Phenotypic and Molecular-Markers-Based Assessment of Jamun (Syzygium cumini) Genotypes from Pakistan
Abstract
:1. Introduction
2. Materials and Methods
2.1. Phenotypic Diversity Estimation
2.2. Genetic Diversity Estimation
2.3. Statistical Analysis
3. Results
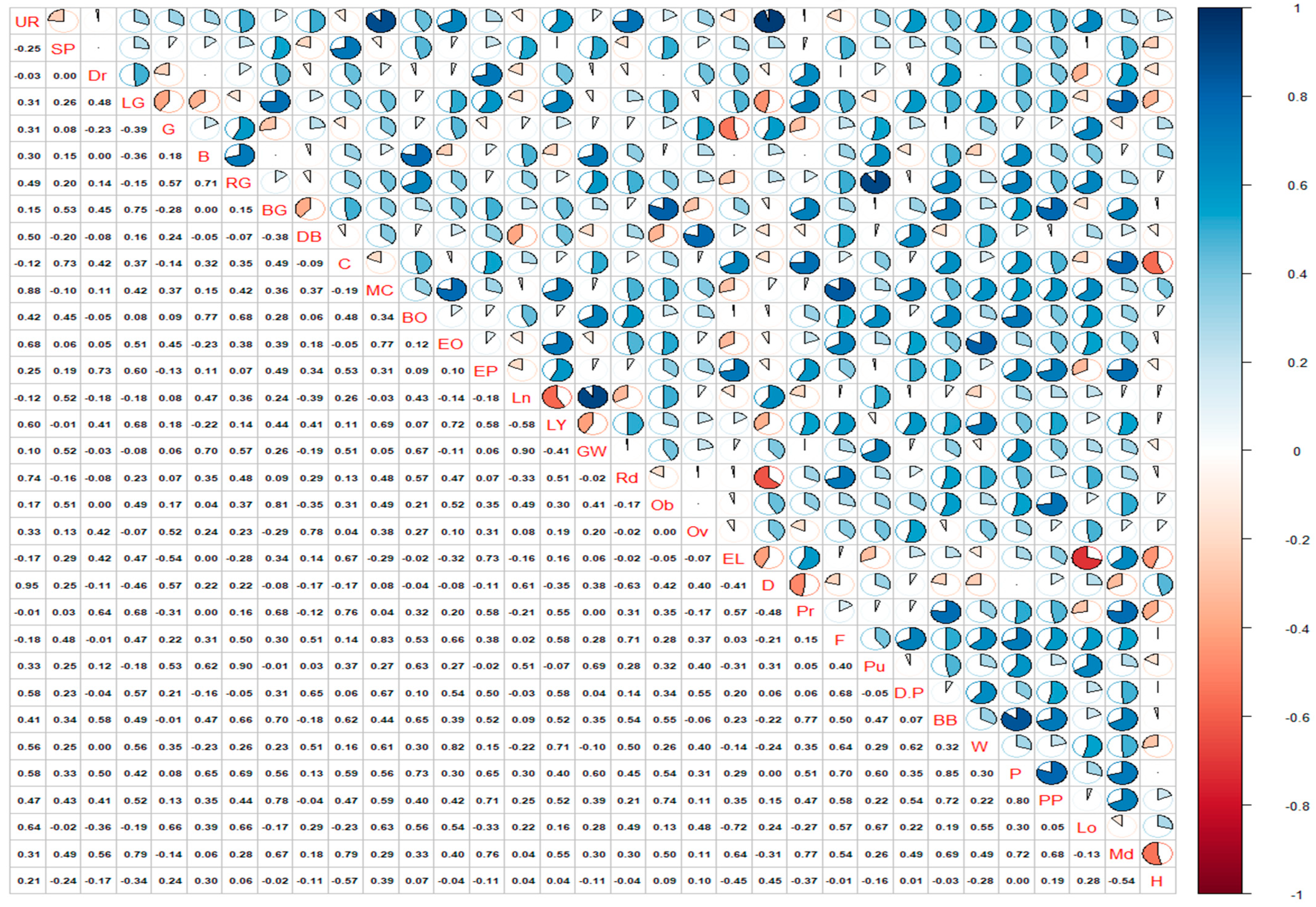
3.1. Frequency Distribution and Correlation of Descriptive Phenotypic Characters among Jamun (S. cumini) Genotypes
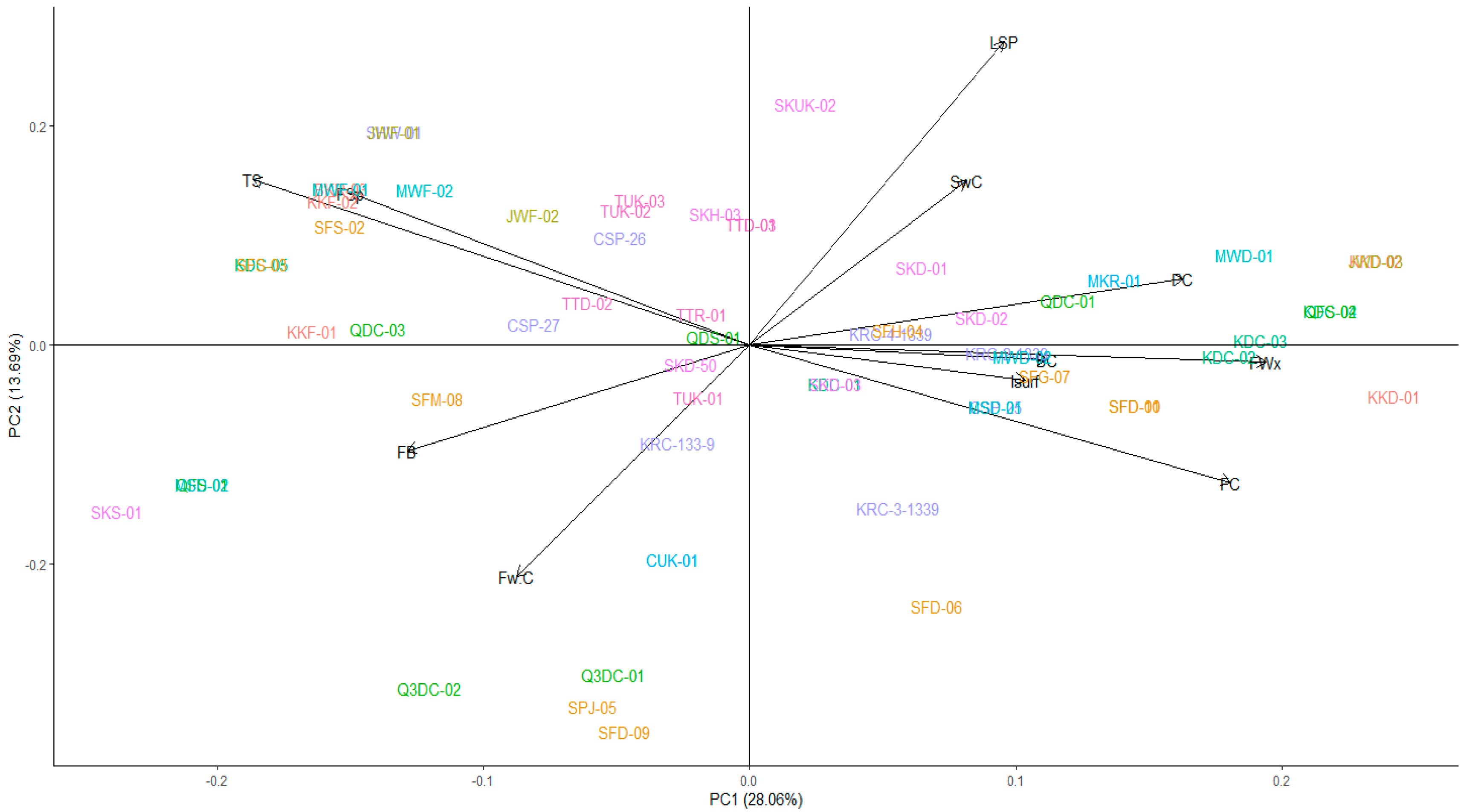
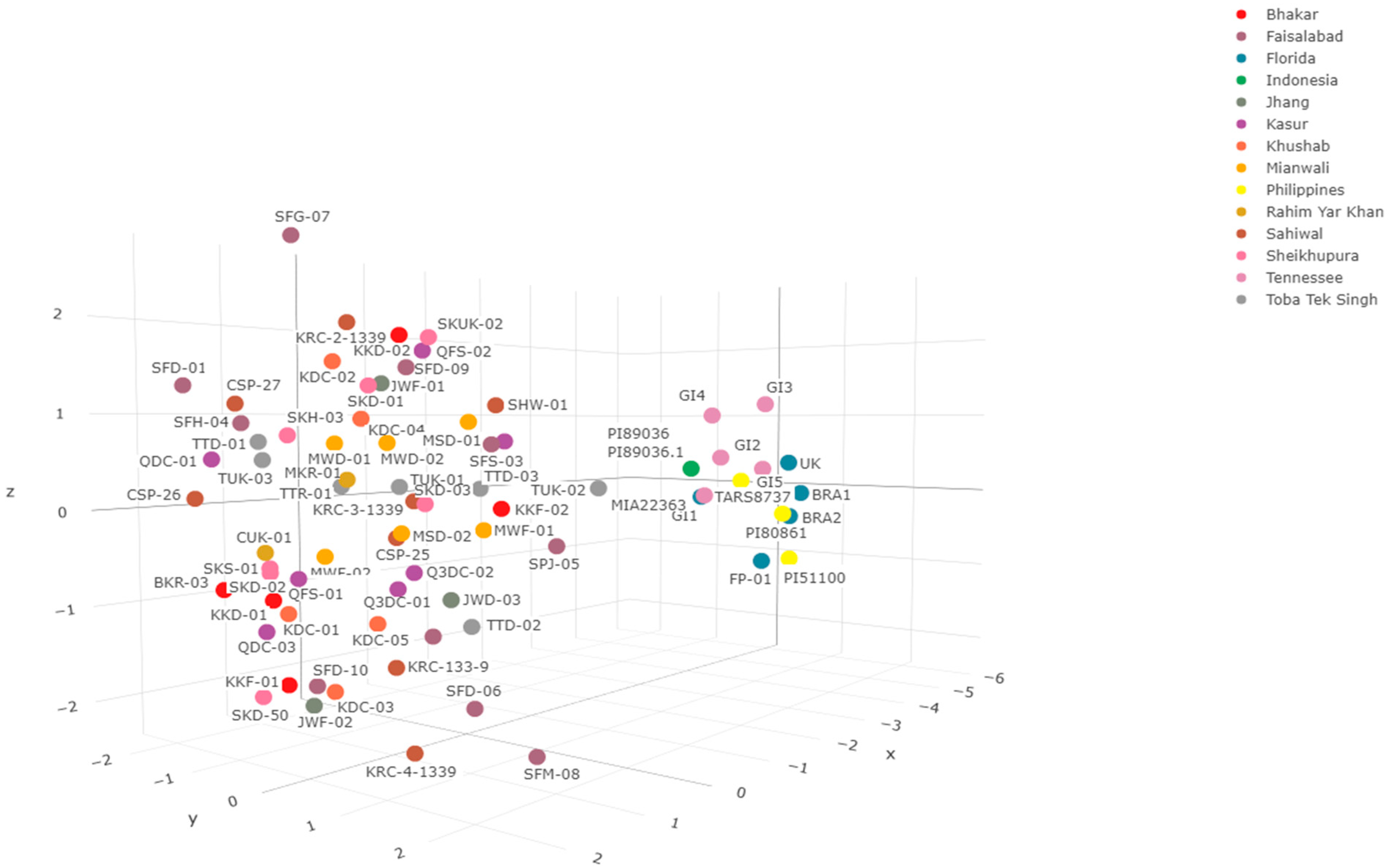
Correspondence Analysis on Phenotypic Bases
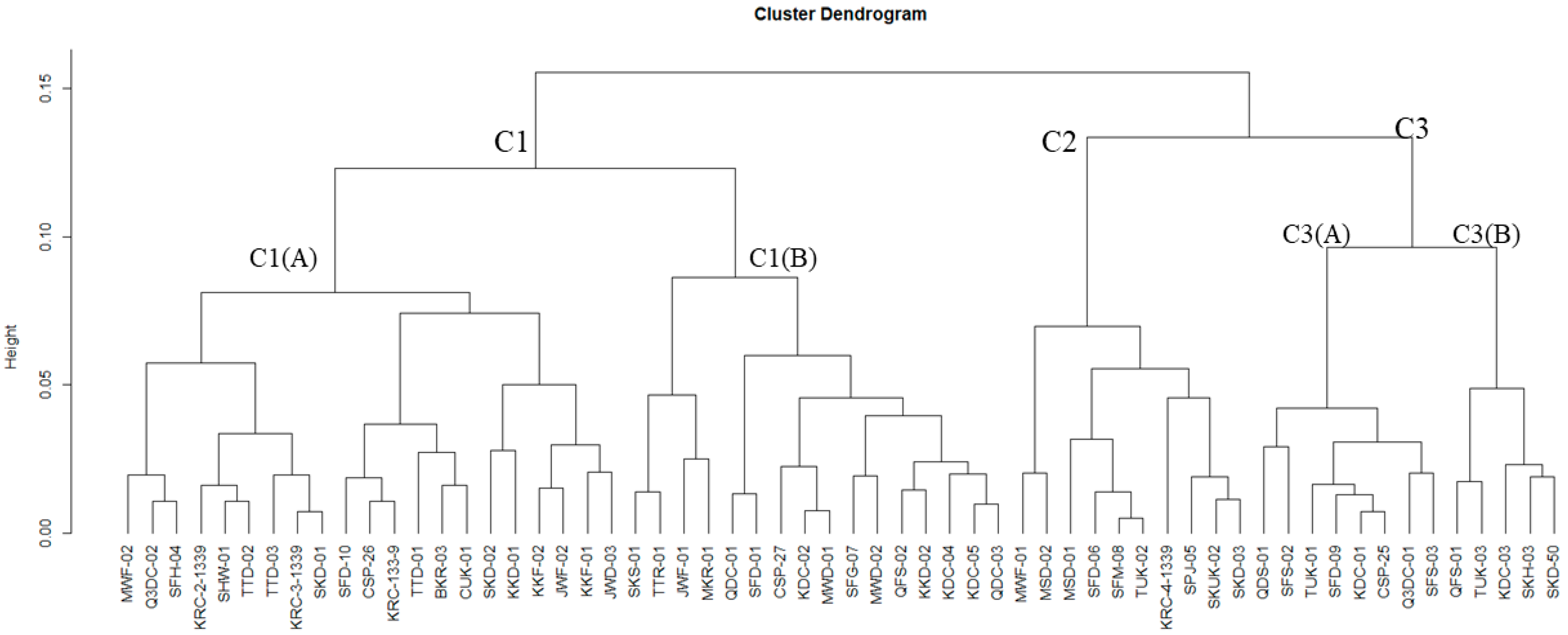
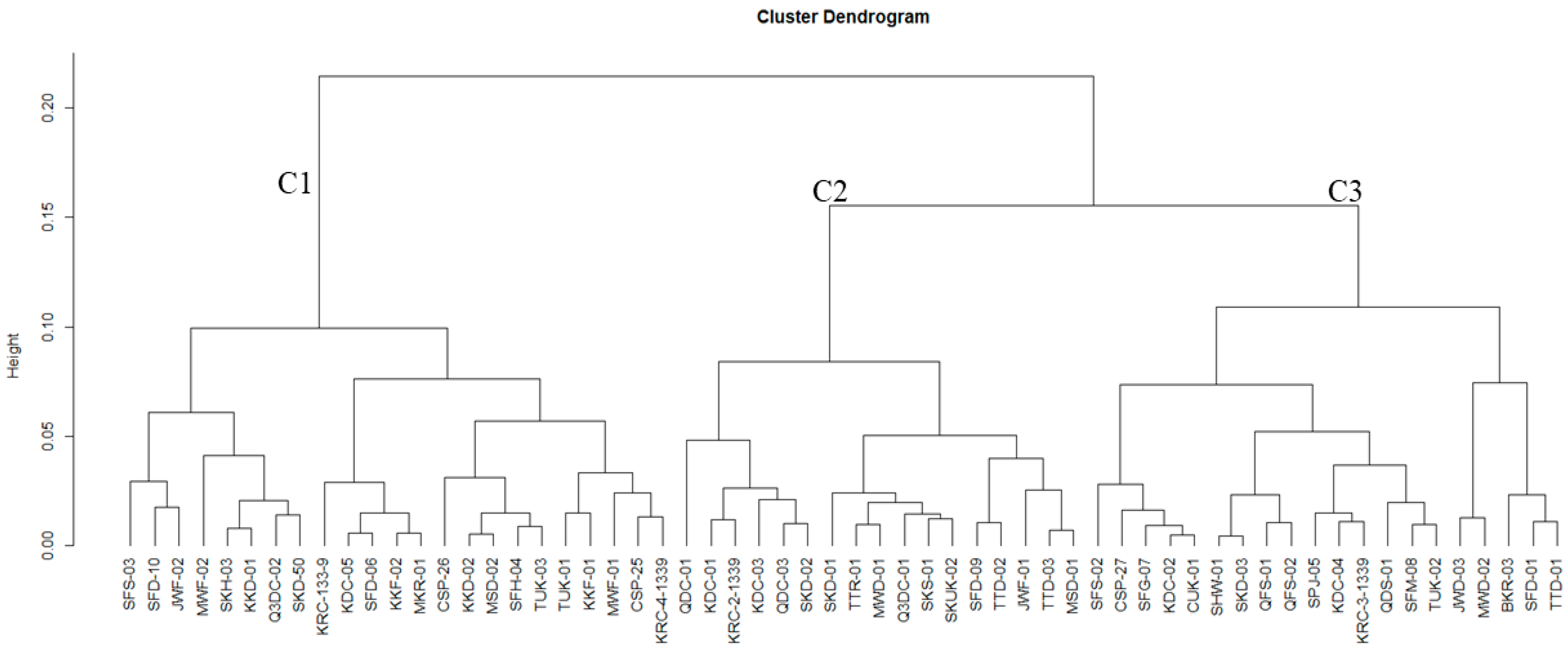
3.2. Polymorphism and Genetic Differentiation among Jamun Genotypes
3.2.1. Genetic Variability Analysis among Jamun Genotypes Using RAPD and ISSR Markers
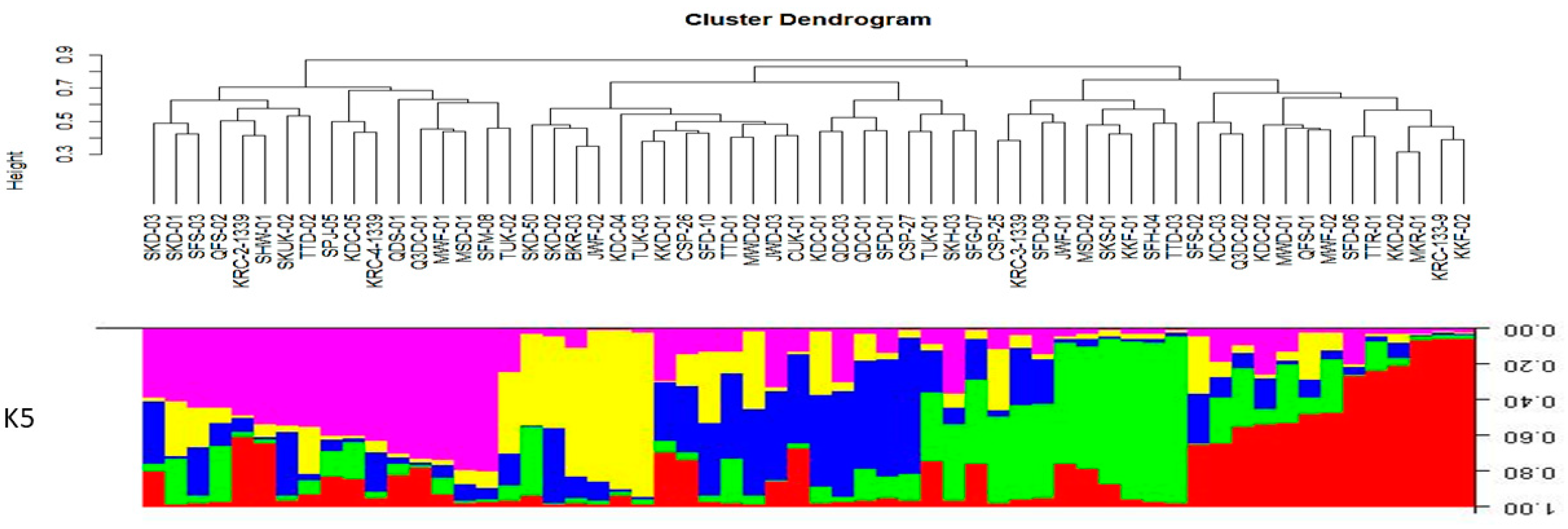
3.2.2. Population Genetic Structure
3.3. Genetic Diversity Comparison between Pakistani and International S. cumini Genotypes
4. Discussion
4.1. Phenotypic Diversity in Jamun Germplasm
4.2. Characterization Based on Genetic Markers
4.3. Genetic Relationships
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nair, K. The Genus Syzygium: Syzygium cumini and Other Underutilized Species; CRC Press: Boca Raton, FL, USA, 2017. [Google Scholar] [CrossRef]
- Parnell, J.A.N.; Craven, L.A.; Biffin, E. Matters of Scale: Dealing with One of the Largest Genera of Angiosperms. In Reconstructing the Tree of Life: Taxonomy and Systematics of Species Rich Taxa; CRC Press LLC: Boca Raton, FL, USA, 2007; ISBN 0849395798. [Google Scholar]
- Gajera, H.P.; Gevariya, S.N.; Patel, S.V.; Golakiya, B.A. Nutritional Profile and Molecular Fingerprints of Indigenous Black Jamun (Syzygium cumini L.) Landraces. J. Food Sci. Technol. 2018, 55, 730–739. [Google Scholar] [CrossRef] [PubMed]
- Rizvi, M.K.; Rabail, R.; Munir, S.; Inam-Ur-Raheem, M.; Qayyum, M.M.N.; Kieliszek, M.; Hassoun, A.; Aadil, R.M. Astounding Health Benefits of Jamun (Syzygium cumini) toward Metabolic Syndrome. Molecules 2022, 27, 7184. [Google Scholar] [CrossRef] [PubMed]
- Tak, Y.; Kaur, M.; Jain, M.C.; Samota, M.K.; Meena, N.K.; Kaur, G.; Kumar, R.; Sharma, D.; Lorenzo, J.M.; Amarowicz, R. Jamun Seed: A Review on Bioactive Constituents, Nutritional Value and Health Benefits. Pol. J. Food Nutr. Sci. 2022, 72, 211–228. [Google Scholar] [CrossRef]
- Ghosh, P.; PRadhaN, R.C.; Mishra, S.; Patel, A.S.; Kar, A. Physicochemical and Nutritional Characterization of Jamun (Syzygium cumini). Curr. Res. Nutr. Food Sci. J. 2017, 5, 25–35. [Google Scholar] [CrossRef]
