagation of a threatened medicinal orchid, Cymbidium aloifolium (L.) Sw. through artificial seed. Asian Pacific J. Trop. Biomed. 2014, 4, 971–976. [Google Scholar] [CrossRef]
- Natta, S.; Mondol, M.S.A.; Pal, K.; Mandal, S.; Sahana, N.; Pal, R.; Pandit, G.K.; Alam, B.K.; Das, S.S.; Biswas, S.S.; et al. Chemical composition, antioxidant activity, and bioactive constituents of six native endangered medicinal orchid species from north-eastern Himalayan region of India. South Afri. Bot. 2022, 150, 248–259. [Google Scholar] [CrossRef]
- Zhang, X.; Chen, W.; Du, Y.; Su, P.; Qiu, Y.; Ning, J.; Liu, M. Phytochemistry and pharmacological activities of Arundina graminifolia (D. Don) Hochr. and other common Orchidaceae medicinal plants. J. Ethnopharmacol. 2021, 276, 114143. [Google Scholar] [CrossRef]
- Chung, H.-H.; Shi, S.-K.; Huang, B.; Chen, J.-T. Enhanced agronomic traits and medicinal constituents of Autotetraploids in Anoectochilus formosanus Hayata, a top-grade medicinal orchid. Molecules 2017, 22, 1907. [Google Scholar] [CrossRef]
- Rosa, P.W.; Machado, M.S.; de Campos-Buzzi, F.; Niero, R.; Monache, F.D.; Filho, V.C. Seasonal and biological variations of Epidendrum mosenii: Quantification of 24-methylenecycloartanol using gas chromatography. Nat. Prod. Res. 2007, 21, 975–981. [Google Scholar] [CrossRef] [PubMed]
- Huang, N.-K.; Chern, Y.; Fang, J.-M.; Lin, C.-I.; Chen, W.-P.; Lin, Y.-L. Neuroprotective principles from Gastrodia elata. J. Nat. Prod. 2007, 70, 571–574. [Google Scholar] [CrossRef] [PubMed]
- Nongdam, P. Ethno-medicinal uses of some orchids of Nagaland, North-east India. Res. J. Med. Plants 2014, 8, 126–139. [Google Scholar] [CrossRef]
- Lam, Y.; Ng, T.B.; Yao, R.M.; Shi, J.; Xu, K.; Sze, S.C.W.; Zhang, K.Y. Evaluation of chemical constituents and important mechanism of pharmacological biology in dendrobium plants. Evid.-Based Complement. Alternat. Med. 2015, 2015, 841752. [Google Scholar] [CrossRef] [PubMed]
- Patil, M.C.; Mahajan, R.T. Ethnobotanical potential of Eulophia species for their possible biological activity. Int. J. Pharm. Sci. Rev. Res. 2013, 21, 297–307. [Google Scholar]
- Bose, B.; Choudhury, H.; Tandon, P.; Kumaria, S. Studies on secondary metabolite profiling, anti-inflammatory potential, in vitro photoprotective and skin-aging related enzyme inhibitory activities of Malaxis acuminata, a threatened orchid of nutraceutical importance. J. Photochem. Photobiol. B Biol. 2017, 173, 686–695. [Google Scholar] [CrossRef]
- Uddin, M.J.; Rahman, M.M.; Abdullah-Al-Mamun, M.; Sadik, G. Vanda roxburghii: An experimental evaluation of antinociceptive properties of a traditional epiphytic medicinal orchid in animal models. BMC Complement. Altern. Med. 2015, 15, 305. [Google Scholar] [CrossRef] [PubMed]
- Ferrara, L. Medicinal and pharmaceutical properties of Vanilla planifolia. Int. J. Med. Rev. 2020, 7, 25–29. [Google Scholar]
- Mishra, A.P.; Saklani, S.; Salehi, B.; Parcha, V.; Sharifi-Rad, M.; Milella, L.; Iriti, M.; Sharifi-Rad, J.; Srivastava, M. Satyrium nepalense, a high-altitude medicinal orchid of Indian Himalayan region: Chemical profile and biological activities of tuber extracts. Cell. Mol. Biol. 2018, 64, 35–53. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Wang, X.; Fang, J.; Zhao, Z.; Huang, L.; Guo, H.; Zheng, X. Bletilla striata: Medicinal uses, phytochemistry and pharmacological activities. J. Ethnopharmacol. 2017, 195, 20–38. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, K.; Tanaka, R.; Sakurai, H.; Iguchi, K.; Yamada, Y.; Hsu, C.S.; Sakuma, C.; Kikuchi, H.; Shibayama, H.; Kawai, T. Structure of Cymbidine A, a monomeric peptidoglycan-related compound with hypotensive and diuretic activities, isolated from a higher plant, Cymbidium goeringii (Orchidaceae). Chem. Pharm. Bull. 2007, 55, 780–783. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Liu, G.T.; Shi, J.G.; Zhang, J.J. Effects of Coeloglossum viride var. bracteatum extract on memory deficits and pathological changes in senescent mice. Basic Clin. Pharmacol. Toxicol. 2006, 98, 55–60. [Google Scholar] [PubMed]
