Characterization and Phylogenetic Analyses of the Complete Chloroplast Genome Sequence in Arachis Species
Abstract
1. Introduction
2. Result
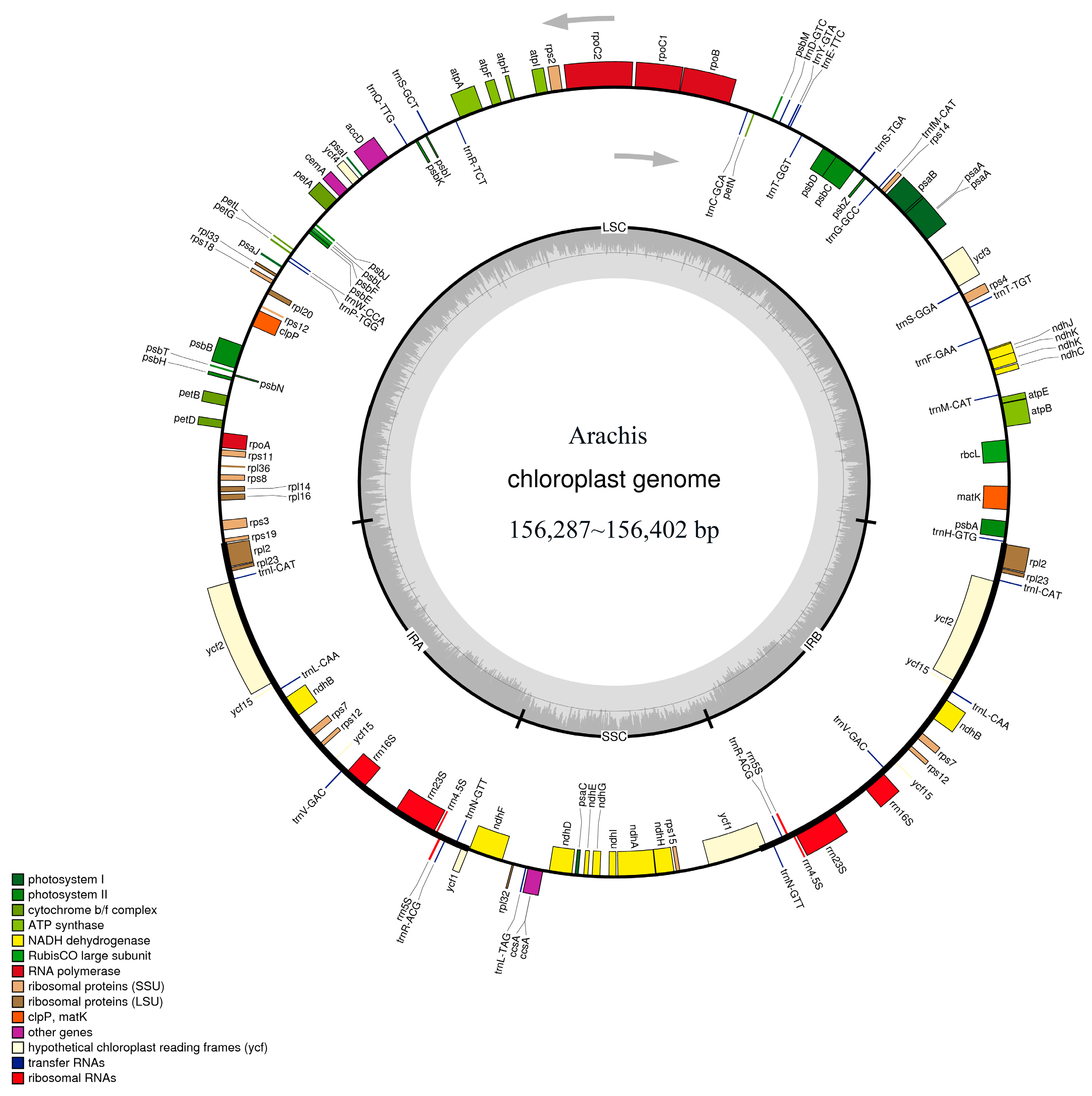
2.1. Basic Characteristics of the Acquired Arachis Chloroplast Genomes
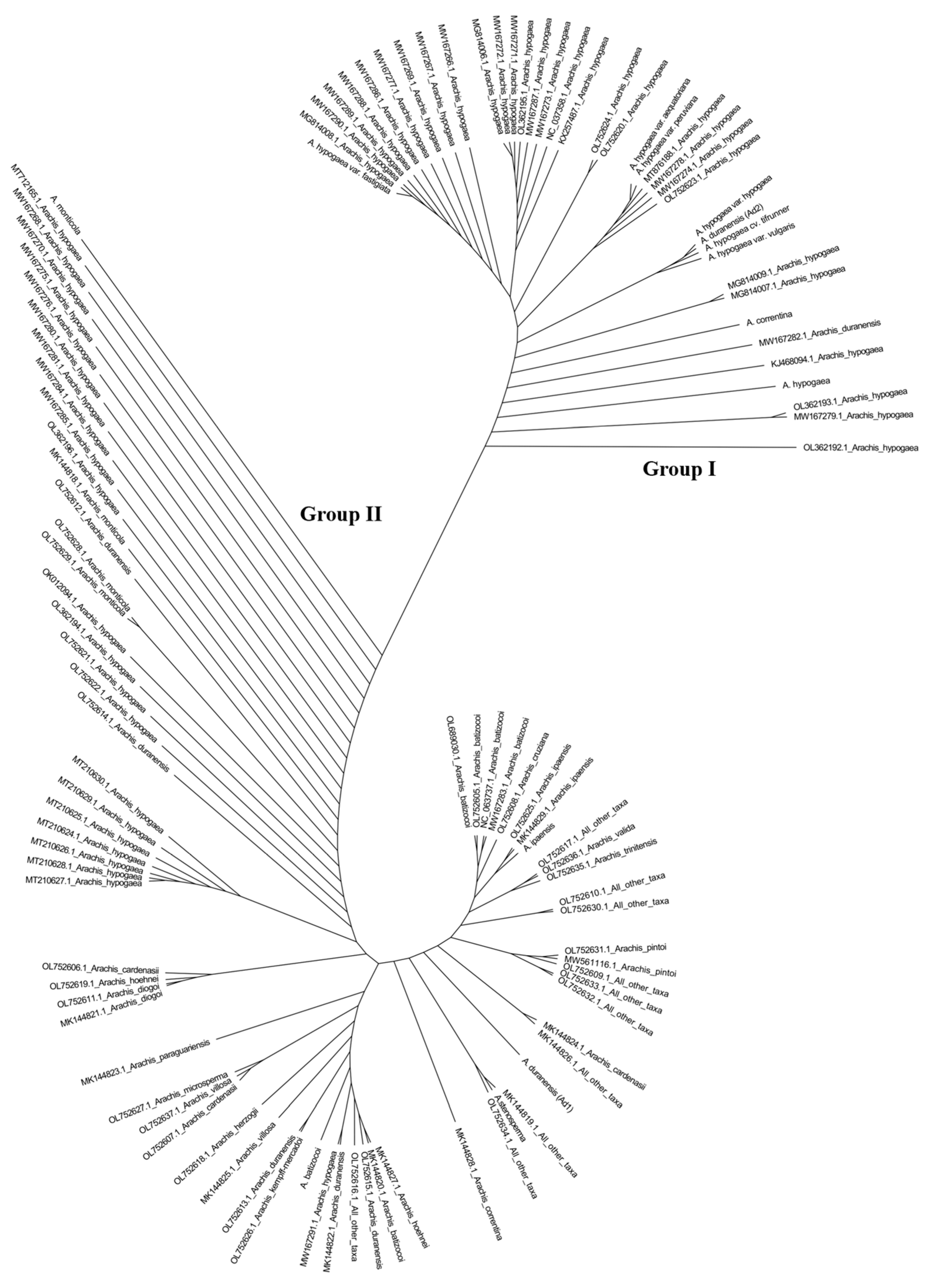
2.2. Phylogenetic Analysis
2.3. Information on Conserved and Variable Genes in the Arachis Chloroplast Genome
2.4. Arachis Chloroplast Genomes Have Diversity
2.5. The Selective Pressure of Arachis Chloroplast Genes Using Codeml
2.6. The Replication of Chloroplast Genes
3. Discussion
4. Materials and Methods
4.1. Plant Material and DNA Extraction
4.2. Genome Assembly and Annotation
4.3. Phylogenetic Analysis
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Gao, L.L.; Hong, Z.H.; Wang, Y.; Wu, G.Z. Chloroplast proteostasis: A story of birth, life, and death. Plant Commun. 2023, 4, 100424. [Google Scholar] [CrossRef] [PubMed]
- Daniell, H.; Lin, C.S.; Yu, M.; Chang, W.J. Chloroplast genomes: Diversity, evolution, and applications in genetic engineering. Genome Biol. 2016, 17, 134. [Google Scholar] [CrossRef] [PubMed]
- Maréchal, E. Primary Endosymbiosis: Emergence of the Primary Chloroplast and the Chromatophore, Two Independent Events. Methods Mol. Biol. 2018, 1829, 3–16. [Google Scholar]
- Bonnett, H.T. On the mechanism of the uptake of Vaucheria chloroplasts by carrot protoplasts treated with polyethylene glycol. Planta 1976, 131, 229–233. [Google Scholar] [CrossRef]
- Howe, C.J.; Barbrook, A.C.; Koumandou, V.L.; Nisbet RE, R.; Symington, H.A.; Wightman, T.F. Evolution of the chloroplast genome. Philos. Trans. R. Soc. B-Biol. Sci. 2003, 358, 99–107. [Google Scholar] [CrossRef] [PubMed]
- Chiba, Y.J.C. Cytochemical Studies on Chloroplasts I:Cytologic demonstration of nucleic acids in chloroplasts. Cytologia 1951, 16, 259–264. [Google Scholar] [CrossRef]
- Ohyama, K.; Fukuzawa, H.; Kohchi, T.; Shirai, H.; Sano, T.; Sano, S.; Umesono, K.; Shiki, Y.; Takeuchi, M.; Chang, Z.J.N. Chloroplast gene organization deduced from complete sequence of liverwort Marchantia polymorpha chloroplast DNA. Nature 1986, 322, 572–574. [Google Scholar] [CrossRef]
- Shinozaki, K.; Ohme, M.; Tanaka, M.; Wakasugi, T.; Hayashida, N.; Matsubayashi, T.; Zaita, N.; Chunwongse, J.; Obokata, J.; Yamaguchi-Shinozaki, K.; et al. The complete nucleotide sequence of the tobacco chloroplast genome: Its gene organization and expression. EMBO J. 1986, 5, 2043–2049. [Google Scholar] [CrossRef]
- Saski, C.; Lee, S.B.; Daniell, H.; Wood, T.C.; Tomkins, J.; Kim, H.G.; Jansen, R.K. Complete chloroplast genome sequence of Gycine max and comparative analyses with other legume genomes. Plant Mol. Biol. 2005, 59, 309–322. [Google Scholar] [CrossRef]
- Hiratsuka, J.; Shimada, H.; Whittier, R.; Ishibashi, T.; Sakamoto, M.; Mori, M.; Kondo, C.; Fan, J.; Zhu, W.Y.; Li, Z.F.; et al. Chloroplast genome sequence of a yellow colored rice (Oryza sativa L.): Insight into the genome structure and phylogeny. Mitochondrial DNA B Resour. 2020, 5, 3650–3652. [Google Scholar]