- Qamar, M.; Akhtar, S.; Ismail, T.; Wahid, M.; Abbas, M.W.; Mubarak, M.S.; Yuan, Y.; Barnard, R.T.; Ziora, Z.M.; Esatbeyoglu, T. Phytochemical Profile, Biological Properties, and Food Applications of the Medicinal Plant Syzygium cumini. Foods 2022, 11, 378. [Google Scholar] [CrossRef]
- Jagetia, G.C. A review on the role of Jamun, Syzygium cumini Skeels in the treatment of diabetes. Int. J. Complement. Altern. Med. 2018, 11, 91–95. [Google Scholar] [CrossRef]
- Mulkalwar, S.; Kulkarni, V.; Deshpande, T.; Bhide, H.; Patel, A.; Tilak, A.V. Antihyperglycemic Activity of Syzygium cumini (Jamun) in Diabetic Rats. J. Pharm. Res. Int. 2021, 33, 12–19. [Google Scholar] [CrossRef]
- Agarwala, P.; Gaurb, P.K.; Tyagia, N.; Purib, D.; Kumarc, N.; Kumard, S.S. An Overview of Phytochemical, Therapeutic, Pharmacological and Traditional Importance of Syzygium cumini. Asian J. Pharmacogn. 2019, 3, 5–17. [Google Scholar]
- Syama, H.P.; Arya, A.D.; Dhanya, R.; Nisha, P.; Sundaresan, A.; Jacob, E.; Jayamurthy, P. Quantification of Phenolics in Syzygium cumini Seed and Their Modulatory Role on Tertiary Butyl-Hydrogen Peroxide-Induced Oxidative Stress in H9c2 Cell Lines and Key Enzymes in Cardioprotection. J. Food Sci. Technol. 2017, 54, 2115–2125. [Google Scholar] [CrossRef]
- Masaenah, E.; Elya, B.; Setiawan, H.; Fadhilah, Z.; Wediasari, F.; Nugroho, G.A.; Mozef, T. Antidiabetic Activity and Acute Toxicity of Combined Extract of Andrographis Paniculata, Syzygium cumini, and Caesalpinia Sappan. Heliyon 2021, 7, e08561. [Google Scholar] [CrossRef]
- Khalique, A.; Rauf, A. An Overview of Jamun (Eugenia jambolana Linn): A Traditional Multipotential Drug. Indian J. Unani Med. IX 2016, 9, 71–75. [Google Scholar]
- Chagas, V.T.; França, L.M.; Malik, S.; Paes, A.M.d.A. Syzygium cumini (L.) Skeels: A Prominent Source of Bioactive Molecules against Cardiometabolic Diseases. Front. Pharmacol. 2015, 6, 259. [Google Scholar] [CrossRef] [PubMed]
- Sarvade, S.; Gautam, D.S.; Bhalawe, S.; Bisen, P.K. An Overview of Potential Multipurpose Agroforestry Tree Species, Syzygium cuminii (L.) Skeels in India. J. Appl. Nat. Sci. 2016, 8, 1714–1719. [Google Scholar] [CrossRef]
- Madani, B.; Mirshekari, A.; Yahia, E.M.; Golding, J.B.; Hajivand, S.; Dastjerdy, A.M. Jamun (Syzygium cumini L. Skeels): A Promising Fruit for the Future. Hortic. Rev. (Am. Soc. Hortic. Sci). 2021, 48, 275–306. [Google Scholar] [CrossRef]
- Tewari, S.K.; Nainwal, R.C.; Singh, D. Potential of Syzygium cumini for Biocontrol and Phytoremediation. In The Genus Syzygium; CRC Press: Boca Raton, FL, USA, 2017; pp. 237–254. ISBN 1315118777. [Google Scholar] [CrossRef]