- Matsuda, H.; Morikawa, T.; Xie, H.; Yoshikawa, M. Antiallergic phenanthrenes and stilbenes from the tubers of Gymnadenia conopsea. Planta Medica 2004, 70, 847–855. [Google Scholar] [CrossRef] [PubMed]
- Arora, M.; Mahajan, A.; Sembi, J.S. A Review on phytochemical and pharmacological potential of family Orchidaceae. Int. Res. J. Pharm. 2017, 8, 9–24. [Google Scholar] [CrossRef]
- Hossain, M.M. Therapeutic orchids: Traditional uses and recent advances-an overview. Fitoterapia 2011, 82, 102–140. [Google Scholar] [CrossRef]
- Akter, S.; Huda, M.K.; Rahman, M.; Hoque, M.M.; Eva, T.A. Phytochemical profiling and bioactivities of Pholidota pallida Lindl. Int. J. Adv. Res. Bot. 2019, 5, 8–13. [Google Scholar]
- Tsering, J.; Tam, N.; Tag, H.; Gogoi, B.J.; Apang, O. Medicinal orchids of Arunachal Pradesh: A review. Bull. Arunachal For. Res. 2017, 32, 1–16. [Google Scholar]
- Singh, A.; Duggal, S. Medicinal Orchids: An overview. Ethnobot. Leafl. 2009, 13, 351–363. [Google Scholar]
- Bhattarai, P.; Adhikari, Y.; Kunwar, R.M.; Bussmann, R.W. Pleione humilis (Sm.) D. Don. Orchidaceae. In Ethnobotany of the Himalayas, Ethnobotany of Mountain Regions, Kunwar, R., Sher, H., Bussmann, R.W., Eds.; Springer: Cham, Switzerland, 2021. [Google Scholar] [CrossRef]
- Chen, C.C.; Wu, L.G.; Ko, F.N.; Teng, C.M. Antiplatelet aggregation principles of Dendrobium loddigesii. J. Nat. Prod. 1994, 57, 1271–1274. [Google Scholar] [CrossRef] [PubMed]
- Chinsamy, M.; Finnie, J.F.; van Staden, J. The ethnobotany of South African medicinal orchids. S. Afr. J. Bot. 2011, 77, 2–9. [Google Scholar] [CrossRef]
- Majumder, P.L.; Pal, S. Rotundatin, a new 9,10-dihydrophenanthrene derivative from Dendrobium rotunatum. Phytochemistry 1992, 31, 3225–3228. [Google Scholar] [CrossRef]
- Honda, C.; Yamaki, M. Phenanthrenes from Dendrobium plicatile. Phytochemistry 2000, 53, 987–990. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Xu, J.K.; Wang, J.; Li, W.N.; Kurihara, H.; Kitanaka, S.; Yao, X.S. Bioactive bibenzyl derivatives and fluorenones from Dendrobium nobile. J. Nat. Prod. 2007, 70, 24–28. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wang, C.L.; Guo, S.X.; Yang, J.S.; Xiao, P.G. Two new compounds from Dendrobium candidum. Chem. Pharm. Bull. 2008, 56, 1477–1479. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Yu, H.; Qing, C.; Zhang, Y.; Liu, Y.; Chen, Y. Two new biphenanthrenes with cytotoxic activity from Bulbophyllum odoratissimum. Fitoterapia 2009, 80, 381–384. [Google Scholar] [CrossRef] [PubMed]
- Lalitharani, S.; Mohan, V.R.; Maruthupandian, A. Pharmacognostic investigations on Bulbophyllum albidum (Wight) Hook. f. Int. J. PharmTech Res. 2011, 3, 556–562. [Google Scholar]
- Majumder, P.L.; Guha, B.N.S.; Ray, P.N.S. Four new stilbenoids from the orchids Coelogyne ochracea and Coelogyne cristata. J. Indian Chem. Soc. 2011, 88, 1293–1304. [Google Scholar]
- Williams, C.A. The leaf flavonoids of the Orchidaceae. Phytochemistry 1979, 18, 803–813. [Google Scholar] [CrossRef]
- Gutierrez, R.M.P.; Sosa, I.A.; Vadillo, C.H.; Victoria, T.C. Effect of flavonoids from Prosthechea michuacana on carbon tetrachloride-induced acute hepatotoxicity in mice. Pharma. Biol. 2011, 49, 1121–1127. [Google Scholar] [CrossRef] [PubMed]