- Skuza, L.; Androsiuk, P.; Gastineau, R.; Paukszto, Ł.; Jastrzębski, J.P.; Cembrowska-Lech, D. Molecular structure, comparative and phylogenetic analysis of the complete chloroplast genome sequences of weedy rye Secale cereale ssp. segetale. Sci. Rep. 2023, 13, 5412. [Google Scholar] [CrossRef] [PubMed]
- Maier, R.M.; Neckermann, K.; Igloi, G.L.; Kössel, H. Complete sequence of the maize chloroplast genome: Gene content, hotspots of divergence and fine tuning of genetic information by transcript editing. J. Mol. Biol. 1995, 251, 614–628. [Google Scholar] [CrossRef] [PubMed]
- Yi, D.K.; Kim, K.J. Complete chloroplast genome sequences of important oilseed crop Sesamum indicum L. PLoS ONE 2012, 7, e35872. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Xiong, G.; Li, P.; He, F.; Huang, Y.; Wang, K.; Li, Z.; Hua, J. Analysis of complete nucleotide sequences of 12 Gossypium chloroplast genomes: Origin and evolution of allotetraploids. PLoS ONE 2012, 7, e37128. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.B.; Kaittanis, C.; Jansen, R.K.; Hostetler, J.B.; Tallon, L.J.; Town, C.D.; Daniell, H. The complete chloroplast genome sequence of Gossypium hirsutum: Organization and phylogenetic relationships to other angiosperms. BMC Genom. 2006, 7, 61. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, R.I.; Azuma, J.; Sakamoto, M. Complete nucleotide sequence of the cotton (Gossypium barbadense L.) chloroplast genome with a comparative analysis of sequences among 9 dicot plants. Genes. Genet. Syst. 2006, 81, 311–321. [Google Scholar] [CrossRef] [PubMed]
- Alekseyev, Y.O.; Fazeli, R.; Yang, S.; Basran, R.; Maher, T.; Miller, N.S.; Remick, D. A Next-Generation Sequencing Primer-How Does It Work and What Can It Do? Acad. Pathol. 2018, 5, 2374289518766521. [Google Scholar] [CrossRef] [PubMed]
- McCombie, W.R.; McPherson, J.D.; Mardis, E.R. Next-Generation Sequencing Technologies. Cold Spring Harb. Perspect. Med. 2019, 9, a036798. [Google Scholar] [CrossRef] [PubMed]
- Carbonell-Caballero, J.; Alonso, R.; Ibañez, V.; Terol, J.; Talon, M.; Dopazo, J. A Phylogenetic Analysis of 34 Chloroplast Genomes Elucidates the Relationships between Wild and Domestic Species within the Genus Citrus. Mol. Biol. Evol. 2015, 32, 2015–2035. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhang, Y.; Song, M.; Guan, Y.; Ma, X. Species Identification of Dracaena Using the Complete Chloroplast Genome as a Super-Barcode. Front. Pharmacol. 2019, 10, 1441. [Google Scholar] [CrossRef]
- Huo, Y.; Gao, L.; Liu, B.; Yang, Y.; Kong, S.; Sun, Y.; Wu, X. Complete chloroplast genome sequences of four Allium species: Comparative and phylogenetic analyses. Sci. Rep. 2019, 9, 12250. [Google Scholar] [CrossRef] [PubMed]
- Van Binh Nguyen, V.B.N.; Vo Ngoc Linh Giang, V.N.L.G.; Waminal, N.E.; Park HyunSeung, P.H.; Kim NamHoon, K.N.; Jang WooJong, J.W.; Yang TaeJin, Y.T. Comprehensive comparative analysis of chloroplast genomes from seven Panax species and development of an authentication system based on species-unique single nucleotide polymorphism markers. J. Ginseng Res. 2020, 44, 135–144. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Hu, Q.; Al-Shehbaz, I.A.; Luo, X.; Zeng, T.; Guo, X.; Liu, J. Species Delimitation and Interspecific Relationships of the Genus Orychophragmus (Brassicaceae) Inferred from Whole Chloroplast Genomes. Front. Plant Sci. 2016, 7, 1826. [Google Scholar] [CrossRef] [PubMed]
