Transcriptional Insights into Lily Stem Bulblet Formation: Hormonal Regulation, Sugar Metabolism, and Transcriptional Networks in LA Lily ‘Aladdin’
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Morphological and Histological Observation
2.3. RNA Extraction and Quality Assessment
2.4. PacBio Iso-Seq Library Preparation, Sequencing, and Analysis
2.5. Illumina Transcriptome Library Preparation, Sequencing, and Analysis
2.6. Functional Annotation of Tanscripts, Identification of Differentially Expressed Genes (DEGs), and Functional Enrichment
2.7. Identification of Co-Expression Network Modules
2.8. Quantitative Real-Time PCR Validation
3. Results
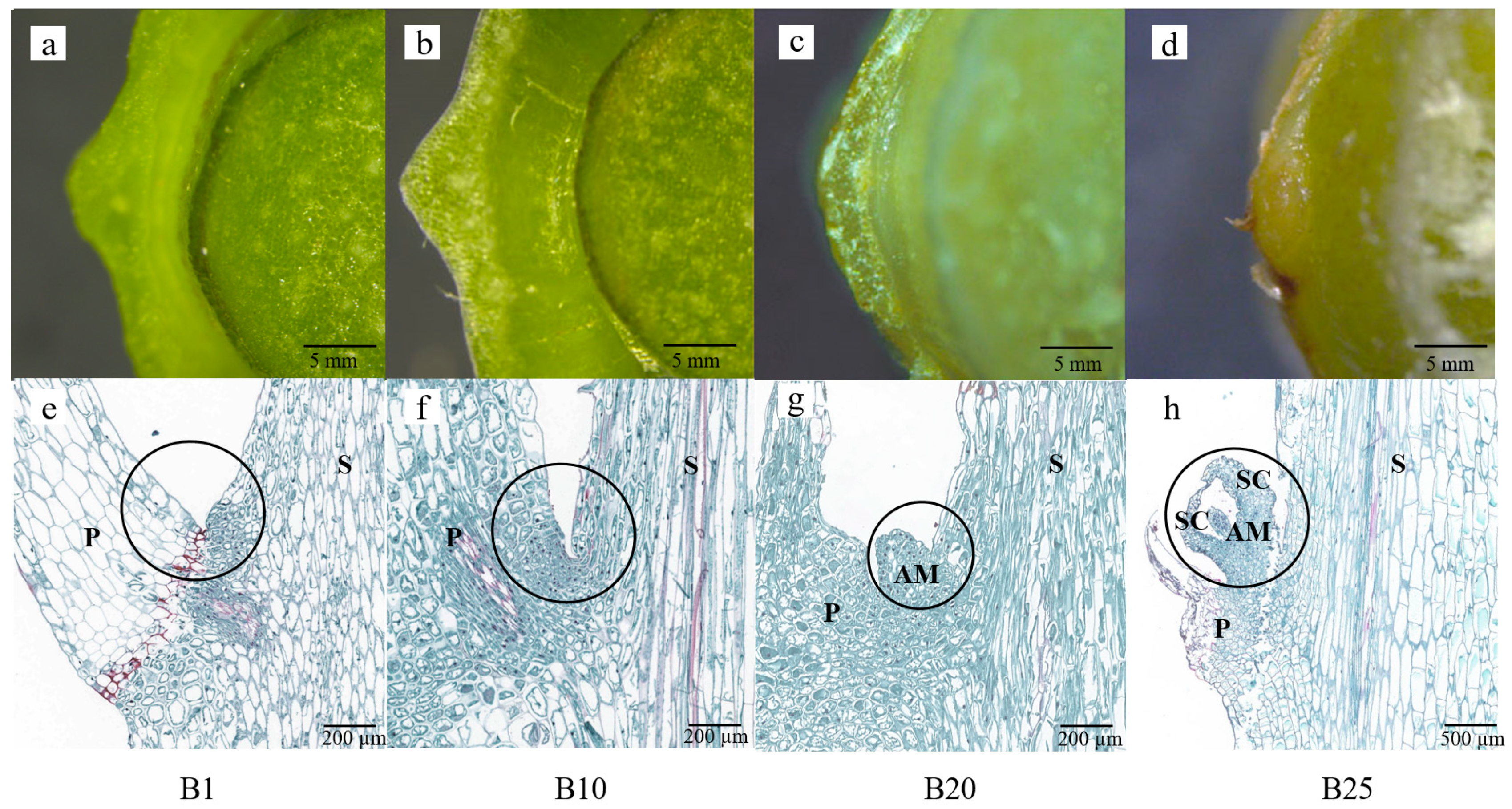
3.1. Morphological and Histological Observations on Bulblet Formation after Application of 6-BA
3.2. RNA-Seq Analysis of L. ‘Aladdin’
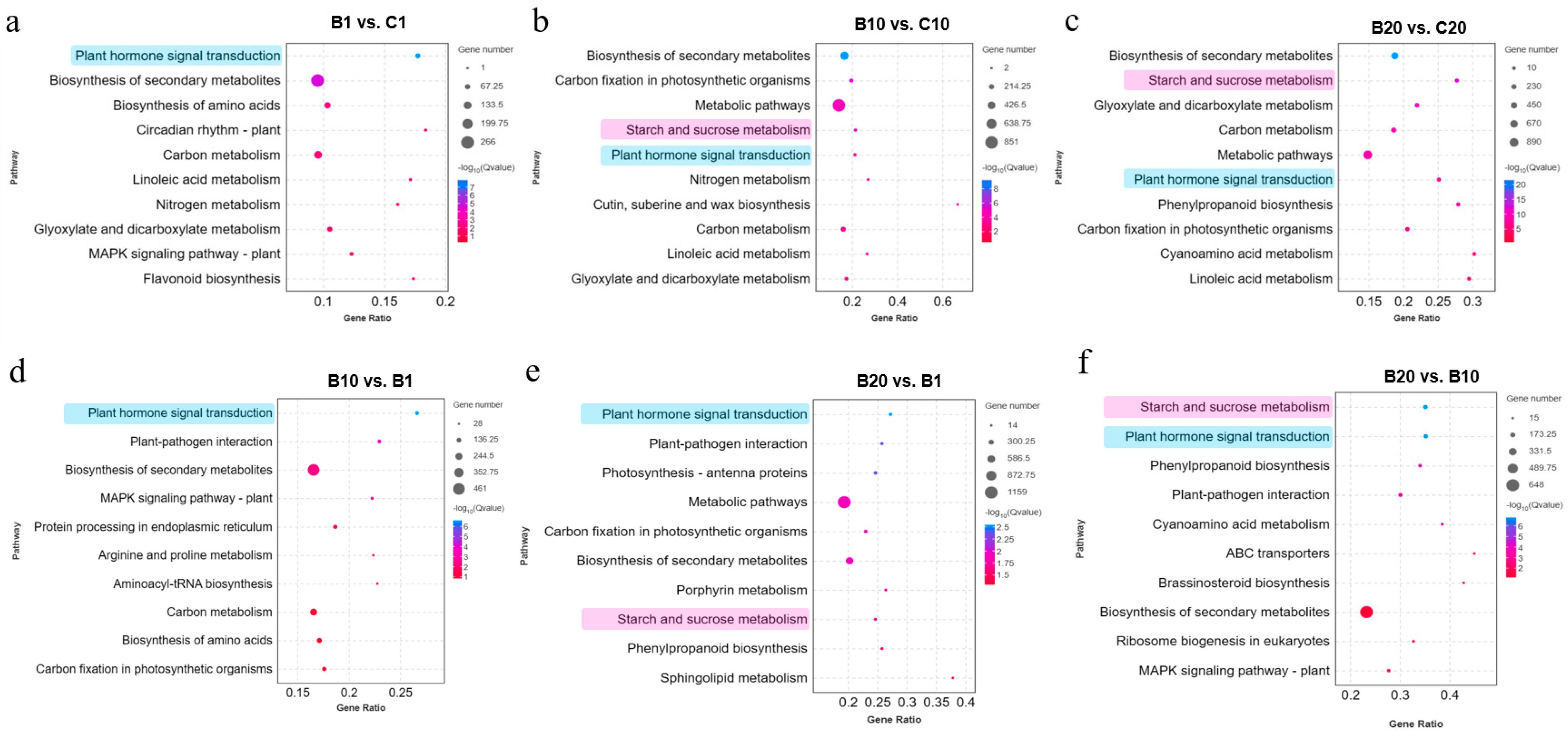
3.3. KEGG Pathway Enrichment Analysis of DEGs
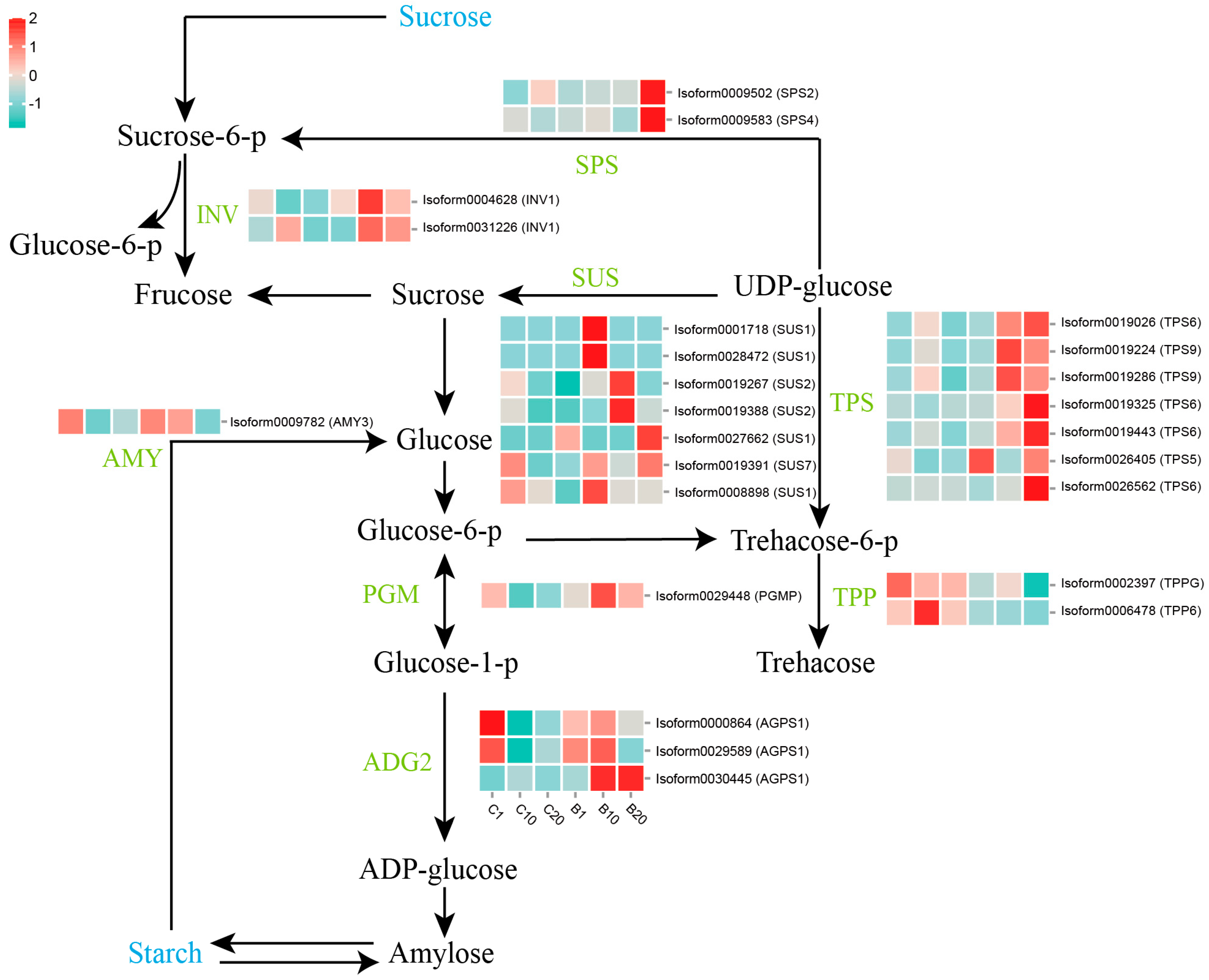
3.4. Expression of Genes Involved in Starch and Sucrose Metabolism Pathways under 6-BA Treatment
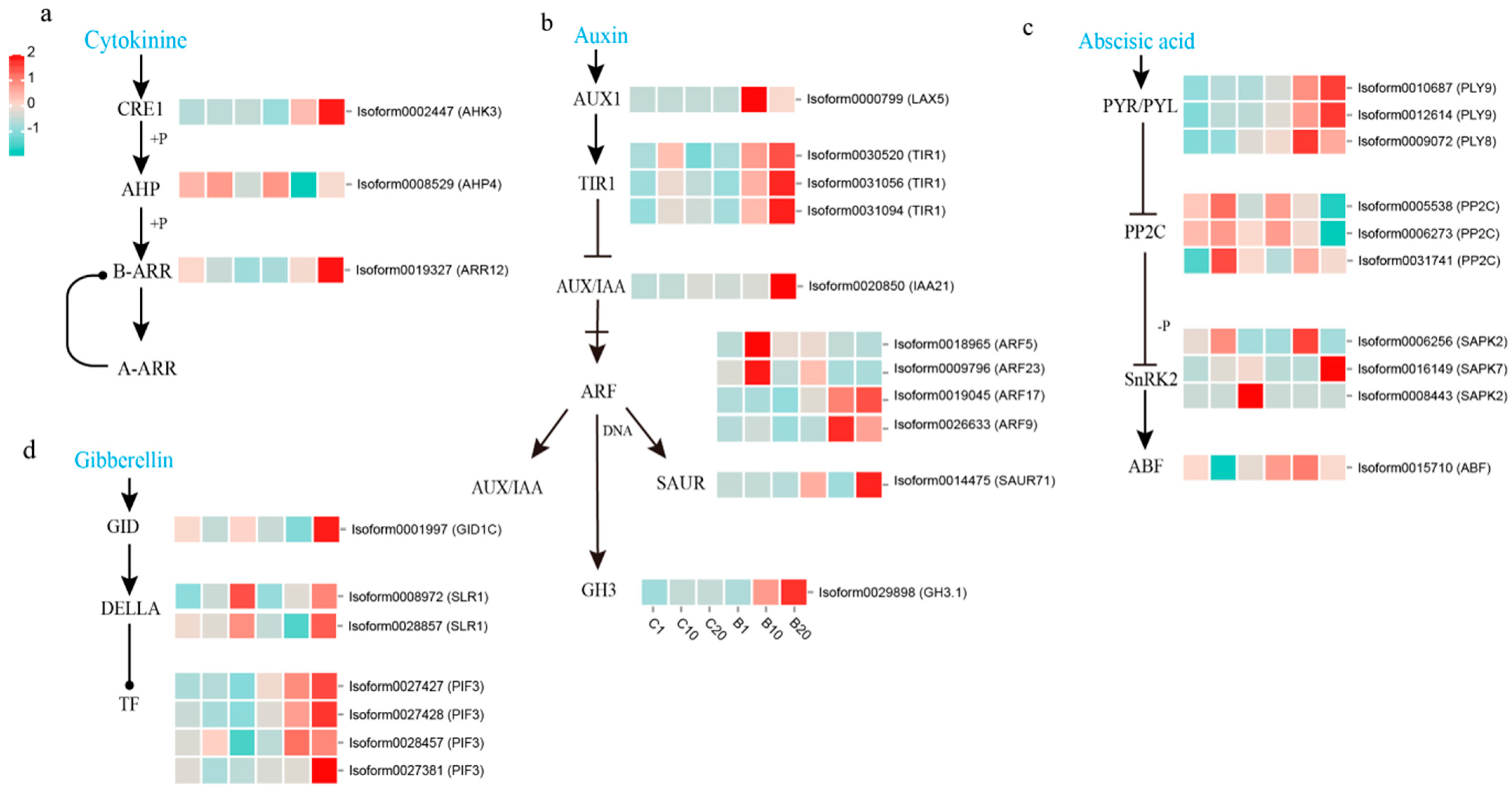
3.5. Analysis of DEGs Involved in Phytohormone Biosynthesis and Signal Transduction Pathway during Stem Bulblet Initiation
3.6. Co-Expression Network Construction and Identification of WGCNA Modules
3.7. Identification of Hub TFs and Network Construction
4. Discussion
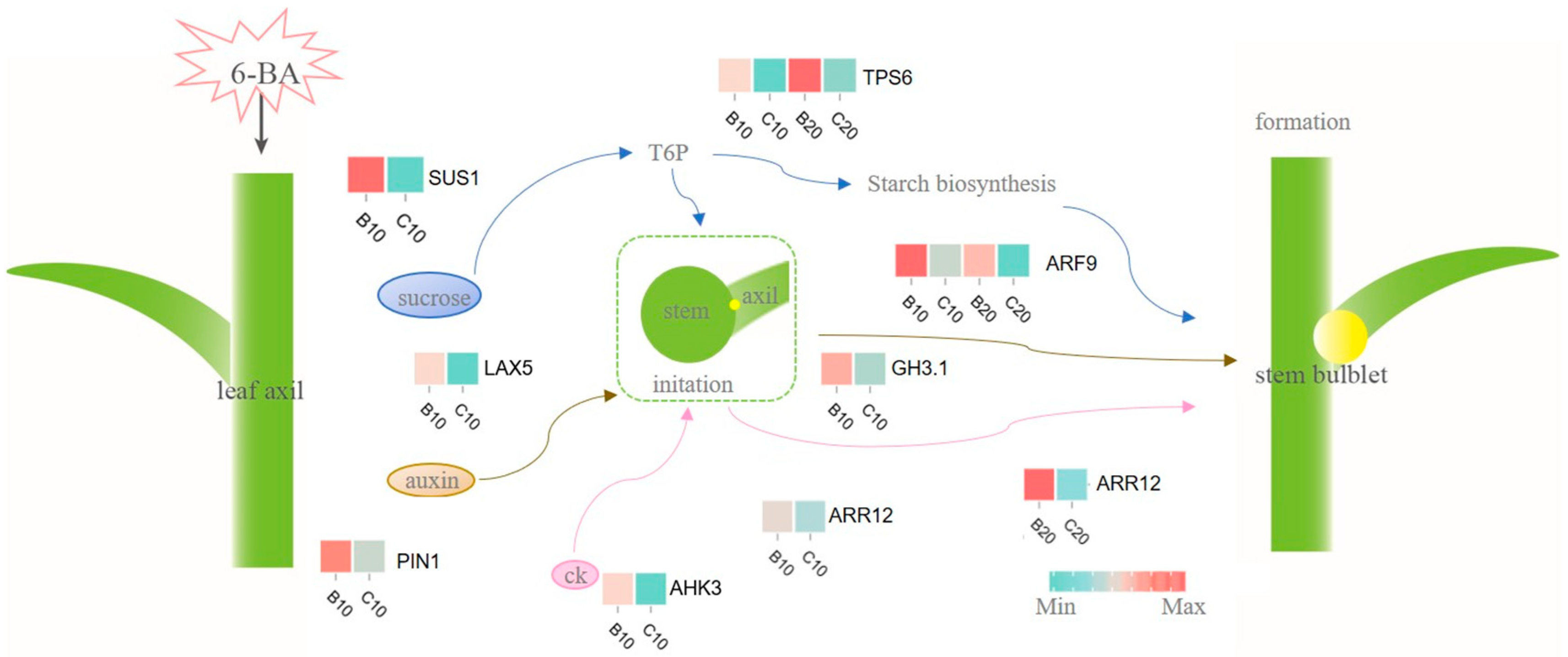
4.1. The Starch and Sucrose Metabolism Mediates Lily Stem Bulblet Initiation
4.2. The Involvement of 6-BA in Plant Hormone Signal Transduction in Stem Bulblet Formation
4.3. TFs Involved in Bulblet Formation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Nhut, D.T. Micropropagation of lily (Lilium longiflorum) via in vitro stem node and pseudo-bulblet culture. Plant Cell Rep. 1998, 17, 913–916. [Google Scholar] [CrossRef]
- Zhang, Y.; Yong, Y.B.; Wang, Q.; Lu, Y.M. Physiological and Molecular Changes during Lily Underground Stem Axillary Bulbils Formation. Russ. J. Plant Physiol. 2018, 65, 372–383. [Google Scholar] [CrossRef]
- Yang, P.; Xu, L.; Xu, H.; Tang, Y.; He, G.; Cao, Y.; Feng, Y.; Yuan, S.; Ming, J. Histological and Transcriptomic Analysis during Bulbil Formation in Lilium lancifolium. Front. Plant Sci. 2017, 8, 1508. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Pachón, N. Mechanisms of Vegetative Propagation in Bulbs: A Molecular Approach. Ph.D. Dissertation, Wageningen University, Wageningen, The Netherlands, 2017. [Google Scholar]
