Localization of S-Locus-Related Self-Incompatibility in Lycium barbarum Based on BSA Analysis
Abstract
:1. Introduction
2. Materials and Methods
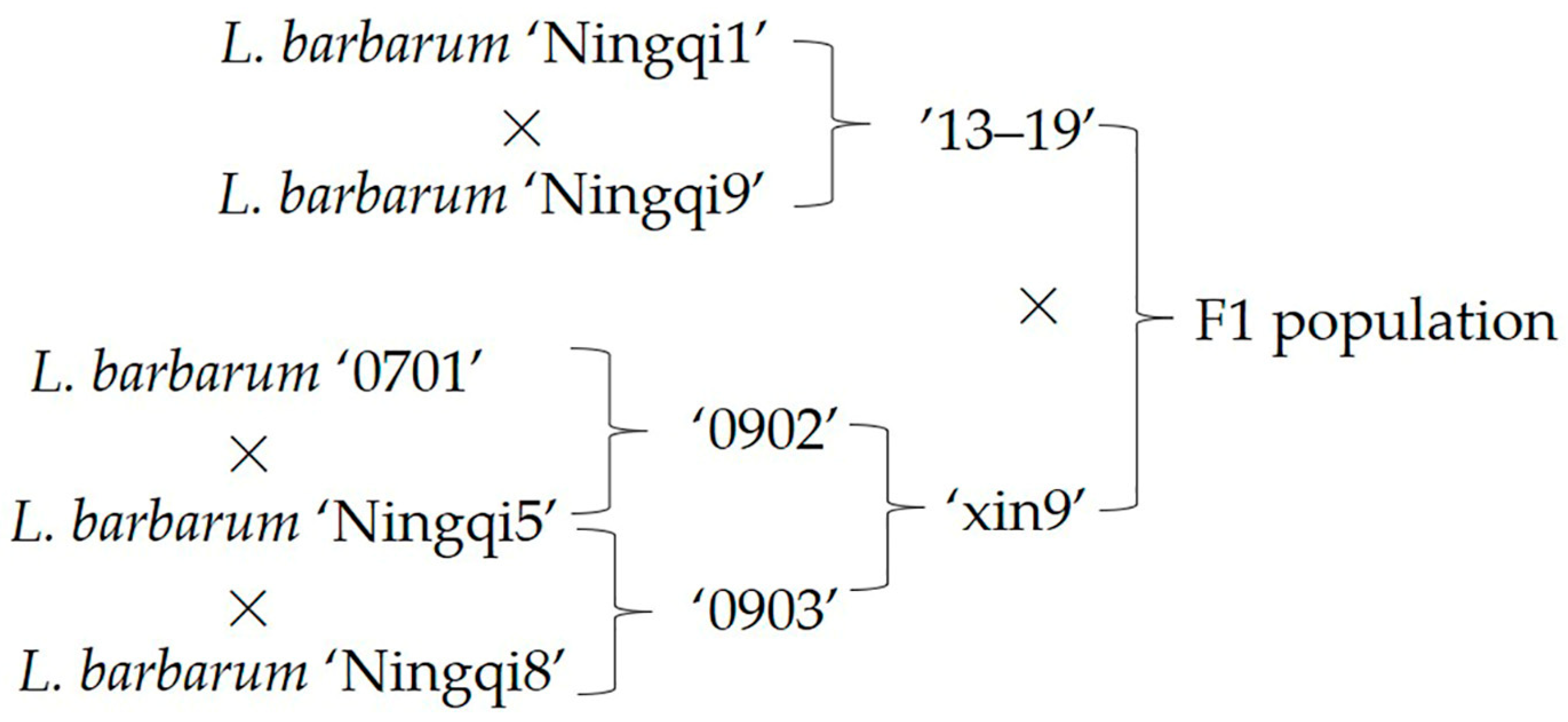
2.1. Materials
2.2. Analysis of Phenotypes Related to Self-Compatibility
2.2.1. Pollination
2.2.2. Determining the Number of Ovules
2.2.3. Calculating the Self-Compatibility Index
2.3. Genomic DNA Extraction, Sequencing, and BSA Pooling Localization
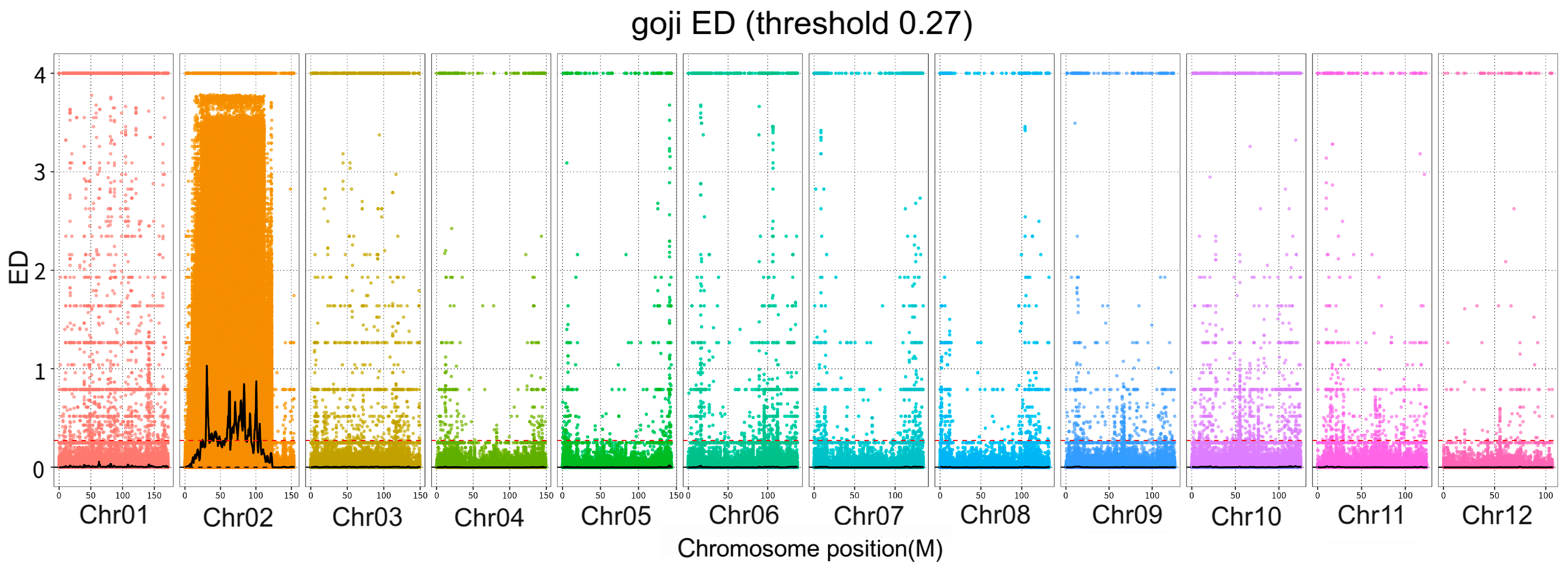
2.3.1. Association Analysis with Euclidean Distance
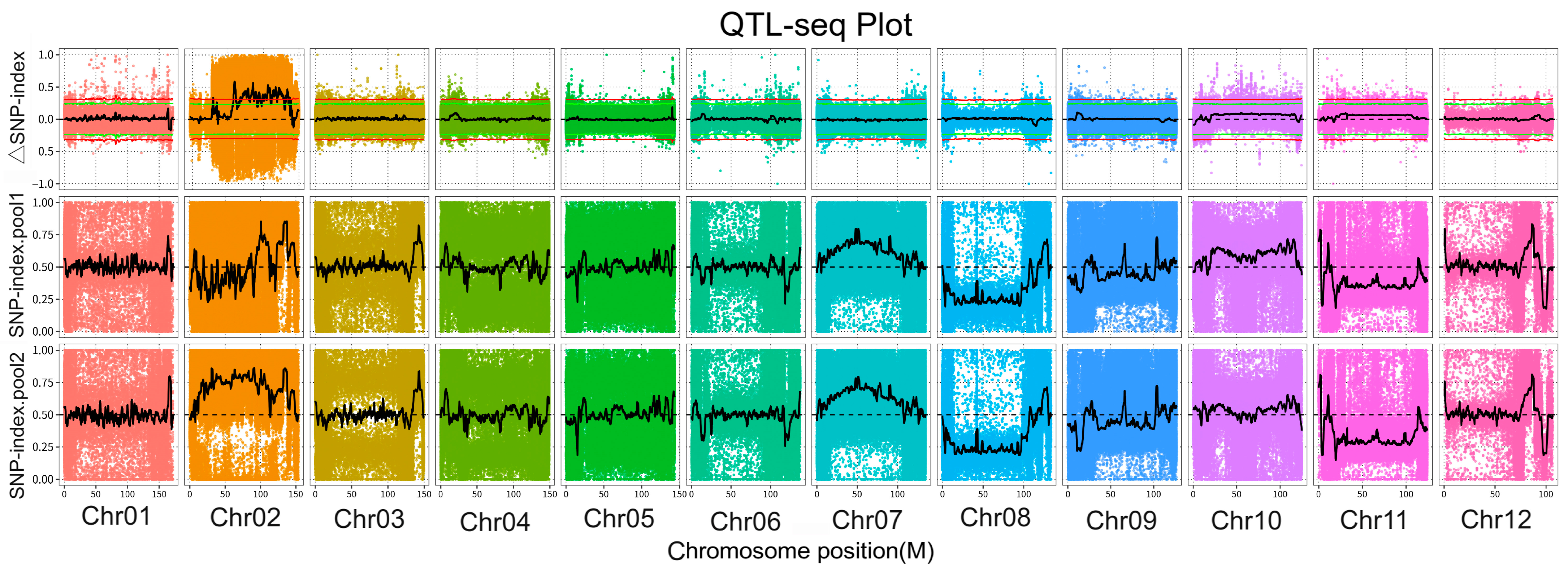
2.3.2. Index-SNP Association Analysis
2.4. Differential Gene Expression Analysis
2.5. S-RNase Gene Analysis
3. Results
3.1. Self-Compatibility-Related Phenotype Analysis
3.2. ED Correlation Analysis
3.3. SNP-Index Association Analysis
3.4. Variation Analysis and Gene Function Annotation within Candidate Regions
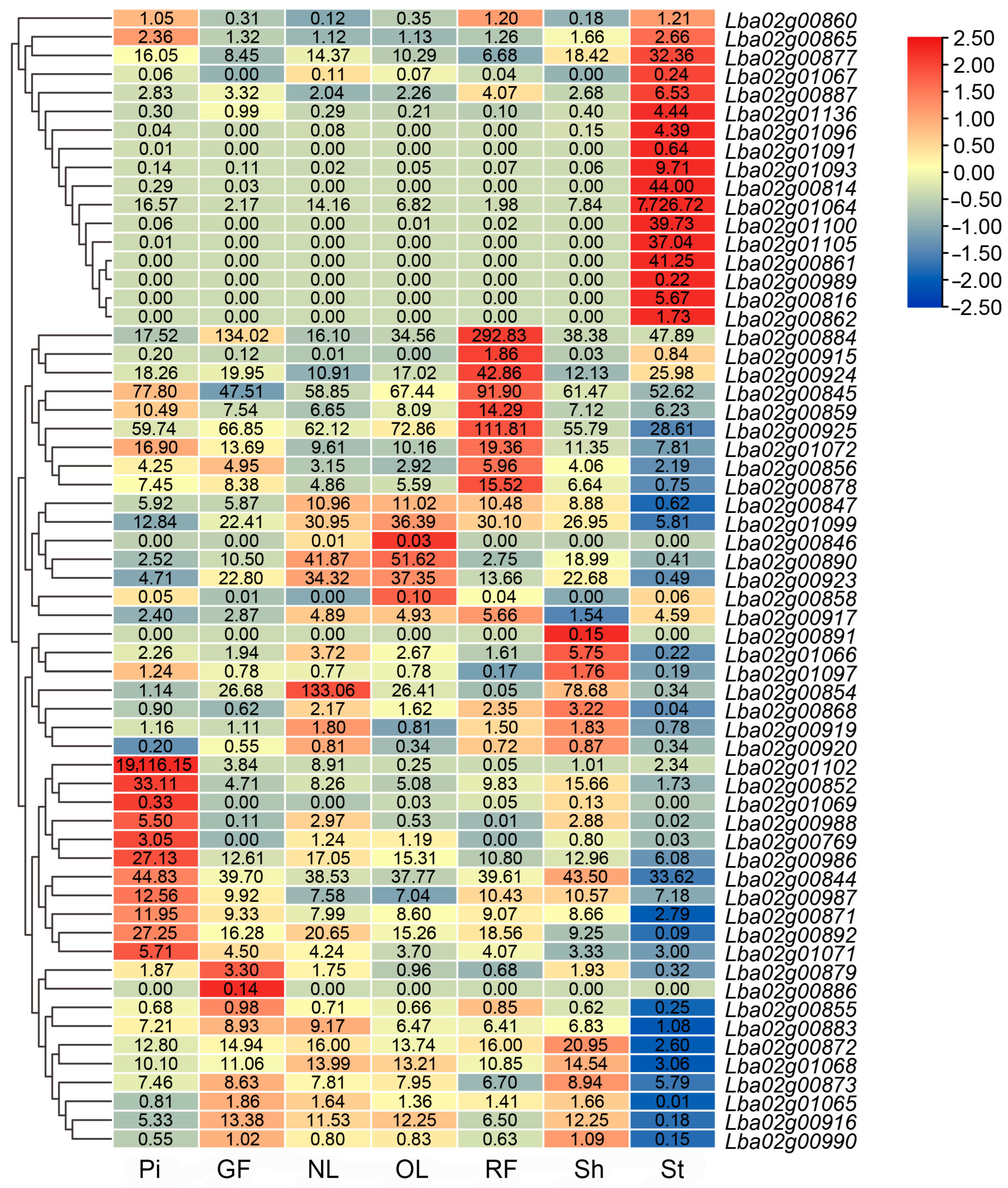
3.5. Differential Expression of Genes within the Localization Interval in Different Organs Based on Transcriptome Analysis
3.6. The Differential Expression of Genes Expressed in the Styles within the Mapped Interval after Self- and Cross-Pollination
3.7. Analysis of S-RNase Genotypes in Parents and Offspring
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Li, S.; Yang, D.; Yeung, C.; Yu, W.; Chang, R.C.; So, K.; Wong, D.; Lo, A.C.Y. Lycium barbarum polysaccharides reduce neuronal damage, blood-retinal barrier disruption and oxidative stress in retinal ischemia/reperfusion injury. PLoS ONE 2011, 6, e16380. [Google Scholar] [CrossRef]
- Jiao, E.; Li, Y.; Wang, B.; Tang, H.; Qin, K. Analysis on self-compatibility of Lycium barbarum L. in Ningxia. Acta Agric. Boreali-Occident. Sin. 2010, 19, 115–119. [Google Scholar]
- Xue, Y.; Carpenter, R.; Dickinson, H.G.; Coen, E.S. Origin of allelic diversity in antirrhinum s locus RNases. Plant Cell 1996, 8, 805–814. [Google Scholar]
- Tovar-mendez, A.; McClure, B. Plant reproduction: Self-incompatibility to go. Curr. Biol. 2016, 26, R115–R117. [Google Scholar] [CrossRef]