- Nie, Y.; Foster, C.S.P.; Zhu, T.; Yao, R.; Duchêne, D.A.; Ho, S.Y.W.; Zhong, B. Accounting for Uncertainty in the Evolutionary Timescale of Green Plants Through Clock-Partitioning and Fossil Calibration Strategies. Syst. Biol. 2020, 69, 1–16. [Google Scholar] [CrossRef]
- Gu, X.; Li, L.; Li, S.; Shi, W.; Zhong, X.; Su, Y.; Wang, T. Adaptive evolution and co-evolution of chloroplast genomes in Pteridaceae species occupying different habitats: Overlapping residues are always highly mutated. BMC Plant Biol. 2023, 23, 511. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.Z.; Liu, Y.L.; Zhang, D.; Li, W.; Gao, J.; Liu, Y.; Li, K.; Shi, C.; Zhao, Y.; Zhao, Y.J.; et al. Evolution of Oryza chloroplast genomes promoted adaptation to diverse ecological habitats. Commun. Biol. 2019, 2, 278. [Google Scholar] [CrossRef] [PubMed]
- Zhai, W.; Duan, X.; Zhang, R.; Guo, C.; Li, L.; Xu, G.; Shan, H.; Kong, H.; Ren, Y. Chloroplast genomic data provide new and robust insights into the phylogeny and evolution of the Ranunculaceae. Mol. Phylogenetics Evol. 2019, 135, 12–21. [Google Scholar] [CrossRef] [PubMed]
- Olejniczak, S.A.; Łojewska, E.; Kowalczyk, T.; Sakowicz, T. Chloroplasts: State of research and practical applications of plastome sequencing. Planta 2016, 244, 517–527. [Google Scholar] [CrossRef] [PubMed]
- Yin, D.; Wang, Y.; Zhang, X.; Ma, X.; He, X.; Zhang, J. Development of chloroplast genome resources for peanut (Arachis hypogaea L.) and other species of Arachis. Sci. Rep. 2017, 7, 11649. [Google Scholar] [CrossRef]
- Teske, D.; Peters, A.; Möllers, A.; Fischer, M. Genomic Profiling: The Strengths and Limitations of Chloroplast Genome-Based Plant Variety Authentication. J. Agric. Food Chem. 2020, 68, 14323–14333. [Google Scholar] [CrossRef]
- Wang, Y.; Yu, J.; Chen, Y.K.; Wang, Z.C. Complete Chloroplast Genome Sequence of the Endemic and Endangered Plant Dendropanax oligodontus: Genome Structure, Comparative and Phylogenetic Analysis. Genes 2022, 13, 2028. [Google Scholar] [CrossRef] [PubMed]
- Drouin, G.; Daoud, H.; Xia, J. Relative rates of synonymous substitutions in the mitochondrial, chloroplast and nuclear genomes of seed plants. Mol. Phylogenetics Evol. 2008, 49, 827–831. [Google Scholar] [CrossRef] [PubMed]
- Bertioli, D.J.; Cannon, S.B.; Froenicke, L.; Huang, G.; Farmer, A.D.; Cannon, E.K.; Liu, X.; Gao, D.; Clevenger, J.; Dash, S.; et al. The genome sequences of Arachis duranensis and Arachis ipaensis, the diploid ancestors of cultivated peanut. Nat. Genet. 2016, 48, 438–446. [Google Scholar] [CrossRef] [PubMed]
- Seijo, G.; Lavia, G.I.; Fernández, A.; Krapovickas, A.; Ducasse, D.A.; Bertioli, D.J.; Moscone, E.A. Genomic relationships between the cultivated peanut (Arachis hypogaea, Leguminosae) and its close relatives revealed by double GISH. Am. J. Bot. 2007, 94, 1963–1971. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Li, X.; Hu, C.; Qiu, X.; Li, J.; Li, X.; Zhu, H.; Wang, J.; Sui, J.; Qiao, L. Identification and characterization of transposable element AhMITE1 in the genomes of cultivated and two wild peanuts. BMC Genom. 2022, 23, 500. [Google Scholar] [CrossRef]