- Chatfield, S.P.; Raizada, M.N. Ethylene and shoot regeneration: Hookless1 modulates de novo shoot organogenesis in Arabidopsis thaliana. Plant Cell Rep. 2008, 27, 655–666. [Google Scholar] [CrossRef] [PubMed]
- Motte, H.; Vereecke, D.; Geelen, D.; Werbrouck, S. The molecular path to in vitro shoot regeneration. Biotechnol. Adv. 2014, 32, 107–121. [Google Scholar] [CrossRef]
- Sang, Y.L.; Cheng, Z.J.; Zhang, X.S. Plant stem cells and de novo organogenesis. New Phytol. 2018, 218, 1334–1339. [Google Scholar] [CrossRef]
- Greb, T.; Clarenz, O.; Schafer, E.; Muller, D.; Herrero, R.; Schmitz, G.; Theres, K. Molecular analysis of the LATERAL SUPPRESSOR gene in Arabidopsis reveals a conserved control mechanism for axillary meristem formation. Genes. Dev. 2003, 17, 1175–1187. [Google Scholar] [CrossRef]
- Zhang, Y.; Zeng, Z.; Yong, Y.B.; Lyu, Y.M. Hormonal Regulatory Patterns of LaKNOXs and LaBEL1 Transcription Factors Reveal Their Potential Role in Stem Bulblet Formation in LA Hybrid Lily. Int. J. Mol. Sci. 2021, 22, 13502. [Google Scholar] [CrossRef]
- He, G.; Yang, P.; Tang, Y.; Cao, Y.; Qi, X.; Xu, L.; Ming, J. Mechanism of exogenous cytokinins inducing bulbil formation in Lilium lancifolium in vitro. Plant Cell Rep. 2020, 39, 861–872. [Google Scholar] [CrossRef]
- Rosspopoff, O.; Chelysheva, L.; Saffar, J.; Lecorgne, L.; Gey, D.; Caillieux, E.; Colot, V.; Roudier, F.; Hilson, P.; Berthomé, R.; et al. Direct conversion of root primordium into shoot meristem relies on timing of stem cell niche development. Development 2017, 144, 1187–1200. [Google Scholar] [CrossRef]
- de Reuille, P.B.; Bohn-Courseau, I.; Ljung, K.; Morin, H.; Carraro, N.; Godin, C.; Traas, J. Computer simulations reveal properties of the cell-cell signaling network at the shoot apex in Arabidopsis. Proc. Natl. Acad. Sci. USA 2006, 103, 1627–1632. [Google Scholar] [CrossRef] [PubMed]
- Heisler, M.G.; Ohno, C.; Das, P.; Sieber, P.; Reddy, G.V.; Long, J.A.; Meyerowitz, E.M. Patterns of auxin transport and gene expression during primordium development revealed by live imaging of the Arabidopsis inflorescence meristem. Curr. Biol. 2005, 15, 1899–1911. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y. The Mechanism of Auxin Involving in Bulbil Formation of Lilium lancifolium; Chinese Academy of Agricultural Sciences: Beijing, China, 2019. [Google Scholar]