- Sharma, V.K.; Sharma, D.; Sharma, R.; Sharma, S.D.; Dhiman, K.; Murkute, A.A. Morpho-Molecular Exploration and Selection of Elite Genotypes from Indigenous Syzygium cumini L. Skeels (Jamun) Diversity of North-Western Indian Himalayas. Plant Genet. Resour. 2023, 21, 312–322. [Google Scholar] [CrossRef]
- Babu, K.D.; Patel, R.K.; Yadav, D.S. Comparative Evaluation of Guava Selections under North Eastern Region of India. Acta Hortic. 2007, 735, 99–103. [Google Scholar] [CrossRef]
- Gajera, H.P.; Gevariya, S.N.; Hirpara, D.G.; Patel, S.V.; Golakiya, B.A. Antidiabetic and Antioxidant Functionality Associated with Phenolic Constituents from Fruit Parts of Indigenous Black Jamun (Syzygium cumini L.) Landraces. J. Food Sci. Technol. 2017, 54, 3180–3191. [Google Scholar] [CrossRef]
- Soh, W.K. Taxonomy of Syzygium. In The Genus Syzygium: Syzygium cumini and Other Underutilized Species; CRC Press: Boca Raton, FL, USA, 2017; pp. 1–5. [Google Scholar] [CrossRef]
- Raza, A.; Ali, M.U.; Nisar, T.; Qasrani, S.A.; Hussain, R.; Sharif, M.N. Proximate Composition of Jamun (Syzygium cumini) Fruit and Seed. Digestion 2015, 15, 1221–1223. [Google Scholar]
- ud Din, S.; Jaskani, M.J.; Naqvi, S.A.; Awan, F.S. Diversity and Divergence in Domesticated and Wild Jamun (Syzygium cumini) Genotypes of Pakistan. Sci. Hortic. 2020, 273, 109617. [Google Scholar] [CrossRef]
- Mas-Gómez, J.; Cantín, C.M.; Moreno, M.Á.; Martínez-García, P.J. Genetic Diversity and Genome-Wide Association Study of Morphological and Quality Traits in Peach Using Two Spanish Peach Germplasm Collections. Front. Plant Sci. 2022, 13, 854770. [Google Scholar] [CrossRef]
- Gerrano, A.S.; Van Rensburg, W.S.J.; Mavengahama, S.; Bairu, M.; Venter, S.; Adebola, P.O. Qualitative Morphological Diversity of Amaranthus Species. J. Trop. Agric. 2017, 55, 12–20. [Google Scholar]
- Azad, R.; Ranawaka, R.; Senanayake, G.; Kumara, K.L.W.; Pushpakumara, D.; Wijesinghe, K.G.G.; Geekiyanage, S. Morphological Variation of Cinnamon (Cinnamomum verum Persl) Germplasm in Matara District of Sri Lanka. Int. J. Minor Fruits Med. Aromat. Plants 2016, 2, 6–14. [Google Scholar]
- Di Guardo, M.; Farneti, B.; Khomenko, I.; Modica, G.; Mosca, A.; Distefano, G.; Bianco, L.; Troggio, M.; Sottile, F.; La Malfa, S. Genetic Characterization of an Almond Germplasm Collection and Volatilome Profiling of Raw and Roasted Kernels. Hortic. Res. 2021, 8, 27–46. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, I.; Bhagat, S.; Simachalam, P.; Srivastava, R.C. Molecular Characterization of Syzygium cuminii (Wild Jamun) from A&N Islands. Indian J. Hortic. 2012, 69, 306–311. [Google Scholar]