- Bertioli, D.J.; Jenkins, J.; Clevenger, J.; Dudchenko, O.; Gao, D.; Seijo, G.; Leal-Bertioli, S.C.M.; Ren, L.; Farmer, A.D.; Pandey, M.K.; et al. The genome sequence of segmental allotetraploid peanut Arachis hypogaea. Nat. Genet. 2019, 51, 877–884. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Lu, Q.; Liu, H.; Zhang, J.; Hong, Y.; Lan, H.; Li, H.; Wang, J.; Liu, H.; Li, S.; et al. Sequencing of Cultivated Peanut, Arachis hypogaea, Yields Insights into Genome Evolution and Oil Improvement. Mol. Plant 2019, 12, 920–934.e39. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Li, H.; Pandey, M.K.; Yang, Q.; Wang, X.; Garg, V.; Li, H.; Chi, X.; Doddamani, D.; Hong, Y.; et al. Draft genome of the peanut A-genome progenitor (Arachis duranensis) provides insights into geocarpy, oil biosynthesis, and allergens. Proc. Natl. Acad. Sci. USA 2016, 113, 6785–6790. [Google Scholar] [CrossRef]
- Wang, J.; Li, Y.; Li, C.; Yan, C.; Zhao, X.; Yuan, C.; Sun, Q.; Shi, C.; Shan, S. Twelve complete chloroplast genomes of wild peanuts: Great genetic resources and a better understanding of Arachis phylogeny. BMC Plant Biol. 2019, 19, 504. [Google Scholar] [CrossRef]
- Prabhudas, S.K.; Prayaga, S.; Madasamy, P.; Natarajan, P. Shallow Whole Genome Sequencing for the Assembly of Complete Chloroplast Genome Sequence of Arachis hypogaea L. Front. Plant Sci. 2016, 7, 1106. [Google Scholar] [CrossRef]
- Wang, J.; Li, C.; Yan, C.; Zhao, X.; Shan, S. A comparative analysis of the complete chloroplast genome sequences of four peanut botanical varieties. PeerJ 2018, 6, e5349. [Google Scholar] [CrossRef] [PubMed]
- Grabiele, M.; Chalup, L.; Robledo, G.; Seijo, G.J.P.S. Evolution, Genetic and geographic origin of domesticated peanut as evidenced by 5S rDNA and chloroplast DNA sequences. Plant Syst. Evol. 2012, 298, 1151–1165. [Google Scholar] [CrossRef]
- Tian, X.; Shi, L.; Guo, J.; Fu, L.; Du, P.; Huang, B.; Wu, Y.; Zhang, X.; Wang, Z. Chloroplast Phylogenomic Analyses Reveal a Maternal Hybridization Event Leading to the Formation of Cultivated Peanuts. Front. Plant Sci. 2021, 12, 804568. [Google Scholar] [CrossRef] [PubMed]
- Brock, J.R.; Mandáková, T.; McKain, M.L.M.A.; Olsen, K.M. Chloroplast phylogenomics in Camelina (Brassicaceae) reveals multiple origins of polyploid species and the maternal lineage of C. sativa. Hortic. Res. 2022, 9, uhab050. [Google Scholar] [CrossRef] [PubMed]
- Meng, J.; Li, X.; Li, H.; Yang, J.; Wang, H.; He, J. Comparative analysis of the complete chloroplast genomes of four aconitum medicinal species. Molecules 2018, 23, 1015. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.X.; Li, R.; Worth, J.R.P.; Li, X.; Li, P.; Cameron, K.M.; Fu, C.X. The complete chloroplast genome of Chinese bayberry (Morella rubra, Myricaceae): Implications for understanding the evolution of fagales. Front. Plant Sci. 2017, 8, 968. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, E.N.; Ruhlman, T.A.; Sabir, J.S.; Hajrah, N.H.; Alharbi, N.S.; Al-Malki, A.L.; Jansen, R.K. Evolution, Plastid genome sequences of legumes reveal parallel inversions and multiple losses of rps16 in papilionoids. J. Syst. Evol. 2015, 53, 458–468. [Google Scholar] [CrossRef]