- Li, J.; Sun, M.; Li, H.; Ling, Z.; Wang, D.; Zhang, J.; Shi, L. Full-length transcriptome-referenced analysis reveals crucial roles of hormone and wounding during induction of aerial bulbils in lily. BMC Plant Biol. 2022, 22, 415. [Google Scholar] [CrossRef]
- Gao, C.; Zhang, L.; Xu, Y.; Liu, Y.; Xiao, X.; Cui, L.; Xia, Y.; Wu, Y.; Ren, Z. Full-length transcriptome analysis revealed that 2,4-dichlorophenoxyacetic acid promoted in vitro bulblet initiation in lily by affecting carbohydrate metabolism and auxin signaling. Front. Plant Sci. 2023, 14, 1236315. [Google Scholar] [CrossRef]
- Gao, M.; Zhang, S.; Luo, C.; He, X.; Wei, S.; Jiang, W.; He, F.; Lin, Z.; Yan, M.; Dong, W. Transcriptome analysis of starch and sucrose metabolism across bulb development in Sagittaria sagittifolia. Gene 2018, 649, 99–112. [Google Scholar] [CrossRef]
- Li, X.Y.; Wang, C.X.; Cheng, J.Y.; Zhang, J.; da Silva, J.A.T.; Liu, X.Y.; Duan, X.; Li, T.L.; Sun, H.M. Transcriptome analysis of carbohydrate metabolism during bulblet formation and development in Lilium davidii var. unicolor. BMC Plant Biol. 2014, 14, 358. [Google Scholar] [CrossRef]
- Wu, Y.; Ren, Z.; Gao, C.; Sun, M.; Li, S.; Min, R.; Wu, J.; Li, D.; Wang, X.; Wei, Y.; et al. Change in Sucrose Cleavage Pattern and Rapid Starch Accumulation Govern Lily Shoot-to-Bulblet Transition In Vitro. Front. Plant Sci. 2020, 11, 564713. [Google Scholar] [CrossRef]
- Ren, Z.M.; Zhang, D.; Jiao, C.; Li, D.Q.; Wu, Y.; Wang, X.Y.; Gao, C.; Lin, Y.F.; Ruan, Y.L.; Xia, Y.P. Comparative transcriptome and metabolome analyses identified the mode of sucrose degradation as a metabolic marker for early vegetative propagation in bulbs of Lycoris. Plant J. 2022, 112, 115–134. [Google Scholar] [CrossRef]
- Wang, Q.; Hasson, A.; Rossmann, S.; Theres, K. Divide et impera: Boundaries shape the plant body and initiate new meristems. New Phytol. 2016, 209, 485–498. [Google Scholar] [CrossRef] [PubMed]
- Tian, C.; Zhang, X.; He, J.; Yu, H.; Wang, Y.; Shi, B.; Han, Y.; Wang, G.; Feng, X.; Zhang, C.; et al. An organ boundary-enriched gene regulatory network uncovers regulatory hierarchies underlying axillary meristem initiation. Mol. Syst. Biol. 2014, 10, 755. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Jiao, Y. Regulation of Axillary Meristem Initiation by Transcription Factors and Plant Hormones. Front. Plant Sci. 2016, 7, 183. [Google Scholar] [CrossRef] [PubMed]