- Zhang, D.; Li, Y.; Zhao, X.; Zhang, C.; Liu, D.; Lan, S.; Yin, W.; Liu, Z. Molecular insights into self-incompatibility systems: From evolution to breeding. Plant Commun. 2023, 5, 100719. [Google Scholar] [CrossRef]
- Darwin, C.R. The Effects of Cross and Self-Fertilisation in the Vegetable Kingdom, 2nd ed.; John Murry: London, UK, 1878. [Google Scholar]
- Zhao, H.; Zhang, Y.; Zhang, H.; Song, Y.; Zhao, F.; Zhang, Y.; Zhu, S.; Zhang, H.; Zhou, Z.; Guo, H.; et al. Origin, loss and regain of self-incompatibility in angiosperms. Plant Cell 2022, 34, 579–596. [Google Scholar] [CrossRef]
- Goring, D.R.; Bosch, M.; Franklin-Tong, V.E. Contrasting self-recognition rejection systems for self-incompatibility in Brassica and Papaver. Curr. Biol. 2023, 33, R530–R542. [Google Scholar] [CrossRef]
- McClure, B.A.; Haring, V.; Ebert, P.R.; Anderson, M.A.; Simpson, R.J.; Sakiyama, F.; Clarke, A.E. Style self-incompatibility gene products of Nicotlana alata are ribonucleases. Nature 1989, 342, 955–957. [Google Scholar] [CrossRef]
- Qiao, H.; Wang, F.; Zhao, L.; Zhou, J.; Lai, Z.; Zhang, Y.; Robbins, T.P.; Xue, Y. The F-box protein AhSLF-S2 controls the pollen function of S-RNase-based self-incompatibility. Plant Cell 2004, 16, 2307–2322. [Google Scholar] [CrossRef]
- Snowman, B.N.; Kovar, D.R.; Shevchenko, G.; Franklin-Tong, V.; Staiger, C.J. Signal-mediated depolymerization of actin in pollen during the self-Incompatibility response. Plant Cell 2002, 14, 2613–2626. [Google Scholar] [CrossRef]
- Matsumoto, D.; Yamane, H.; Abe, K.; Tao, R. Identification of a Skp1-like protein interacting with SFB, the pollen S determinant of the gametophytic self-Incompatibility in Prunus. Plant Physiol. 2012, 159, 1252–1262. [Google Scholar] [CrossRef]
- Zeng, B.; Wang, J.; Hao, Q.; Yu, Z.; Abudukayoumu, A.; Tang, Y.L.; Zhang, X.F.; Ma, X. Identification of a novel SBP1-containing SCF SFB complex in wild dwarf almond (Prunus tenella). Front. Genet. 2019, 10, 1019. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, P.; Cai, Y.; Li, Y.; Tang, C.; Zhu, N.; Wang, P.; Zhang, S.; Wu, J. PbrBZR1 interacts with PbrARI2.3 to mediate brassinosteroid-regulated pollen tube growth during self-incompatibility signaling in pear. Plant Physiol. 2023, 92, 2356–2373. [Google Scholar] [CrossRef]