- Kuo, C.T.; Chen, B.C.; Yu, C.C.; Weng, C.M.; Hsu, M.J.; Chen, C.C.; Chen, M.C.; Teng, C.M.; Pan, S.L.; Bien, M.Y.; et al. Apoptosis signal-regulating kinase 1 mediates denbinobin-induced apoptosis in human lung adenocarcinoma cells. J. Biomed. Sci. 2009, 16, 43. [Google Scholar] [CrossRef]
- Khouri, N.A.; Nawasreh, M.; Al-hussain, S.M.; Alkofahi, A.S. Effects of orchids (Orchis anatolica) on reproductive function and fertility in adult male mice. Reprod. Med. Biol. 2006, 5, 269–276. [Google Scholar] [CrossRef] [PubMed]
- Saleem, U. Investigating the medicinal properties of orchids. Columbia Sci. Rev. 2007, 4, 22–23. [Google Scholar]
- Maridass, M. Anti diarrhoeal activity of rare orchid Eulophia epidendraea (Retz.) Fisher. Nat. Pharm. Technol. 2011, 1, 5–10. [Google Scholar]
- Maridass, M.; Raju, G.; Ghanthikumar, S. Tissue-regenerative responses on tuber extracts of Eulophia epidendraea (Retz.) Fischer in Wistar rat. Pharmacology 2008, 3, 631–636. [Google Scholar]
- Ng, T.B.; Liu, J.; Wong, J.H.; Ye, X.; Wing Sze, S.C.; Tong, Y.; Zhang, K.Y. Review of research on Dendrobium, a prized folk medicine. Appl. Microbiol. Biotechnol. 2012, 93, 1795–1803. [Google Scholar] [CrossRef]
- Liu, C.L.; Liu, M.C.; Zhu, R.L. Determination of gastrodin, ρ-hydroxybenzyl alcohol, vanillyl alcohol, ρ-hydroxylbenzaldehyde, and vanillin in tall gastrodia tuber by high-performance liquid chromatography. Chromatographia 2002, 55, 317–320. [Google Scholar] [CrossRef]
- Lee, J.Y.; Jang, Y.W.; Kang, H.S.; Moon, H.; Sim, S.S.; Kim, C.J. Anti-inflammatory action of phenolic compounds from Gastrodia elata root. Arch. Pharmacal Res. 2006, 29, 849–858. [Google Scholar] [CrossRef]
- Szlachetko, D.L. Genera et species Orchidalium. Pol. Bot. J. 2001, 46, 11–26. [Google Scholar]
- Fonmboh, D.J.; Fokunang, T.E.; Ndasi, N.P.; Ngangmou, N.T.; Herve, B.; Tita, B.L.; Nubia, K.C.; Awah, T.M.; Aba, E.R.; Fokunang, C.N. An overview of the ethnobotanic, ethnopharmacological and medicinal importance of edible wild root tuber orchids in Cameroon. Asian J. Biotechnol. Bioresour. Technol. 2021, 7, 11–24. [Google Scholar] [CrossRef]
- Dikanda, P.C. Contribution à l’Étude des Plantes Médicinales de New-Malimba dans l’Arrondissement d’Edéa au Cameroun. Master’s Thesis, Université de Yaoundé I, Yaoundé, Cameroon, 2000; 81p. [Google Scholar]
- Ekole, O.D. Etude de Quelques Plantes Camerounaises Pouvant Être Utilisées Comme Médicinales; Mémoire de DIPES II; Université de Yaoundé I: Yaoundé, Cameroon, 1994; 61p. [Google Scholar]
- Simo, M.; Droissart, V.; Sonke, B.; Stevart, T. The orchid flora of the Mbam Minkom Hills (Yaounde, Cameroon). Belg. J. Bot. 2009, 142, 111–123. [Google Scholar]
- Kasulo, L.; Mwabumba, B.; Munthali, C. A review of edible orchids in Malawi. J. Hortic. For. 2009, 1, 133–139. [Google Scholar]
- Suh, C.E.; Sparks, R.S.J.; Fitton, J.G.; Ayonghe, S.N.; Annen, C.; Nana, R.; Luckman, A. The 1999 and 2000 eruptions of Mt. Cameroon; eruption, behaviour, and petrochemistry of lava. Bull. Volcanicity 2003, 65, 267–281. [Google Scholar] [CrossRef]
- Lalika, M.C.S.; Mende, D.H.; Urio, P.; Gimbi, D.M.; Mwanyika, S.J.; Donati, G. Domestication potential and nutrient composition of wild orchids from two southern regions in Tanzania. Time J. Biol. Sci. Technol. 2013, 1, 1–11. [Google Scholar]
- POWO. Vanilla planifolia Andrews. Plants of the World Online. Royal Botanic Gardens, Kew. 2023. Available online: https://powo.science.kew.org/taxon/urn:lsid:ipni.org:names:262578-2 (accessed on 15 January 2024).