- Abraham-Juárez, M.J.; Martínez-Hernández, A.; Leyva-González, M.A.; Herrera-Estrella, L.; Simpson, J. Class I KNOX genes are associated with organogenesis during bulbil formation in Agave tequilana. J. Exp. Bot. 2010, 61, 4055–4067. [Google Scholar] [CrossRef] [PubMed]
- Yang, P.; Xu, H.; Xu, L.; Tang, Y.; He, G.; Cao, Y.; Yuan, S.; Ren, J.; Ming, J. Cloning and expression analysis of LlAGO1 in Lilium lancifolium. Acta Hortic. Sin. 2018, 45, 784–794. [Google Scholar] [CrossRef]
- He, G.; Cao, Y.; Wang, J.; Song, M.; Bi, M.; Tang, Y.; Xu, L.; Ming, J.; Yang, P. WUSCHEL-Related Homeobox Genes Cooperate with Cytokinin to Promote Bulbil Formation in Lilium lancifolium. Plant Physiol. 2022, 190, 387–402. [Google Scholar] [CrossRef] [PubMed]
- Fang, S.; Yang, C.; Ali, M.M.; Lin, M.; Tian, S.; Zhang, L.; Chen, F.; Lin, Z. Transcriptome Analysis Reveals the Molecular Regularity Mechanism Underlying Stem Bulblet Formation in Oriental Lily ‘Siberia’; Functional Characterization of the LoLOB18 Gene. Int. J. Mol. Sci. 2022, 23, 15246. [Google Scholar] [CrossRef] [PubMed]
- Gordon, S.P.; Tseng, E.; Salamov, A.; Zhang, J.; Meng, X.; Zhao, Z.; Kang, D.; Underwood, J.; Grigoriev, I.V.; Figueroa, M.; et al. Widespread Polycistronic Transcripts in Fungi Revealed by Single-Molecule mRNA Sequencing. PLoS ONE 2015, 10, e0132628. [Google Scholar] [CrossRef] [PubMed]
- Salmela, L.; Rivals, E. LoRDEC: Accurate and efficient long read error correction. Bioinformatics 2014, 30, 3506–3514. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Yu, C.; Li, Y.; Lam, T.W.; Yiu, S.M.; Kristiansen, K.; Wang, J. SOAP2: An improved ultrafast tool for short read alignment. Bioinformatics 2009, 25, 1966–1967. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef]
- Zhang, B.; Horvath, S. A general framework for weighted gene co-expression network analysis. Stat. Appl. Genet. Mol. Biol. 2005, 4, 17. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Liu, Q.; Wei, C.; Zhang, M.F.; Jia, G.X. Evaluation of putative reference genes for quantitative real-time PCR normalization in Lilium regale during development and under stress. Peerj 2016, 4, e1837. [Google Scholar] [CrossRef]