- Khachtib, Y.; Zinelabidine, L.H.; Bouda, S.; Hamdali, H.; Hammada, S.; Haddioui, A. Genetic Characterization of Cultivated Apple (Malus x domestica Borkh.) in Morocco Using Microsatellite (SSR) Markers. Ecol. Genet. Genom. 2022, 23, 100122. [Google Scholar] [CrossRef]
- Hiremath, C.; Philip, R.; Sundaresan, V. Molecular Characterization of India Ginseng Withania somnifera (L) Using ISSR Markers. Mol. Biol. Rep. 2021, 48, 3971–3977. [Google Scholar] [CrossRef] [PubMed]
- Mir, M.A.; Mansoor, S.; Sugapriya, M.; Alyemeni, M.N.; Wijaya, L.; Ahmad, P. Deciphering Genetic Diversity Analysis of Saffron (Crocus sativus L.) Using RAPD and ISSR Markers. Saudi J. Biol. Sci. 2021, 28, 1308–1317. [Google Scholar] [CrossRef] [PubMed]
- Soliman, E.R.S.; El-Shazly, H.H.; Börner, A.; Badr, A. Genetic Diversity of a Global Collection of Maize Genetic Resources in Relation to Their Subspecies Assignments, Geographic Origin, and Drought Tolerance. Breed. Sci. 2021, 71, 313–325. [Google Scholar] [CrossRef]
- Venkatesan, J.; Ramu, V.; Sethuraman, T.; Sivagnanam, C.; Doss, G. Molecular Marker for Characterization of Traditional and Hybrid Derivatives of Eleusine coracana (L.) Using ISSR Marker. J. Genet. Eng. Biotechnol. 2021, 19, 1–12. [Google Scholar] [CrossRef]
- Tao, J.; Qiao, G.; Wen, X.-P.; Gao, G.-L.; Liu, T.; Peng, Z.-J.; Cai, Y.-Q.; Chen, N.; Yan, F.-X.; Zhang, B.-X. Characterization of Genetic Relationship of Dragon Fruit Accessions (Hylocereus spp.) by Morphological Traits and ISSR Markers. Sci. Hortic. 2014, 170, 82–88. [Google Scholar] [CrossRef]
- Singh, A.; Verma, S.K.; Prasad, G.; Kumar, A.; Sharma, P.C.; Singh, A.K. Elucidating Genetic Diversity and Population Structure in Jamun [Syzygium cumini L.(Skeels)] Using Morpho-Physiological Traits and CAAT Box-Derived Polymorphism. S. Afr. J. Bot. 2022, 151, 454–465. [Google Scholar] [CrossRef]
- Bashir, M.H.; AM, N.M.S.; Khan, A.Z.; Aziz, S.; Ullah, F.; Qasim, M. Characterization and Advancement of Microsatellite (SSR) Markers for Various Stresses in Wheat. Int. J. Agric. Biosci. 2022, 11, 8. [Google Scholar] [CrossRef]
- Luz, G.C.; Strioto, D.K.; Mangolin, C.A.; Maria, d.F. ISSR Markers to Assess Genetic Diversity of Cultivated Populations from Artificial Selection of Stevia rebaudiana (Bert.) Bertoni. Breed. Sci. 2020, 70, 508–514. [Google Scholar] [CrossRef] [PubMed]
- Demirel, S.; Pehluvan, M.; Aslantaş, R. Evaluation of Genetic Diversity and Population Structure of Peach (Prunus persica L.) Genotypes Using Inter-Simple Sequence Repeat (ISSR) Markers. Genet. Resour. Crop Evol. 2024, 71, 1301–1312. [Google Scholar] [CrossRef]
- Talang, H.; Nandha, A.; Rekha, A.; Ravishankar, K.V.; Talukdar, M. Identification of Nucellar and Zygotic Seedlings of Jamun (Syzygium cumini Skeels.) Using RAPD Marker. J. Pharmacogn. Phytochem. 2022, 11, 101–103. [Google Scholar]