- Bernacchi, D.; Tanksley, S.D. An interspecific backcross of Lycopersicon esculentum x L. hirsutum: Linkage analysis and a QTL study of sexual compatibility factors and floral traits. Genetics 1997, 147, 861–877. [Google Scholar] [CrossRef]
- Gandhi, S.D.; Heesacker, A.F.; Freeman, C.A.; Argyris, J.; Bradford, K.; Knapp, S.J. The self-incompatibility locus (S) and quantitative trait loci for self-pollination and seed dormancy in sunflower. Theor. Appl. Genet. 2005, 111, 619–629. [Google Scholar] [CrossRef]
- Do, C.J.; Studer, B.; Frei, U.; Lübberstedt, T. Fine mapping a self-fertility locus in perennial ryegrass. Theor. Appl. Genet. 2018, 131, 817–827. [Google Scholar]
- Cropano, C.; Manzanares, C.; Yates, S.; Copetti, D.; Canto, J.D.; Lübberstedt, T.; Koch, M.; Studer, B. Identification of candidate genes for self-compatibility in perennial ryegrass (Lolium perenne L.). Front. Plant Sci. 2021, 12, 707901. [Google Scholar] [CrossRef]
- Sun, Q.; Wu, J.; Cheng, F.; Wang, X.; Liang, J.; Shen, S. QTL Mapping of self-compatibility, silique and seeds-associated traits in Brassica rapa. Sci. Agric. Sin. 2016, 49, 2449–2458. [Google Scholar]
- Gong, H.; Rehman, F.; Yang, T.; Li, Z.; Zeng, S.; Pan, L.; Li, Y.; Wang, L. Construction of the first high-density genetic map and QTL mapping for photosynthetic traits in Lycium barbarum L. Mol. Breed. 2019, 39, 106. [Google Scholar] [CrossRef]
- Rehman, F.; Gong, H.; Li, Z.; Zeng, S.; Yang, T.; Ai, P.; Pan, L.; Huang, H.; Wang, Y. Identification of fruit size associated quantitative trait loci featuring SLAF based high-density linkage map of goji berry (Lycium spp.). BMC Plant Biol. 2020, 20, 474. [Google Scholar] [CrossRef]