- Qi, N.N.; Hou, X.M.; Wang, C.L.; Li, C.X.; Huang, D.J.; Li, Y.H.; Wang, N.; Liao, W.B.A. Methane-rich water induces bulblet formation of scale cuttings in Lilium davidii var. unicolor by regulating the signal transduction of phytohormones and their levels. Physiol. Plant. 2021, 172, 1919–1930. [Google Scholar] [CrossRef] [PubMed]
- Hou, X.; Qi, N.; Wang, C.; Li, C.; Huang, D.; Li, Y.; Wang, N.; Liao, W. Hydrogen-rich water promotes the formation of bulblets in Lilium davidii var. unicolor through regulating sucrose and starch metabolism. Planta 2021, 254, 106. [Google Scholar] [CrossRef]
- Schmitz, G.; Theres, K. Shoot and inflorescence branching. Curr. Opin. Plant Biol. 2005, 8, 506–511. [Google Scholar] [CrossRef] [PubMed]
- Roman, H.; Girault, T.; Barbier, F.; Péron, T.; Brouard, N.; Pěnčík, A.; Novák, O.; Vian, A.; Sakr, S.; Lothier, J.; et al. Cytokinins Are Initial Targets of Light in the Control of Bud Outgrowth. Plant Physiol. 2016, 172, 489–509. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Sanz, A.; Brenner, M.L.; Smith, A. Sucrose Synthase, Starch Accumulation, and Tomato Fruit Sink Strength. Plant Physiol. 1993, 101, 321–327. [Google Scholar] [CrossRef] [PubMed]
- Hao, C.; Yang, P.; Qu, Y.; Hao, Z.; Yin, X.; Tang, Y.; Bi, M.; Xu, L.; Hu, F.; Ming, J. Sucrose function on the bulbil formation of Lilium lancifolium. Sci. Hortic. 2024, 323, 112538. [Google Scholar] [CrossRef]
- Xu, J.X.; Li, Q.Z.; Yang, L.Y.; Li, X.; Wang, Z.; Zhang, Y.C. Changes in carbohydrate metabolism and endogenous hormone regulation during bulblet initiation and development in Lycoris radiata. BMC Plant Biol. 2020, 20, 180. [Google Scholar] [CrossRef]
- Wu, Y.; Sun, M.; Li, S.; Min, R.; Gao, C.; Lyu, Q.; Ren, Z.; Xia, Y. Molecular cloning, characterization and expression analysis of three key starch synthesis-related genes from the bulb of a rare lily germplasm, Lilium brownii var. giganteum. J. Zhejiang Univ. Sci. B 2021, 22, 476–491. [Google Scholar] [CrossRef]
- Yadav, U.P.; Ivakov, A.; Feil, R.; Duan, G.Y.; Walther, D.; Giavalisco, P.; Piques, M.; Carillo, P.; Hubberten, H.M.; Stitt, M.; et al. The sucrose-trehalose 6-phosphate (Tre6P) nexus: Specificity and mechanisms of sucrose signalling by Tre6P. J. Exp. Bot. 2014, 65, 1051–1068. [Google Scholar] [CrossRef]
- Wu, Z.G.; Jiang, W.; Tao, Z.M.; Pan, X.J.; Yu, W.H.; Huang, H.L. Morphological and stage-specific transcriptome analyses reveal distinct regulatory programs underlying yam (Dioscorea alata L.) bulbil growth. J. Exp. Bot. 2020, 71, 1899–1914. [Google Scholar] [CrossRef]