- Chen, M.; He, X.; Zhang, Y.; Lu, T.; Yang, J.; Huang, X.; Zhu, J.; Yu, H.; Luo, C. Genetic Diversity and Relationship Analyses of Mango (Mangifera indica L.) Germplasm Resources with ISSR, SRAP, CBDP and CEAP Markers. Sci. Hortic. 2022, 301, 111146. [Google Scholar] [CrossRef]
- Jena, R.C.; Chand, P.K. Multiple DNA Marker-Assisted Diversity Analysis of Indian Mango (Mangifera indica L.) Populations. Sci. Rep. 2021, 11, 10345. [Google Scholar] [CrossRef] [PubMed]
- Mariano, L.C.; Zchonski, F.L.; da Silva, C.M.; Da-Silva, P.R. Genetic Variability in a Brazilian Apple Germplasm Collection with Low Chilling Requirements. PeerJ 2019, 6, e6265. [Google Scholar] [CrossRef] [PubMed]
- Giancarla, V.; Madoşă, E.; Adriana, C.; Ciulca, S.; Camen, D.; Mihaela, M.; Carmen, L. Genetic Diversity of Some Apple Genotypes from West Part of Romania Based on ISSR Markers. J. Hortic. For. Biotechnol. 2021, 25, 64–68. [Google Scholar]
- Khan, S.; Vaishali; Sharma, V. Genetic Differentiation and Diversity Analysis of Medicinal Tree Syzygium cumini (Myrtaceae) from Ecologically Different Regions of India. Physiol. Mol. Biol. Plants 2010, 16, 149–158. [Google Scholar] [CrossRef]
- Sharif, N.; Jaskani, M.J.; Abbas Naqvi, S.; Awan, F.S. Exploitation of Diversity in Domesticated and Wild Ber (Ziziphus mauritiana Lam.) Germplasm for Conservation and Breeding in Pakistan. Sci. Hortic. 2019, 249, 228–239. [Google Scholar] [CrossRef]
- Mahajan, R.K.; Gangopadhyay, K.K.; Gunjeet Kumar, D.V.K.; Srivastava, U.; Gupta, P.N. Minimal Descriptors of Agrihorticultural Crops; Part III fruit Crops; National Bureau of Plant Genetic Resources: New Delhi, India, 2002. [Google Scholar]
- Rasul, K.S.; Grundler, F.M.W.; Tahir, A. Genetic Diversity and Population Structure Assessment of Iraqi Tomato Accessions Using Fruit Characteristics and Molecular Markers. Hortic. Environ. Biotechnol. 2022, 63, 523–538. [Google Scholar] [CrossRef]
- Shakya, R.; Siddiqui, S.A.; Srivatawa, N.; Bajpai, A. Molecular Characterization of Jamun (Syzygium cumini L. Skeels) Genetic Resources. Int. J. Fruit Sci. 2010, 10, 29–39. [Google Scholar] [CrossRef]
- Singh, J.P.; Singh, A.K.; Bajpai, A.; Ahmad, I.Z. Comparative Analysis of DNA Polymorphisms and Phylogenetic Relationships among Syzygium cumini Skeels Based on Phenotypic Characters and RAPD Technique. Bioinformation 2014, 10, 201. [Google Scholar] [CrossRef]
- Greenacre, M. Correspondence Analysis in Practice; Chapman and Hall/CRC: Boca Raton, FL, USA, 2017; ISBN 1315369982. [Google Scholar] [CrossRef]