- Zhao, J.; Li, H.; Xu, Y.; Yin, Y.; Huang, T.; Zhang, B.; Wang, Y.; Li, Y.; Cao, Y.; An, W. A consensus and saturated genetic map provides insight into genome anchoring, synteny of Solanaceae and leaf- and fruit-related QTLs. in wolfberry (Lycium Linn.). BMC Plant Biol. 2021, 21, 350. [Google Scholar] [CrossRef]
- Li, Z.; Xu, Y. Bulk segregation analysis in the NGS era: A review of its teenage years. Plant J. 2022, 109, 1355–1374. [Google Scholar] [CrossRef]
- Boutet, E.; Lieberherr, D.; Tognolli, M.; Schneider, M.; Bansal, P.; Bridge, A.; Poux, S.; Bougueleret, L.; Xenarios, I. UniProtKB/Swiss-prot, the manually annotated section of the UniProt knowledgebase: How to use the entry view. Plant Bioinform. 2015, 1374, 23–54. [Google Scholar]
- Kato, N.; Esaka, M. Expansion of transgenic tobacco protoplasts expressing pumpkin ascorbate oxidase is more rapid than that of wild-type protoplasts. Planta 2000, 210, 1018–1022. [Google Scholar] [CrossRef]
- Pignocchi, C.; Foyer, C.H. Apoplastic ascorbate metabolism and its role in the regulation of cell signaling. Curr. Opin. Plant Biol. 2003, 6, 379–389. [Google Scholar] [CrossRef]
- Pignocchi, C.; Kiddle, G.; Hernández, I.; Foster, S.J.; Asensi, A.; Taybi, T.; Barnes, J.D.; Foyer, C.H. Ascorbate oxidase-dependent changes in the redox state of the apoplast modulate gene transcription leading to modified hormone signaling and orchestration of defense processes in tobacco. Plant Physiol. 2006, 141, 423–435. [Google Scholar] [CrossRef]
- Sanmartin, M.; Pateraki, I.; Chatzopoulou, F.; Kanellis, A.K. Differential expression of the ascorbate oxidase multigene family during fruit development and in response to stress. Planta 2007, 225, 873–885. [Google Scholar] [CrossRef]
- Hwang, J.Y.; Mannowetz, N.; Zhang, Y.D.; Everley, R.A.; Gygi, S.P.; Bewersdorf, J.; Lishko, P.V.; Chung, J.J. Dual sensing of physiologic pH and Calcium by EFCAB9 regulates sperm motility. Cell 2019, 177, 1480–1494. [Google Scholar] [CrossRef]
- Gao, W.; Long, L.; Tian, X.; Jin, J.; Liu, H.; Zhang, H.; Xu, F.; Song, C. Genome-wide identification and expression analysis of stress-associated proteins (SAPs) containing A20/AN1 zinc finger in cotton. Mol. Genet. Genom. 2016, 2919, 2199–2213. [Google Scholar] [CrossRef]