- Dobrogojski, J.; Adamiec, M.; Luciński, R. The chloroplast genome: A review. Acta Physiol. Plant. 2020, 42, 98. [Google Scholar] [CrossRef]
- Zhang, Y.; Tian, L.; Lu, C. Chloroplast Gene Expression: Recent Advances and Perspectives. Plant Commun. 2023, 4, 100611. [Google Scholar] [CrossRef]
- Spielman, S.J.; Wilke, C.O. The relationship between dN/dS and scaled selection coefficients. Mol. Biol. Evol. 2015, 32, 1097–1108. [Google Scholar] [CrossRef]
- Nielsen, R.; Yang, Z. Likelihood models for detecting positively selected amino acid sites and applications to the HIV-1 envelope gene. Genetics 1998, 148, 929–936. [Google Scholar] [CrossRef] [PubMed]
- Kosakovsky Pond, S.L.; Frost, S.D. Not so different after all: A comparison of methods for detecting amino acid sites under selection. Mol. Biol. Evol. 2005, 22, 1208–1222. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Raina, S.N.; Rajpal, V.R.; Singh, A.K. Seed protein fraction electrophoresis in peanut (Arachis hypogaea L.) accessions and wild species. Physiol. Mol. Biol. Plants Int. J. Funct. Plant Biol. 2018, 24, 465–481. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Mittal, N.; Leamy, L.J.; Barazani, O.; Song, B.H. Back into the wild-Apply untapped genetic diversity of wild relatives for crop improvement. Evol. Appl. 2017, 10, 5–24. [Google Scholar] [CrossRef] [PubMed]
- Dempewolf, H.; Baute, G.; Anderson, J.; Kilian, B.; Smith, C.; Guarino, L.J.C.S. Past and Future Use of Wild Relatives in Crop Breeding. Crop Sci. 2017, 57, 1070–1082. [Google Scholar] [CrossRef]
- Zheng, Z.; Sun, Z.; Fang, Y.; Qi, F.; Liu, H.; Miao, L.; Du, P.; Shi, L.; Gao, W.; Han, S.; et al. Genetic diversity, population structure, and botanical variety of 320 global peanut accessions revealed through tunable genotyping-by-sequencing. Sci. Rep. 2018, 8, 14500. [Google Scholar] [CrossRef] [PubMed]
- Otyama, P.I.; Kulkarni, R.; Chamberlin, K.; Ozias, A.P.; Chu, Y.; Lincoln, L.M.; MacDonald, G.E.; Anglin, N.L.; Dash, S.; Bertioli, D.J.; et al. Genotypic characterization of the U.S. peanut core collection. G3-Genes. Genomes Genet. 2020, 10, 4013–4026. [Google Scholar] [CrossRef] [PubMed]
- Luo, R.; Liu, B.; Xie, Y.; Li, Z.; Huang, W.; Yuan, J.; He, G.; Chen, Y.; Pan, Q.; Liu, Y.; et al. SOAPdenovo2: An empirically improved memory-efficient short-read de novo assembler. GigaScience 2012, 1, 18. [Google Scholar] [CrossRef] [PubMed]
- Saha, S.; Bridges, S.; Magbanua, Z.V.; Peterson, D.G. Empirical comparison of ab initio repeat finding programs. Nucleic Acids Res. 2008, 36, 2284–2294. [Google Scholar] [CrossRef]
- Behboudi, R.; Nouri-Baygi, M.; Naghibzadeh, M. RPTRF: A rapid perfect tandem repeat finder tool for DNA sequences. Biosystems 2023, 226, 104869. [Google Scholar] [CrossRef]