- Abdi, H.; Béra, M. Correspondence Analysis. In Encyclopedia of Social Network Analysis and Mining; Springer: New York, NY, USA, 2014. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of Population Structure Using Multilocus Genotype Data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef]
- Earl, D.A.; VonHoldt, B.M. STRUCTURE HARVESTER: A Website and Program for Visualizing STRUCTURE Output and Implementing the Evanno Method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the Number of Clusters of Individuals Using the Software STRUCTURE: A Simulation Study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.A.; Iqbal, M.S.; Ahmad, K.S.; Akbar, M.; Mehmood, A.; Hussain, S.A.; Arshad, N.; Munir, S.; Masood, H.; Ahmad, T. Plant Species Diversity Assessment and Monitoring in Catchment Areas of River Chenab, Punjab, Pakistan. PLoS ONE 2022, 17, e0272654. [Google Scholar] [CrossRef]
- Shinwari, M.I.; Shinwari, M.I. Botanical Diversity in Pakistan; Past Present and Future. World Environ. 2010, 1, 85–104. [Google Scholar]
- Muhammad, S.; Sheikh, S.A.; Nizamani, S.M.; Bhanger, M.I.; Imran, A.; Ejaz, A. A Study on the Determination of Mineral Elements in Jamun Fruit (Eugenia jambolana) Products. Pak. J. Nutr. 2012, 11, 181–186. [Google Scholar] [CrossRef]
- Devi, C.A.; Sharma, G.L.; Singh, P.; Panigrahi, H.K.; Sharma, H.G.; Nag, J.L.; Paikra, P.S. Studies on Variability in Fruit Characters of Jamun (Syzygium cumini Skeels) Genotypes. Pharma Innov. J. 2019, 8, 391–393. [Google Scholar]
- Dissanayake, P.K.; Wekumbura, W.G.C.; Wijeratne, A.W.; Wijesundara, D.S.A. Morphological Characterization, Antioxidant Capacity and Diversity of Syzygium cumini Trees from Sri Lanka. Hortic. Plant J. 2022, 8, 53–67. [Google Scholar] [CrossRef]
- Guo, Q.; Liu, J.; Li, J.; Cao, S.; Zhang, Z.; Zhang, J.; Zhang, Y.; Deng, Y.; Niu, D.; Su, L. Genetic Diversity and Core Collection Extraction of Robinia pseudoacacia L. Germplasm Resources Based on Phenotype, Physiology, and Genotyping Markers. Ind. Crops Prod. 2022, 178, 114627. [Google Scholar] [CrossRef]
- Khadivi, A.; Mirheidari, F.; Moradi, Y.; Paryan, S. Morphological Variability of Wild Pomegranate (Punica granatum L.) Accessions from Natural Habitats in the Northern Parts of Iran. Sci. Hortic. 2020, 264, 109165. [Google Scholar] [CrossRef]
- Nikhil, H.N.; Goswami, A.K.; Singh, S.K.; Kumar, C.; Goswami, S.; Singh, R.; Bharadwaj, C.; Maurya, N.K. Assessment of Morpho-Genetic Diversity of Guava (Psidium guajava) Hybrids and Genotypes. Indian J. Agric. Sci. 2021, 91, 1640–1645. [Google Scholar] [CrossRef]