- Zhang, N.; Li, R.; Shen, W.; Jiao, S.; Zhang, J.; Xu, W. Genome-wide evolutionary characterization and expression analyses of major latex protein (MLP) family genes in Vitis vinifera. Mol. Genet. Genom. 2018, 293, 1061–1075. [Google Scholar] [CrossRef]
- Fujii, S.; Kubo, K.; Takayama, S. Non-self and self-recognition models in plant self-incompatibility. Nat. Plants 2016, 2, 16130. [Google Scholar] [CrossRef]
- Lee, H.K.; Goring, D.R. Two subgroups of receptor-like kinases promote early compatible pollen responses in the Arabidopsis thaliana pistil. J. Exp. Bot. 2020, 72, 1198–1211. [Google Scholar] [CrossRef]
- Yan, J.; Su, P.; Meng, X.; Liu, P. Phylogeny of the plant receptor-like kinase(RLK) gene family and expression analysis of wheat RLK genes in response to biotic and abiotic stresses. BMC Genom. 2023, 24, 224. [Google Scholar] [CrossRef]
- Harkness, A.; Brandvain, Y. Non-self recognition-based self-incompatibility can alternatively promote or prevent introgression. New Phytol. 2021, 231, 1630–1643. [Google Scholar] [CrossRef]
- Zou, C.; Wang, P.; Xu, Y. Bulked sample analysis in genetics, genomics and crop improvement. Plant Biotechnol. J. 2016, 14, 1941–1955. [Google Scholar] [CrossRef]
- Gillmor, C.S.; Roedera, H.K.; Sieber, P.; Somerville, C.; Lukowitz, W.A. Genetic screen for mutations affecting cell division in the Arabidopsis thaliana embryo identifies seven loci required for cytokinesis. PLoS ONE 2016, 11, e0146492. [Google Scholar] [CrossRef]
- Abe, A.; Kosugi, S.; Yoshida, K.; Natsume, S.; Takagi, H.; Kanzaki, H.; Matsumura, H.; Yoshida, K.; Mitsuoka, C.; Tamiru, M.; et al. Genome sequencing reveals agronomically important loci in rice using MutMap. Nat. Biotechnol. 2012, 30, 174–178. [Google Scholar] [CrossRef]
- Li, C.; Xiang, X.; Huang, Y.; Zhou, Y.; An, D.; Dong, J.; Zhao, C.; Liu, H.; Li, Y.; Wang, Q.; et al. Long-read sequencing reveals genomic structural variations that underlie creation of quality protein maize. Nat. Commun. 2020, 11, 17. [Google Scholar] [CrossRef]