- Mo, J.L.; Qu, Y.X.; He, G.R.; Yang, P.P.; Wang, L.H.; Zhang, L.N.; Wu, X.W.; Zhang, D.; Li, L.L.; Ming, J. Effect of exogenous 6-BA induced bulblets formation in aerial cultivation. Sci. Hortic. 2023, 309, 111644. [Google Scholar] [CrossRef]
- Li, G.; Tan, M.; Cheng, F.; Liu, X.; Qi, S.; Chen, H.; Zhang, D.; Zhao, C.; Han, M.; Ma, J. Molecular role of cytokinin in bud activation and outgrowth in apple branching based on transcriptomic analysis. Plant Mol. Biol. 2018, 98, 261–274. [Google Scholar] [CrossRef]
- Luo, L.; Zeng, J.; Wu, H.; Tian, Z.; Zhao, Z. A Molecular Framework for Auxin-Controlled Homeostasis of Shoot Stem Cells in Arabidopsis. Mol. Plant 2018, 11, 899–913. [Google Scholar] [CrossRef] [PubMed]
- Abraham Juárez, M.J.; Hernández Cárdenas, R.; Santoyo Villa, J.N.; O’Connor, D.; Sluis, A.; Hake, S.; Ordaz-Ortiz, J.; Terry, L.; Simpson, J. Functionally different PIN proteins control auxin flux during bulbil development in Agave tequilana. J. Exp. Bot. 2015, 66, 3893–3905. [Google Scholar] [CrossRef] [PubMed]
- Guo, C.; Li, J.; Li, M.; Xu, X.; Chen, Y.; Chu, J.; Yao, X. Regulation Mechanism of Exogenous Brassinolide on Bulbil Formation and Development in Pinellia ternata. Front. Plant Sci. 2021, 12, 809769. [Google Scholar] [CrossRef]
- Ravnikar, M.; El, J.; Plaper, I.; Pacapan, A. Jasmonic acid stimulates shoot and bulb formation of garlic in vitro. J. Plant Growth Regul. 1993, 12, 73–77. [Google Scholar] [CrossRef]
- Hamant, O.; Pautot, V. Plant development: A TALE story. Comptes Rendus Biol. 2010, 333, 371–381. [Google Scholar] [CrossRef]
- Bürglin, T.R. Analysis of TALE superclass homeobox genes (MEIS, PBC, KNOX, Iroquois, TGIF) reveals a novel domain conserved between plants and animals. Nucleic Acids Res. 1997, 25, 4173–4180. [Google Scholar] [CrossRef]
- Bellaoui, M.; Pidkowich, M.S.; Samach, A.; Kushalappa, K.; Kohalmi, S.E.; Modrusan, Z.; Crosby, W.L.; Haughn, G.W. The Arabidopsis BELL1 and KNOX TALE homeodomain proteins interact through a domain conserved between plants and animals. Plant Cell 2001, 13, 2455–2470. [Google Scholar] [CrossRef] [PubMed]
- Robinson-Beers, K.; Pruitt, R.E.; Gasser, C.S. Ovule Development in Wild-Type Arabidopsis and Two Female-Sterile Mutants. Plant Cell 1992, 4, 1237–1249. [Google Scholar] [CrossRef] [PubMed]