- Vega, M.; Hernández, M.; Herrera-Cabrera, B.E.; Wegier, A. Vanilla planifolia. In IUCN Red List of Threatened Species; E.T103090930A172970359; International Union for Conservation of Nature and Natural Resources: Gland, Switzerland, 2020. [Google Scholar] [CrossRef]
- de Oliveira, R.T.; da Silva Oliveira, J.P.; Macedo, A.F. Vanilla beyond Vanilla planifolia and Vanilla × tahitensis: Taxonomy and historical notes, reproductive biology, and metabolites. Plants 2022, 11, 3311. [Google Scholar] [CrossRef] [PubMed]
- Hasing, T.; Tang, H.; Brym, M.; Khazi, F.; Huang, T.; Chambers, A.H. A phased Vanilla planifolia genome enables genetic improvement of flavor and production. Nat. Food 2020, 1, 811–819. [Google Scholar] [CrossRef] [PubMed]
- Balilashaki, K.; Zakizadeh, H.; Olfati, J.-A.; Vahedi, M.; Kumar, A.; Indracanti, M. Recent advances in Phalaenopsis orchid improvement using Omics approaches. Plant Tissue Cult. Biotechnol. 2019, 29, 133–214. [Google Scholar] [CrossRef]
- Leitch, I.J.; Kahandawala, I.; Suda, J.; Hanson, L.; Ingrouille, M.J.; Chase, M.W.; Fay, M.F. Genome size diversity in orchids: Consequences and evolution. Ann. Bot. 2009, 104, 469–481. [Google Scholar] [CrossRef]
- Lin, S.; Lee, H.C.; Chen, W.H.; Chen, C.C.; Kao, Y.Y.; Fu, Y.M.; Chen, Y.H.; Lin, T.Y. Nuclear DNA contents of Phalaenopsis sp. and Doritis pulcherrima. J. Am. Soc. Hortic. Sci. 2001, 126, 195–199. [Google Scholar] [CrossRef]
- Fattmah; Sukma, D. Development of sequence-based microsatellite marker for Phalaenopsis orchid. HAYATI J. Biosci. 2011, 18, 71–76. [Google Scholar] [CrossRef]
- Ko, Y.Z.; Shih, H.C.; Tsai, C.C.; Ho, H.H.; Liao, P.C.; Chiang, Y.C. Screening transferable microsatellite markers across genus Phalaenopsis (Orchidaceae). Bot. Stud. 2017, 58, 48. [Google Scholar] [CrossRef]
- Ai, Y.; Zheng, Q.D.; Wang, M.J.; Xiong, L.W.; Li, P.; Guo, L.T.; Wang, M.Y.; Peng, D.H.; Lan, S.R.; Liu, Z.J. Molecular mechanism of different flower color formation of Cymbidium ensifolium. Plant Mol. Biol. 2023, 113, 193–204. [Google Scholar] [CrossRef]
- Zhang, J.; Wu, K.; Zeng, S.; Teixeira da Silva, J.A.; Zhao, X.; Tian, C.E.; Xia, H.; Duan, J. Transcriptome analysis of Cymbidium sinense and its application to the identification of genes associated with floral development. BMC Genom. 2013, 14, 279. [Google Scholar] [CrossRef]
- Aceto, S.; Sica, M.; De Paolo, S.; D’Argenio, V.; Cantiello, P.; Salvatore, F.; Gaudio, L. The analysis of the inforescence miRNome of the orchid Orchis italica reveals a DEF-Like MADS-box gene as a new miRNA target. PLoS ONE 2014, 9, e97839. [Google Scholar] [CrossRef]
- Pontrelli, P.; Accetturo, M.; Gesualdo, L. miRNome analysis using real-time PCR. Methods Mol. Biol. 2014, 1186, 201–232. [Google Scholar]
- Kawahara, Y.; Oono, Y.; Wakimoto, H.; Ogata, J.; Kanamori, H.; Sasaki, H.; Mori, S.; Matsumoto, T.; Itoh, T. TENOR: Database for comprehensive mRNA-Seq experiments in rice. Plant Cell Physiol. 2016, 57, e7. [Google Scholar] [CrossRef]
- Szczesniak, M.W.; Rosikiewicz, W.; Makalowska, I. CANTATAdb: A collection of plant long non-coding RNAs. Plant Cell Physiol. 2016, 57, e8. [Google Scholar] [CrossRef]