- Ren, X.; Li, H.; Yin, Y.; Duan, L.; Wang, Y.; Liang, X.; Wan, R.; Huang, T.; Zhang, B.; Xi, W.; et al. Genetic analysis of fruit traits in wolfberry (Lycium L.) by the major gene plus polygene model. Agronomy 2022, 12, 1403. [Google Scholar] [CrossRef]
- Yin, Y.; An, W.; Zhao, J.; Li, Y.; Fan, F.; Chen, J.; Cao, Y.; Zhan, X. Constructing the wolfberry (Lycium spp.) genetic linkage map using AFLP and SSR markers. J. Integr. Agric. 2022, 21, 131–138. [Google Scholar] [CrossRef]
- Price, J.H.; Raduski, A.R.; Brandvain, Y.; Tassel, D.L.V.; Smith, K.P. Development of first linkage map for Silphium integrifolium (Asteraceae) enables identification of sporophytic self-incompatibility locus. Heredity 2022, 128, 304–312. [Google Scholar] [CrossRef]
- Sangduen, N.; Sorensen, E.L.; Liang, G.H. Pollen germination and pollen tube growth following self-pollination and intra- and interspecific pollination of Medicago species. Euphytica 1983, 32, 527–534. [Google Scholar] [CrossRef]
- Sijacic, P.; Wang, X.; Skirpan, A.L.; Wang, Y.; Dowd, P.E.; Mccubbin, A.G.; Huang, S.; Kao, T. Identification of the pollen determinant of S-RNase-mediated self-incompatibility. Nature 2004, 429, 302–305. [Google Scholar] [CrossRef]
- Xu, K.; Wu, N.; Yao, W.; Li, X.; Zhou, Y.; Li, H. The biological function and roles in phytohormone signaling of the F-box protein in plants. Agronomy 2021, 11, 2360. [Google Scholar] [CrossRef]
- Sun, P.L.; Li, S.; Lu, D.; Williams, J.S.; Kao, T. Pollen S-locus F-box proteins of Petunia involved in S-RNase-based self-incompatibility are themselves subject to ubiquitin-mediated degradation. Plant J. 2015, 83, 213–223. [Google Scholar] [CrossRef]
- Kubo, K.; Entani, T.; Takara, A.; Wang, N.; Fields, A.M.; Hua, Z.; Toyoda, M.; Kawashima, S.; Ando, T.; Isogai, A.; et al. Collaborative non-self recognition system in S-RNase-based self-incompatibility. Science 2010, 330, 796–799. [Google Scholar] [CrossRef]