- Smith, H.M.; Hake, S. The interaction of two homeobox genes, BREVIPEDICELLUS and PENNYWISE, regulates internode patterning in the Arabidopsis inflorescence. Plant Cell 2003, 15, 1717–1727. [Google Scholar] [CrossRef]
- Kanrar, S.; Onguka, O.; Smith, H.M. Arabidopsis inflorescence architecture requires the activities of KNOX-BELL homeodomain heterodimers. Planta 2006, 224, 1163–1173. [Google Scholar] [CrossRef] [PubMed]
- Bhatt, A.M.; Etchells, J.P.; Canales, C.; Lagodienko, A.; Dickinson, H. VAAMANA—A BEL1-like homeodomain protein, interacts with KNOX proteins BP and STM and regulates inflorescence stem growth in Arabidopsis. Gene 2004, 328, 103–111. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; You, S.; Taylor-Teeples, M.; Li, W.L.; Schuetz, M.; Brady, S.M.; Douglas, C.J. BEL1-LIKE HOMEODOMAIN6 and KNOTTED ARABIDOPSIS THALIANA7 interact and regulate secondary cell wall formation via repression of REVOLUTA. Plant Cell 2014, 26, 4843–4861. [Google Scholar] [CrossRef]
- Licausi, F.; Ohme-Takagi, M.; Perata, P. APETALA2/Ethylene Responsive Factor (AP2/ERF) transcription factors: Mediators of stress responses and developmental programs. New Phytol. 2013, 199, 639–649. [Google Scholar] [CrossRef]
- Mao, J.L.; Miao, Z.Q.; Wang, Z.; Yu, L.H.; Cai, X.T.; Xiang, C.B. Arabidopsis ERF1 Mediates Cross-Talk between Ethylene and Auxin Biosynthesis during Primary Root Elongation by Regulating ASA1 Expression. PLoS Genet. 2016, 12, e1005760. [Google Scholar] [CrossRef]
- Cai, X.T.; Xu, P.; Zhao, P.X.; Liu, R.; Yu, L.H.; Xiang, C.B. Arabidopsis ERF109 mediates cross-talk between jasmonic acid and auxin biosynthesis during lateral root formation. Nat. Commun. 2014, 5, 5833. [Google Scholar] [CrossRef]
- Heyman, J.; Cools, T.; Canher, B.; Shavialenka, S.; Traas, J.; Vercauteren, I.; Van den Daele, H.; Persiau, G.; De Jaeger, G.; Sugimoto, K.; et al. The heterodimeric transcription factor complex ERF115-PAT1 grants regeneration competence. Nat. Plants 2016, 2, 16165. [Google Scholar] [CrossRef]
- Mehrnia, M.; Balazadeh, S.; Zanor, M.I.; Mueller-Roeber, B. EBE, an AP2/ERF transcription factor highly expressed in proliferating cells, affects shoot architecture in Arabidopsis. Plant Physiol. 2013, 162, 842–857. [Google Scholar] [CrossRef]
- Zhang, C.; Fan, L.; Le, B.H.; Ye, P.; Mo, B.; Chen, X. Regulation of ARGONAUTE10 Expression Enables Temporal and Spatial Precision in Axillary Meristem Initiation in Arabidopsis. Dev. Cell 2020, 55, 603–616.e605. [Google Scholar] [CrossRef]