- Chao, Y.-T.; Yen, S.-H.; Yeh, J.-H.; Chen, W.-C.; Shih, M.-C. Orchidstra 2.0—A transcriptomics resource for the orchid family. Plant Cell Physiol. 2017, 58, e9. [Google Scholar] [CrossRef]
- Aya, K.; Kobayashi, M.; Tanaka, J.; Ohyanagi, H.; Suzuki, T.; Yano, K.; Matsuoka, M. De novo transcriptome assembly of a fern, Lygodium japonicum, and a web resource database, Ljtrans DB. Plant Cell Physiol. 2015, 56, e5. [Google Scholar] [CrossRef]
- Ohyanagi, H.; Ebata, T.; Huang, X.; Gong, H.; Fujita, M.; Mochizuki, T.; Toyoda, A.; Fujiyama, A.; Kaminuma, E.; Nakamura, Y.; et al. Oryza Genome: Genome diversity database of wild Oryza species. Plant Cell Physiol. 2016, 57, e1. [Google Scholar] [CrossRef]
- Liu, S.-S.; Chen, J.; Li, S.-C.; Zeng, X.; Meng, Z.-X.; Guo, S.-X. Comparative transcriptome analysis of genes involved in GA-GID1-DELLA regulatory module in symbiotic and asymbiotic seed germination of Anoectochilus roxburghii (Wall.) Lindl. (Orchidaceae). Int. J. Mol. Sci. 2015, 16, 30190–30203. [Google Scholar] [CrossRef]
- Valadares, R.B.S.; Perotto, S.; Santos, E.C.; Lambais, M.R. Proteome changes in Oncidium sphacelatum (Orchidaceae) at different trophic stages of symbiotic germination. Mycorrhiza 2014, 24, 349–360. [Google Scholar] [CrossRef]
- Wang, W.; Yu, H.; Li, T.; Li, L.; Zhang, G.; Liu, Z.; Huang, T.; Zhang, Y. Comparative proteomics analyses of pollination response in endangered orchid species Dendrobium chrysanthum. Int. J. Mol. Sci. 2017, 18, 2496. [Google Scholar] [CrossRef]
- Sedeek, K.E.; Qi, W.; Schauer, M.A.; Gupta, A.K.; Poveda, L.; Xu, S.; Liu, Z.J.; Grossniklaus, U.; Schiestl, F.P.; Schlüter, P.M. Transcriptome and proteome data reveal candidate genes for pollinator attraction in sexually deceptive orchids. PLoS ONE 2013, 8, e64621. [Google Scholar] [CrossRef]
- Li, X.; Xu, W.; Chowdhury, M.R.; Jin, F. Comparative proteomic analysis of labellum and inner lateral petals in Cymbidium ensifolium flowers. Int. J. Mol. Sci. 2014, 15, 19877–19897. [Google Scholar] [CrossRef]
- Tsai, W.C.; Pan, Z.J.; Hsiao, Y.Y.; Jeng, M.F.; Wu, T.F.; Chen, W.H.; Chen, H.H. Interactions of B-class complex proteins involved in tepal development in Phalaenopsis orchid. Plant Cell Physiol. 2008, 49, 814–824. [Google Scholar] [CrossRef]
- Chen, J.-C.; Wei, M.-J.; Fang, S.-C. Expression analysis of fertilization/early embryogenesis-associated genes in Phalaenopsis orchids. Plant Signal. Behav. 2016, 11, e1237331. [Google Scholar] [CrossRef]
- He, C.M.; Si, C.; Teixeira da Silva, J.A.; Li, M.Z.; Duan, J. Genome-wide identification and classification of MIKC-type MADS-box genes in Streptophyte lineages and expression analyses to reveal their role in seed germination of orchids. BMC Evol. Biol. 2019, 19, 223. [Google Scholar] [CrossRef]
- Williams, S.J.; Gale, S.W.; Hinsley, A.; Gao, J.; St. John, F.A.V. Using consumer preferences to characterize the trade of wild-collected ornamental orchids in China. Conserv. Lett. 2018, 11, e12569. [Google Scholar] [CrossRef]
- Hinsley, A. The Role of Online Platforms in the Illegal Orchid Trade from Southeast Asia; The Global Initiative against Transnational Organized Crime: Geneva, Switzerland, 2018; Available online: www.GlobalInitiative.net (accessed on 25 December 2023).