- Hien, P.T.H.I.T.H.U.; LE Thi, T.C.; Hang, V.U.T.H.I.T. Genetic Diversity of Guava Varieties (Psidium guajava L.) Based on Morphology and ISSR Molecular Markers. Biodivers. J. Biol. Divers. 2024, 25, 1037–1045. [Google Scholar] [CrossRef]
- Mbovora, S.M.; Musvosvi, C.; Gasura, E. Morphological Diversity among Accessions of Apple Tree (Malus× domestica Borkh). Adv. Agric. 2021, 2021, 7705856. [Google Scholar] [CrossRef]
- Dar, J.A.; Wani, A.A.; Dhar, M.K. Assessment of Apple (Malus× domestica Bark.) Germplasm of Kashmir Using RAPD Markers. Int. J. Fruit Sci. 2020, 20, 635–645. [Google Scholar] [CrossRef]
- Carvalho-Junior, G.d.N.C.; Costa, M.F.; Nunes, J.A.R.; de Almeida Lopes, Â.C.; Ferreira-Gomes, R.L. Estimation of the Extent of Phenotypic Diversity among Cashew (Anacardium occidentale L.) Genotypes Based on Agro-Morphological and Physicochemical Traits. Plant Genet. Resour. 2022, 20, 230–238. [Google Scholar]
- N’Djolossè, K.; Adoukonou-Sagbadja, H.; Gbèmavo, C.D.S.J.; Kodjo, S.; Badou, A.; Maliki, R.; Adjovi, N.R.A. Agro-Morphological Characterization of Preselected Cashew (Anacardium occidentale L.) Mother Trees in Benin Farmer’s Plantations. J. Agric. Environ. Int. Dev. 2019, 113, 17–34. [Google Scholar]
- Hasanbegovic, J.; Hadizabulic, S.; Kurtović, M.; Gaši, F.; Ercisli, S.; Dorbić, B.; Durul, M.S. Genetic and Morphological Diversity of Introduced Cultivars of Almonds (Prunus amygdalus L.) in Bosnia and Herzegovina. Cell. Mol. Biol. 2024, 70, 106–114. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Sánchez, R.; Morales-Corts, M.R. Agromorphological Characterization and Nutritional Value of Traditional Almond Cultivars Grown in the Central-Western Iberian Peninsula. Agronomy 2021, 11, 1238. [Google Scholar] [CrossRef]
- Hu, Y.; Sun, D.; Hu, H.; Zuo, X.; Xia, T.; Xie, J. A Comparative Study on Morphological and Fruit Quality Traits of Diploid and Polyploid Carambola (Averrhoa carambola L.) Genotypes. Sci. Hortic. 2021, 277, 109843. [Google Scholar] [CrossRef]
- Palei, S.; Dasmohapatra, R.; Samal, S.; Rout, G.R. Cashew Nut (Anacardium occidentale L.) Breeding Strategies. In Advances in Plant Breeding Strategies: Nut and Beverage Crops: Volume 4; Springer: Cham, Switzerland, 2019; pp. 77–104. [Google Scholar] [CrossRef]
- Khadivi, A.; Mirheidari, F.; Saeidifar, A.; Moradi, Y. Selection of the Promising Accessions of Jamun (Syzygium cumini (L.) Skeels) Based on Pomological Characterizations. Food Sci. Nutr. 2023, 11, 470–480. [Google Scholar] [CrossRef] [PubMed]
- Meena, R.K.; Bhatnagar, P.; Singh, J.; Chopra, R.; Chhipa, H.R. Correlation Dynamics among Growth, Development and Soil Parameters in Response to Vermicompost and PSB on Jamun Cv. Goma Priyanka in Vertisols of Jhalawar District. J. Pharmacogn. Phytochem. 2020, 9, 1950–1952. [Google Scholar]