- Zhu, H.L.; Hu, F.Q.; Wang, R.H.; Zhou, X.; Sze, S.H.; Liou, L.W.; Barefoot, A.; Dickman, M.; Zhang, X.R. Arabidopsis Argonaute10 Specifically Sequesters miR166/165 to Regulate Shoot Apical Meristem Development. Cell 2011, 145, 242–256. [Google Scholar] [CrossRef]
- Ariel, F.D.; Manavella, P.A.; Dezar, C.A.; Chan, R.L. The true story of the HD-Zip family. Trends Plant Sci. 2007, 12, 419–426. [Google Scholar] [CrossRef] [PubMed]
- Prigge, M.J.; Clark, S.E. Evolution of the class III HD-Zip gene family in land plants. Evol. Dev. 2006, 8, 350–361. [Google Scholar] [CrossRef] [PubMed]
- Preston, J.C.; Hileman, L.C. Functional Evolution in the Plant SQUAMOSA-PROMOTER BINDING PROTEIN-LIKE (SPL) Gene Family. Front. Plant Sci. 2013, 4, 80. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, S.; Grande, A.V.; Bujdoso, N.; Saedler, H.; Huijser, P. The microRNA regulated SBP-box genes SPL9 and SPL15 control shoot maturation in Arabidopsis. Plant Mol. Biol. 2008, 67, 183–195. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Liu, R.; Chen, Y.; Cui, J.; Ge, W.; Zhang, K. Molecular identification and functional verification of SPL9 and SPL15 of Lilium. Mol. Genet. Genom. 2022, 297, 63–74. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, K.; Lyu, T.; Lyu, Y. Transcriptional Insights into Lily Stem Bulblet Formation: Hormonal Regulation, Sugar Metabolism, and Transcriptional Networks in LA Lily ‘Aladdin’. Horticulturae 2024, 10, 171. https://doi.org/10.3390/horticulturae10020171
Zhang K, Lyu T, Lyu Y. Transcriptional Insights into Lily Stem Bulblet Formation: Hormonal Regulation, Sugar Metabolism, and Transcriptional Networks in LA Lily ‘Aladdin’. Horticulturae. 2024; 10(2):171. https://doi.org/10.3390/horticulturae10020171
Chicago/Turabian StyleZhang, Kewen, Tong Lyu, and Yingmin Lyu. 2024. "Transcriptional Insights into Lily Stem Bulblet Formation: Hormonal Regulation, Sugar Metabolism, and Transcriptional Networks in LA Lily ‘Aladdin’" Horticulturae 10, no. 2: 171. https://doi.org/10.3390/horticulturae10020171
APA StyleZhang, K., Lyu, T., & Lyu, Y. (2024). Transcriptional Insights into Lily Stem Bulblet Formation: Hormonal Regulation, Sugar Metabolism, and Transcriptional Networks in LA Lily ‘Aladdin’. Horticulturae, 10(2), 171. https://doi.org/10.3390/horticulturae10020171