- Irawati, A. Conservation of orchids, the gems of the tropics. In Conservation of Tropical Plant Species; Normah, M., Chin, H., Reed, B., Eds.; Springer: New York, NY, USA, 2013; pp. 171–187. [Google Scholar]
- Schuiteman, A.; Discovering New Orchids. Royal Botanic Gardens Kew. Available online: https://www.kew.org/read-and-watch/discovering-new-orchids (accessed on 25 December 2023).
- Hinsley, A.; Lee, T.E.; Harrison, J.R.; Roberts, D.L. Estimating the extent and structure of trade in horticultural orchids via social media. Conserv. Biol. 2016, 30, 1038–1047. [Google Scholar] [CrossRef] [PubMed]
- Soda, N.; Verma, L.; Giri, J. CRISPR-Cas9 based plant genome editing: Significance, opportunities, and recent advances. Plant Physiol. Biochem. 2018, 131, 2–11. [Google Scholar] [CrossRef]

| Orchid Genus | Biotechnological Interventions | Research Outcome | Reference |
|---|---|---|---|
| Oncidium sp. | A. tumefaciens-mediated transformation of OMADS1 gene RNAi-induced silencing of PSY gene Particle bombardment of construct pCB199 plasmid | Increased flowering and more flowers White flowers Disease resistance | [9] [10] [11] |
| Phalaenopsis sp. | RNAi-mediated silencing (virus-induced gene silencing) of the PeUFGT3 gene | Decreased anthocyanin and variation in flower color | [12] |
| Phalaenopsis amabilis Coelogyne pandurata Lindley | A. tumefaciens-mediated transformation of the orchid A. tumefaciens-mediated transformation of the KNAT1 gene | Transgenic orchids with the highest frequency of shooting Micropropagation of orchid species | [13] [14] |
| Cymbidium sinense | A. tumefaciens-mediated transformation of CsFT gene | Early flowering | [15] |
| Vanda sp. | Sonication-assisted Agrobacterium-mediated transformation (SAAT) | Enhanced disease resistance | [16] |
| Dendrobium Phalaenopsis | Agrobacterium-mediated transformation of gusA, nptII, hptII marker genes | Successful insertion of genes in transgenic orchid | [17] |
| Phalaenopsis sp. | A. tumefaciens-mediated transformation of eGFP (fluorescent) gene Particle bombardment of EgTCTP gene | Hygromycin resistance, early initiation of primordial shoots in the transformed PLBs | [18,19] |
| Cymbidium niveo-marginum | A. tumefaciens-mediated transformation of gfp, hptII, ORSV CP genes | Enhanced virus resistance | [20] |
| Phalaenopsis sp. | Genetic transformation of orchid with Vitreoscilla hemoglobin (vhb) gene via injection of DNA solution into immature capsules | Transgene with better metabolism and growth | [21] |
| Erycina pusilla | A. tumefaciens-mediated transformation of MSRB7 gene | Enhanced disease resistance | [22] |
| Dendrobium Sonia ‘Earsakul’ | RNAi-induced silencing of DseDFR and DseCHS-B genes | Anthocyanin accumulation was restricted in transgenic orchid | [23] |
| Cattleya sp. | Agrobacterium-mediated genetic transformation with an Odontoglossum ringspot virus replicase gene | Enhanced virus resistance | [24] |
| P. amabilis | Genetic transformation of lipid transfer protein-encoding gene | Improved adaptation to cold stress | [25] |
| Phalaenopsis aphrodite | A. tumefaciens-mediated transformation of Pha21 gene | Enhanced virus resistance | [26] |
| Phalaenopsis sp. | Genetic transformation of Phalaenopsis via pollen tube pathway | Transgenic orchid with improved traits | [27] |
| Dendrobium sp. | Electro-injection of foreign DNA into protocorms | Transgenic orchid with improved traits | [28] |