- Kemena, C.; Dohmen, E.; Bornberg-Bauer, E. DOGMA: A web server for proteome and transcriptome quality assessment. Nucleic Acids Res. 2019, 47, W507–W510. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Smith, D.R. An overview of online resources for intra-species detection of gene duplications. Front. Genet. 2022, 13, 1012788. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S.; Hattori, M.; Aoki-Kinoshita, K.F.; Itoh, M.; Kawashima, S.; Katayama, T.; Araki, M.; Hirakawa, M. From genomics to chemical genomics: New developments in KEGG. Nucleic Acids Res. 2006, 34, D354–D357. [Google Scholar] [CrossRef] [PubMed]
- Huckvale, E.; Moseley, H.N.B. kegg_pull: A software package for the RESTful access and pulling from the Kyoto Encyclopedia of Gene and Genomes. BMC Bioinform. 2023, 24, 78. [Google Scholar] [CrossRef]
- Galperin, M.Y.; Wolf, Y.I.; Makarova, K.S.; Vera Alvarez, R.; Landsman, D.; Koonin, E.V. COG database update: Focus on microbial diversity, model organisms, and widespread pathogens. Nucleic Acids Res. 2021, 49, D274–D281. [Google Scholar] [CrossRef]
- Tatusov, R.L.; Fedorova, N.D.; Jackson, J.D.; Jacobs, A.R.; Kiryutin, B.; Koonin, E.V.; Krylov, D.M.; Mazumder, R.; Mekhedov, S.L.; Nikolskaya, A.N.; et al. The COG database: An updated version includes eukaryotes. BMC Bioinform. 2003, 4, 41. [Google Scholar] [CrossRef] [PubMed]
- Gene Ontology Consortium. Gene Ontology Consortium: Going forward. Nucleic Acids Res. 2015, 43, D1049–D1056. [Google Scholar] [CrossRef] [PubMed]
- The Gene Ontology Consortium. The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res. 2019, 47, D330–D338. [Google Scholar] [CrossRef]
- Magrane, M. UniProt Knowledgebase: A hub of integrated protein data. Database J. Biol. Databases Curation 2011, 2011, bar009. [Google Scholar] [CrossRef]
| Species | Variety Name | Genome Type | Ploidy | |
|---|---|---|---|---|
| Domesticated varieties | ||||
| A. hypogaea | Luhua11 | AABB | 4X | |
| A. hypogaea var. fastigiata | Silihong | AABB | 4X | |
| A. hypogaea var. vulgaris | Baisha1016 | AABB | 4X | |
| A. hypogaea var. peruviana | Yunnanqicai | AABB | 4X | |
| A. hypogaea var. aequatoriana | Chidao1 | AABB | 4X | |
| A. hypogaea cv. tifrunner | Tifrunner | AABB | 4X | |
| A. hypogaea var. hypogaea | Xiaohongmao | AABB | 4X | |
| Wild allotetraploid species | ||||
| A. monticola | Monticola | AABB | 4X | |
| Wild diploid species | ||||
| A. batizocoi | Ba-1 | KK | 2X | |
| A. ipaensis | Ip-1 | BB | 2X | |
| A. duranensis | Ad-1 | AA | 2X | |
| A. duranensis | Ad-2 | AA | 2X | |
| A.stenosperma | St-1 | AA | 2X | |
| A. correntina | Correntina | AA | 2X |
| Species | Variety Name | Raw Reads | Genome Size (bp) | Gene Number | GC Content (%) | Total Protein | LSC (bp) | SSC (bp) | IR (bp) | rRNA | tRNA |
|---|---|---|---|---|---|---|---|---|---|---|---|
| M1 | Luhua11 | 934 | 156,359 | 88 | 36.4 | 88 | 85,910 | 18,787 | 25,831 | 8 | 43 |
| M2 | Silihong | 1006 | 156,391 | 88 | 36.4 | 88 | 85,913 | 18,794 | 25,842 | 8 | 43 |
| M3 | Baisha1016 | 897 | 156,355 | 88 | 36.4 | 88 | 85,906 | 18,769 | 25,840 | 8 | 43 |
| M4 | Yunnanqicai | 842 | 156,395 | 88 | 36.4 | 88 | 85,918 | 18,789 | 25,844 | 8 | 43 |
| M5 | Tifrunner | 930 | 156,395 | 88 | 36.4 | 88 | 85,924 | 18,803 | 25,834 | 8 | 43 |