- Li, S.; Williams, J.S.; Sun, P.L.; Kao, T.H. All 17 S-locus F-box proteins of the S2- and S3-haplotypes of Petunia inflata are assembled into similar SCF complexes with a specific function in self-incompatibility. Plant J. 2016, 87, 606–616. [Google Scholar] [CrossRef]
- Wu, L.; Williams, J.S.; Sun, L.H.; Kao, T.H. Sequence analysis of the Petunia inflata S-locus region containing 17 S-Locus F-Box genes and the S-RNase gene involved in self-incompatibility. Plant J. 2020, 104, 1348–1368. [Google Scholar] [CrossRef]



| S-Genotype | Forward Primer | Reverse Primer | Product Length/bp |
|---|---|---|---|
| S1 | ATGTTTAAATCACGACTCATGACTG | TCACCCTGGAAAATAAATTAAAGTG | 810 |
| S2 | GCTACCCAAAAGTGGGTTATAA | GAGTTTGCCATTAATTATGCTTAGC | 666 |
| S8 | GTGGGCAACCAAAAAGTGGGTTATG | GCCTTTTCGATATCATGAAGCA | 400 |
| S11 | GCTACCCAGAAGTGAGTTATGA | CAGCTCACTTATTTTTCTGGCATCC | 169 |
| Traits | Compatibility Index (CI) | Compared Compatibility Index (CCI) | Fruit Rate (FR) | Average Fruit Weight (AFW) |
|---|---|---|---|---|
| Compatibility index (CI) | 1 | |||
| Compared compatibility index (CCI) | 0.974 ** | 1 | ||
| Fruit rate (FR) | 0.818 ** | 0.816 ** | 1 | |
| Average fruit weight (AFW) | 0.817 ** | 0.794 ** | 0.889 ** | 1 |
| Chromosome ID | Start Site | End Site | Size/Mb |
|---|---|---|---|
| Chr02 | 24,700,000 | 24,900,000 | 0.2 |
| Chr02 | 29,100,000 | 34,000,000 | 4.9 |
| Chr02 | 34,700,000 | 35,400,000 | 0.7 |
| Chr02 | 35,800,000 | 36,900,000 | 1.1 |
| Chr02 | 37,100,000 | 39,800,000 | 2.7 |
| Chr02 | 40,200,000 | 45,300,000 | 5.1 |
| Chr02 | 46,000,000 | 46,200,000 | 0.2 |
| Chr02 | 47,600,000 | 49,500,000 | 1.9 |
| Chr02 | 56,700,000 | 57,500,000 | 0.8 |
| Chr02 | 57,800,000 | 58,000,000 | 0.2 |
| Chr02 | 58,300,000 | 58,600,000 | 0.3 |
| Chr02 | 58,900,000 | 63,700,000 | 4.8 |
| Chr02 | 64,700,000 | 85,800,000 | 21.1 |
| Chr02 | 86,100,000 | 87,200,000 | 1.1 |
| Chr02 | 87,500,000 | 93,400,000 | 5.9 |
| Chr02 | 97,700,000 | 102,000,000 | 4.3 |
| Chr02 | 103,900,000 | 106,300,000 | 2.4 |
| Chromosome ID | Start Site | End Site | Size/Mb |
|---|---|---|---|