| Oncidium sp. | Genetic transformation of pflp gene | Resistance against E. carotovora pathogen | [29] |
| Dendrobium sp. | Genetic transformation of Firefly Luciferase gene | Transgenic orchid glows in the dark | [30] |
| Dendrobium sp. | Sense and anti-sense constructs used for genetic transformation | Enhanced vase life of transgenic orchid | [30] |
| Phalaenopsis sp. | Genetic transformation of ß-1,3-endoglucanase gene | Transgenic orchid is resistant to fungus | [31] |
| Calanthe sp. | Seed imbibition; electroporation of GUS, NPT II gene | Transgenic orchid with improved traits | [32] |
| Orchid Genus | Gene Name(s) | Functional Role(s) | Reference |
|---|---|---|---|
| Dendrobium | DOH1: HOMEOBOX1 | Floral transition and flower development | [104] |
| Dendrobium | DOSOC1 | Promotes early flowering | [105] |
| Dendrobium | DnVRN1 | Floral induction | [106] |
| Dendrobium | DnAGL19 | Flowering regulation | [107] |
| Dendrobium | DOFT | Inflorescence and flower development | [108] |
| Dendrobium | DOAP1 | Formation of floral meristems | [109] |
| Dendrobium | DOFTIP1 | Promotes flowering | [108] |
| Doritaenopsis | DhEFL4 | Requirement for photoperiod perception and circadian function | [110] |
| Oncidium | OMADS1 | Induced precocious flowering | [111] |
| Oncidium | OnTFL1 | Encoding floral activator | [112] |
| Phalaenopsis | PeMADS6 | Flower longevity and ovary development | [113] |
| Phalaenopsis | PhalCOL | Early flowering phenotype | [54] |
| Phalaenopsis nation | PeSEP | Floral organ determination | [114] |
| Phalaenopsis | PaFT1 | Precocious flowering | [115] |
| Phalaenopsis | PhapLFY | Flower initiation | [116] |
| Phalaenopsis ‘Formosa rose’ | ORAP11 and ORAP13 | Both genes are highly expressed during the early stages of floral buds and vegetative organs | [117] |
| P. aphrodite | PaAP1-1 and PaAP1-2 | PaAP1-1 is expressed in the inner whorls of the pollinia and pedicel and PaAP1-2 is expressed in the pedicel only | [118] |
| Cymbidium ensifolium | CeMADS | Reproductive organ development such as stamen and carpel development and function in the meristem | [119] |
| Scientific Name | Found in | Habitat | Plant Part | Medicinal Attributes | Reference |
|---|---|---|---|---|---|
| Aerides multiflora Roxb. | India | Epiphytic | Roots, Leaves | Wound and cuts, Antibacterial | [143] |
| Calanthe triplicata | India | Terrestrial | Flowers, roots | Anti-inflammatory, Diarrhea, Gastric disorders | [144] |
| Brachycortis obcordata (Lindl.) Summerh. | ---- | Terrestrial | Roots | Dysentery | [66] |
| Dendrobium chrysanthum | China | Epiphytic | Leaves | Skin diseases, Anti-pyretic, Immunoregulatory | [145] |
| Bulbophyllum umbellatum Lindl. | Asia | Epiphytic | All plant parts | Increase congeniality | [66] |
| Orchis latifolia L. | India Iran Afghanistan | Terrestrial | Roots | Diabetes, Dysentry, Malnutrition, Diarrhea | [146] |
| Maxillaria densa | Mexico | Epiphytic | All plant parts | Analgesic, Relaxant | [147] |
| Cymbidium aloifolium (L.) Sw. | Asia | Epiphytic | Bulbs, Rhizomes | Dislocated bones and fracture | [148] |
| Acampe papillosa | India | Epiphytic | Roots | Rheumatism, Syphilis Neuralgia | [149] |
| Arundina graminifolia (D. Don) Hochr. | Nepal Thailand China Japan | Terrestrial | Roots | Bodyache | [150] |
| Anoectochilus formosanus Hayata | Taiwan | Terrestrial | Tubers | Abdominal pain, Nephritis, Hypertension, Anti-inflammatory | [151] |
| Epidendrum mosenii | Korea China | Mostly epiphytes, some terrestrial | Stem | Antinociceptive | [152] |