| M6 | Monticola | 1053 | 156,395 | 88 | 36.4 | 88 | 85,924 | 18,803 | 25,834 | 8 | 43 |
| M7 | Ip-1 | 936 | 156,399 | 90 | 36.4 | 90 | 85,938 | 18,793 | 25,834 | 8 | 43 |
| M8 | Ad-1 | 1323 | 156,343 | 91 | 36.4 | 91 | 85,902 | 18,805 | 25,818 | 8 | 29 |
| M9 | Ba-1 | 1807 | 156,402 | 91 | 36.4 | 91 | 85,922 | 18,812 | 25,834 | 8 | 29 |
| M10 | St-1 | 1797 | 156,303 | 91 | 36.4 | 91 | 85,853 | 18,804 | 25,823 | 8 | 29 |
| M11 | Ad-2 | 2122 | 156,359 | 91 | 36.4 | 91 | 85,953 | 18,760 | 25,823 | 8 | 29 |
| M12 | Correntina | 1934 | 156,373 | 91 | 36.4 | 91 | 85,930 | 18,797 | 25,823 | 8 | 29 |
| M13 | Chidao1 | 1553 | 156,373 | 91 | 36.4 | 91 | 85,930 | 18,797 | 25,823 | 8 | 29 |
| M14 | Xiaohongmao | 1789 | 156,287 | 91 | 36.4 | 91 | 85,843 | 18,798 | 25,823 | 8 | 29 |
| Gene Categories | Conserved Gene | Synonymous Mutations | Amino Acid Mutations |
|---|---|---|---|
| Photosystem I | psaC, psaI, psaJ | psaB | psaA |
| Photosystem II | psbC, psbE, psbF, psbH, psbI, psbJ, psbK, psbL, psbM, psbN, psbZ, psi_psbT | psbA, psbB, psbD, psbT | |
| RuBisCO large subunit | rbcL | ||
| Cytochrome b/f complex | petL, petN | petB, petG | petA, petD |
| c-type cytochrome | ccsA | ||
| ATP synthase | atpB, atpE, | atpI | atpA, atpF, atpH |
| NADH dehydrogenase | ndhI, ndhJ | ndhD, ndhE | ndhA, ndhB, ndhC, ndhF, ndhG, ndhH, ndhK |
| Assembly/stability of photosystem I | ycf3, ycf4 | ||
| RNA polymerase genes | rpoA, rpoB, rpoC1, rpoC2 | ||
| Ribosomal protein | rps12 *, rps14, rps18, rpl23 *, rpl32, rpl33 | rps4, rps7 *, rps11, rpl14, rpl20, | rps2, rps3, rps8, rps15, rps19, rpl2 *, rpl16, rpl36 |
| Ribosomal RNA | rrn16 *, rrn5 *, rrn4.5 * | rrn23 * | |
| Transfer RNA | trnAUGC *, trnCGCA, trnDGUC, trnEUUC, trnFGAA, trnGGCC, trnGUCC, trnHGUG, trnICAU *, trnIGAU *, trnKUUU, trnLCAA, trnLUAA *, trnLUAG, trnMCAU, trnNGUU *, trnQUUG, trnRACG *, trnRUCU, trnSGCU, trnSGGA, trnSUGA, trnTGGU, trnTUGU, trnVGAC *, trnWCCA, trnYGUA | trnMCAU, trnPUGG, trnWCCA | |
| Acetyl-CoA carboxylase subunit | accD | ||
| Proteolysis subunit | clpP | ||
| Carbon metabolism | cemA | ||
| Maturase | matK | ||
| Conserved reading frames | ycf68, orf56, orf42 | ycf1, ycf2 * |
| Gene Categories | Gene | M1 vs. M2 | M7 vs. M8 |
|---|---|---|---|
| Photosystem I | psaA | None | 16 L |
| Cytochrome b/f complex | petA | None | 176 V |
| petD | None | 137 P | |
| c-type cytochrome | ccsA | None | 61 I, 121 I, 284 F |
| ATP synthase | atpA | 391 S | 391 S |
| atpF | 1 S, 2 F, 3 S, 4 F, 5 G, 6 F, 7 N, 8 T, 9 D, 10 I, 11 L, 12 A | 1 S, 2 F, 3 S, 4 F, 5 G, 6 F, 7 N, 8 T, 9 D, 10 I, 11 L, 12 A | |
| atpH | 11 V | 11 V | |
| NADH dehydrogenase | ndhA | 185 L | 185 L |
| ndhB | 4 E, 5 M, 6 A, 7 L, 8 T, 10 F, 11 L, 13 F, 14 Y, 15 N, 16 S, 20 P, 21 D, 22 Y, 24 G | 4 E, 5 M, 6 A, 7 L, 8 T, 10 F, 11 L, 13 F, 14 Y, 15 N, 16 S, 20 P, 21 D, 22 Y, 24 G | |
| ndhF | 21 L, 186 L, 332 M, 476 Y, 490 N, 582 L, 586 S, 601 Q, 689 F | 21 L, 186 L, 332 M, 476 Y, 490 N, 582 L, 586 S, 601 Q, 689 F | |
| ndhG | 30 T, 166 A | 30 T, 166 A | |
| ndhH | 292 I, 301 P | 292 I, 301 P | |