| Chr02 | 11,100,000 | 11,300,000 | 0.2 |
| Chr02 | 12,700,000 | 13,400,000 | 0.7 |
| Chr02 | 13,800,000 | 19,100,000 | 5.3 |
| Chr02 | 21,000,000 | 52,300,000 | 31.3 |
| Chr02 | 52,800,000 | 57,400,000 | 4.6 |
| Chr02 | 59,000,000 | 83,900,000 | 24.9 |
| Chr02 | 84,300,000 | 85,300,000 | 1 |
| Chr02 | 87,600,000 | 92,900,000 | 5.3 |
| Chr02 | 115,500,000 | 115,800,000 | 0.3 |
| Chr02 | 120,700,000 | 121,700,000 | 1 |
| Chromosome ID | Start Site | End Site | Size/Mb |
|---|---|---|---|
| Chr02 | 24,700,000 | 24,900,000 | 0.2 |
| Chr02 | 30,800,000 | 34,000,000 | 3.2 |
| Chr02 | 34,700,000 | 35,400,000 | 0.7 |
| Chr02 | 35,800,000 | 36,900,000 | 1.1 |
| Chr02 | 37,100,000 | 38,300,000 | 1.2 |
| Chr02 | 40,200,000 | 45,300,000 | 5.1 |
| Chr02 | 47,600,000 | 49,200,000 | 2.6 |
| Chr02 | 56,700,000 | 57,200,000 | 0.5 |
| Chr02 | 59,600,000 | 63,100,000 | 3.5 |
| Chr02 | 70,500,000 | 81,100,000 | 10.6 |
| Chr02 | 88,800,000 | 92,300,000 | 3.5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, C.; Wu, J.; Gao, Y.; Dai, G.; Shang, X.; Ma, H.; Zhang, X.; Xu, W.; Qin, K. Localization of S-Locus-Related Self-Incompatibility in Lycium barbarum Based on BSA Analysis. Horticulturae 2024, 10, 190. https://doi.org/10.3390/horticulturae10020190
Wang C, Wu J, Gao Y, Dai G, Shang X, Ma H, Zhang X, Xu W, Qin K. Localization of S-Locus-Related Self-Incompatibility in Lycium barbarum Based on BSA Analysis. Horticulturae. 2024; 10(2):190. https://doi.org/10.3390/horticulturae10020190
Chicago/Turabian StyleWang, Cuiping, Jiali Wu, Yan Gao, Guoli Dai, Xiaohui Shang, Haijun Ma, Xin Zhang, Wendi Xu, and Ken Qin. 2024. "Localization of S-Locus-Related Self-Incompatibility in Lycium barbarum Based on BSA Analysis" Horticulturae 10, no. 2: 190. https://doi.org/10.3390/horticulturae10020190
APA StyleWang, C., Wu, J., Gao, Y., Dai, G., Shang, X., Ma, H., Zhang, X., Xu, W., & Qin, K. (2024). Localization of S-Locus-Related Self-Incompatibility in Lycium barbarum Based on BSA Analysis. Horticulturae, 10(2), 190. https://doi.org/10.3390/horticulturae10020190