| Gastrodia elata | Asia | Heterotrophic | All plant parts | Epilepsy, Tetanus, Neuroprotective | [153] |
| Habenaria pectinata D. Don | India | Terrestrial | Tubers | Snake-bite treatment, Arthritis | [154] |
| Cypripedium elegan Reichenb .f. Nep | Asia America | Terrestrial | Roots | Epilepsy, Spasms, Rheumatism | [66] |
| Dendrobium densiflorum Lindl. | India | Epiphytic | Pseudobulbs | Skin diseases | [155] |
| Eulophia nuda Landl. | India | Terrestrial | Tubers | Bronchitis, Tumors | [156] |
| Malaxis acuminta D. Don | India | Terrestrial | Pseudobulbs | Antioxidant, Anti-aging | [157] |
| Vanda roxburghii | India | Epiphytic | Leaves | Anti-pyretic, Sciatica, Bronchitis | [158] |
| Vanilla planifolia | Mexico | Epiphytic | Sheath | Rheumatism, Hysteria, High-fever | [159] |
| Satyrium nepalense | India Nepal | Terrestrial | Tubers | Malaria, Dysentry | [160] |
| Bletilla striata | Taiwan Nepal China | Terrestrial | Tubers | Cancer, blood disorders, Tuberculosis | [161] |
| Cymbidium goeringii | Asia Australia | Epiphytic | Whole plant | Diuretic | [162] |
| Coeloglossum viride | England | Terrestrial | Rhizome | Neuroprotective | [163] |
| Goodyera discolor | Asia | Terrestrial | Whole plant | Antihepatotoxic | [149] |
| Gymnadenia conopsea | Europe Asia | Terrestrial | Tubers | Anti-allergic, Aphrodisiac | [164] |
| Pholidota yunnanensis | China | Epiphytic | --- | Antioxidant | [165] |
| Orchis laxiflora Lam. | Europe, Africa, Asia | Heterotrophic | Bulbs | Bronchitis, Diarrhea | [166] |
| Luisia zeylanica Lindl. | India, Sri lanka Thailand | Epiphytic | Leaves | Wound healing, Treating burns | [145] |
| Pholidota pallida Lindl. | India | Epiphytic | Roots Pseudobulbs | Analgesic | [167] |
| Thunia alba (Lindl.) Rchb. F | India Myanmar Thailand | Epiphytic | All plant | Treatment of dislocated bones | [66] |
| Zeuxine strateumatica (L.) Schltr. | China Japan India | Terrestrial | Tubers Roots | Tonic | [168] |
| Trudelia cristata (Lindl.) Senghas | India Bangladesh Bhutan | Epiphytic | Leaves Roots | Wound healing, Treatment of dislocated bones | [66] |
| Vanda tessellata (Roxb.) Rchb. f. | India Sri lanka Burma | Epiphytic | Leaves Roots | Rheumatism, Anti-pyretic | [169] |
| Platanthera sikkimensis (Hook. f.) Kraenzlin. | India | Terrestrial | Pseudobulbs Bulbs | Analgesic | [66] |
| Pleione humilis (Sm.) D. Don | India | Epiphytic | Pseudobulbs | Wound healing, Tonic | [170] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tiwari, P.; Sharma, A.; Bose, S.K.; Park, K.-I. Advances in Orchid Biology: Biotechnological Achievements, Translational Success, and Commercial Outcomes. Horticulturae 2024, 10, 152. https://doi.org/10.3390/horticulturae10020152
Tiwari P, Sharma A, Bose SK, Park K-I. Advances in Orchid Biology: Biotechnological Achievements, Translational Success, and Commercial Outcomes. Horticulturae. 2024; 10(2):152. https://doi.org/10.3390/horticulturae10020152
Chicago/Turabian StyleTiwari, Pragya, Abhishek Sharma, Subir Kumar Bose, and Kyeung-Il Park. 2024. "Advances in Orchid Biology: Biotechnological Achievements, Translational Success, and Commercial Outcomes" Horticulturae 10, no. 2: 152. https://doi.org/10.3390/horticulturae10020152
APA StyleTiwari, P., Sharma, A., Bose, S. K., & Park, K.-I. (2024). Advances in Orchid Biology: Biotechnological Achievements, Translational Success, and Commercial Outcomes. Horticulturae, 10(2), 152. https://doi.org/10.3390/horticulturae10020152