| ndhK | 2 S, 6 L, 8 P, 10 P, 11 K, 12 Y, 13 V, 15 A, 16 M, 18 A, 19 C, 22 T, 25 M, 26 F, 29 D, 30 S, 31 Y, 33 P, 34 G, 35 C, 36 P, 37 P, 41 A, 44 D, 48 T, 51 K, 52 K, 53 Y, 54 K, 55 K | 1 P, 2 S, 6 L, 8 P, 10 P, 11 K, 12 Y, 13 V, 15 A, 16 M, 18 A, 19 C, 20 T, 21 I, 22 T, 24 G, 25 M, 26 F, 27 S, 29 D, 30 S, 31 Y, 32 L, 33 P, 34 G, 35 C, 36 P, 37 P, 38 K, 40 E, 41 A, 44 D, 45 A, 47 T, 48 T, 51 K, 52 K, 53 Y, 54 K, 55 K | |
| Assembly/stability of photosystem I | ycf3 | 40 R, 41 D, 43 M, 77 N | 40 R, 77 N |
| ycf4 | None | 3 W, 118 I | |
| RNA polymerase genes | rpoA | None | 111 N, 133 T, 234 A, 269 L |
| rpoB | None | 44 L, 210 D, 646 F | |
| rpoC1 | 1 F, 3 I, 4 D, 5 P, 6 L, 9 S, 11 P, 12 N, 449 K | 1 F, 3 I, 4 D, 5 P, 6 L, 9 S, 11 P, 12 N, 449 K | |
| rpoC2 | 430 L, 469 P, 634 P, 660 E, 675 L, 697 K, 773 L, 824 H, 912 K, 996 S, 998 E, 1000 L, 1001 K, 1002 G, 1003 K, 1004 L, 1013 L, 1014 K, 1015 K, 1017 C, 1193 I, 1335 K | 430 L, 469 P, 634 P, 660 E, 675 L, 697 K, 773 L, 824 H, 912 K, 996 S, 998 E, 1000 L, 1001 K, 1002 G, 1003 K, 1004 L, 1013 L, 1014 K, 1015 K, 1017 C, 1193 I, 1335 K | |
| Ribosomal protein | rpl2 | 131 N, 133 G, 134 V, 135 N, 138 E, 139 G, 140 R, 141 A, 143 I, 144 K, 146 A, 147 T | 131 N, 133 G, 134 V, 135 N, 138 E, 139 G, 140 R, 141 A, 143 I, 144 K, 146 A, 147 T |
| rpl16 | 104 M, 126 Q | 104 M, 126 Q | |
| rpl36 | 24 L | 24 L | |
| rps2 | None | 198 N | |
| rps3 | 153 Q | 153 Q | |
| rps8 | None | 28 C | |
| rps15 | 18 N | 18 N | |
| rps19 | 1 K, 2 K | 1 K, 2 K | |
| Acetyl-CoA carboxylase subunit | accD | 4 G, 50 P, 119 L, 199 G, 286 M, 399 N | 4 G, 50 P, 119 L, 199 G, 286 M, 399 N |
| Proteolysis subunit | clpP | 1 I, 99 R | 1 I, 99 R |
| Conserved reading frames | ycf1 | 162 K, 181 F, 226 V, 233 D, 241 F, 242 K, 257 H, 264 I, 301 A, 309 K, 349 T, 362 S, 371 Q, 379 S, 405 L, 406 S, 407 N | 162 K, 181 F, 226 V, 233 D, 241 F, 242 K, 257 H, 264 I, 301 A, 309 K, 349 T, 362 S, 371 Q, 379 S, 405 L, 406 S, 407 N |
| ycf2 | 1293 K | 169 W, 531 S, 532 E, 538 N, 751 H, 1206 R, 1223 L, 1292 W, 1293 K, 1294 T |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, X.; Liang, T.; Guo, Y.; Liang, Y.; Zou, X.; Si, T.; Ni, Y.; Zhang, X. Characterization and Phylogenetic Analyses of the Complete Chloroplast Genome Sequence in Arachis Species. Horticulturae 2024, 10, 464. https://doi.org/10.3390/horticulturae10050464
Yu X, Liang T, Guo Y, Liang Y, Zou X, Si T, Ni Y, Zhang X. Characterization and Phylogenetic Analyses of the Complete Chloroplast Genome Sequence in Arachis Species. Horticulturae. 2024; 10(5):464. https://doi.org/10.3390/horticulturae10050464
Chicago/Turabian StyleYu, Xiaona, Tianzhu Liang, Yi Guo, Yan Liang, Xiaoxia Zou, Tong Si, Yu Ni, and Xiaojun Zhang. 2024. "Characterization and Phylogenetic Analyses of the Complete Chloroplast Genome Sequence in Arachis Species" Horticulturae 10, no. 5: 464. https://doi.org/10.3390/horticulturae10050464
APA StyleYu, X., Liang, T., Guo, Y., Liang, Y., Zou, X., Si, T., Ni, Y., & Zhang, X. (2024). Characterization and Phylogenetic Analyses of the Complete Chloroplast Genome Sequence in Arachis Species. Horticulturae, 10(5), 464. https://doi.org/10.3390/horticulturae10050464