- Samarina, L.S.; Malyarovskaya, V.I.; Reim, S.; Koninskaya, N.G.; Matskiv, A.O.; Tsaturyan, G.A.; Rakhmangulov, R.S.; Shkhalakhova, R.M.; Shurkina, E.S.; Kulyan, R.V. Genetic Diversity in Diospyros Germplasm in the Western Caucasus Based on SSR and ISSR Polymorphism. Biology 2021, 10, 341. [Google Scholar] [CrossRef]
- Zhang, Y.; Cao, Y.-F.; Huo, H.-L.; Xu, J.-Y.; Tian, L.-M.; Dong, X.-G.; Dan, Q.I.; Chao, L.I.U. An Assessment of the Genetic Diversity of Pear (Pyrus L.) Germplasm Resources Based on the Fruit Phenotypic Traits. J. Integr. Agric. 2022, 21, 2275–2290. [Google Scholar] [CrossRef]
- Dev, R.; Singh, S.K.; Singh, R.; Singh, A.K.; Patel, V.B.; Alizadeh, M.; Motha, K.; Kumar, K. Assessment of Genetic Diversity of Grape Mutants Based on RAPD and SSR Markers. Indian J. Hortic. 2021, 78, 17–24. [Google Scholar] [CrossRef]
- Hussein, M.A.A.; Eid, M.; Rahimi, M.; Filimban, F.Z.; Abd El-Moneim, D. Comparative Assessment of SSR and RAPD Markers for Genetic Diversity in Some Mango Cultivars. PeerJ 2023, 11, e15722. [Google Scholar] [CrossRef]
- EL-Tarawy, O.M. Genetic Identification of Some Guava Seedling Trees. Ann. Agric. Sci. Moshtohor 2020, 58, 1039–1050. [Google Scholar] [CrossRef]
- Liliana, B.; Anca-Amalia, U.; Cornelia, B.A.; Mihaela, I. Genetic Diversity Analysis among Several Apple Genotypes Using ISSR Markers. Fruit Grow. Res. 2022, 38, 40–44. [Google Scholar] [CrossRef]
- Singh, Y.S.; Shira, V.D.; Swamy, G.S.K. Genetic Variation for Morphological and Physicochemical Traits in Jamun (Syzygium cuminii Skeels). Asian J. Hortic. 2016, 11, 163–167. [Google Scholar] [CrossRef]
- Chesnokov, Y.V.; Artemyeva, A.M. Evaluation of the Measure of Polymorphism Information of Genetic Diversity. Сельскoхoзяйственная биoлoгия 2015, 50, 571–578. [Google Scholar] [CrossRef]
- Ho, V.T.; Ngo, T.K.A.; Phan, T.H.T.; Ta, T.T.T.; Tran, T.K.P. Genetic Diversity among Passion Fruit (Passiflora edulis) Accessions of Southern Vietnam Using RAPD and ISSR Markers. SABRAO J. Breed. Genet. 2021, 53, 1–14. [Google Scholar]
- Adhiena, M.; Muez, M. Multivariate Analysis for Yield and Yield Related Traits in Durum Wheat Genotypes in Southern Tigray, North, Ethiopia. Int. J. Agric. Biosci. 2019, 8, 36–39. [Google Scholar]
- Gajera, H.P.; Tomar, R.S.; Patel, S.V.; Viradia, R.R.; Golakiya, B.A. Comparison of RAPD and ISSR Markers for Genetic Diversity Analysis among Different Endangered Mangifera indica Genotypes of Indian Gir Forest Region. J. Plant Biochem. Biotechnol. 2011, 20, 217–223. [Google Scholar] [CrossRef]
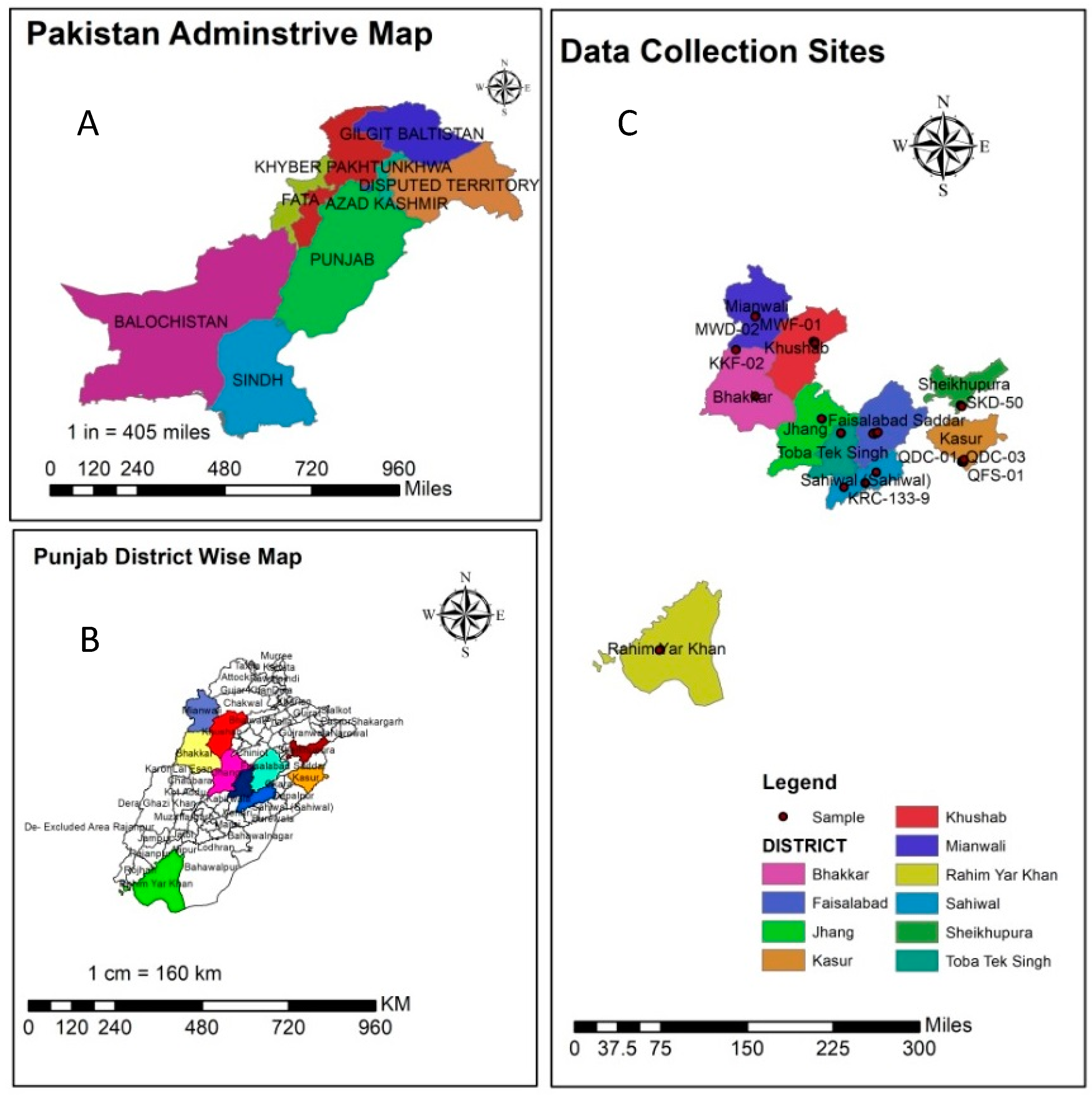

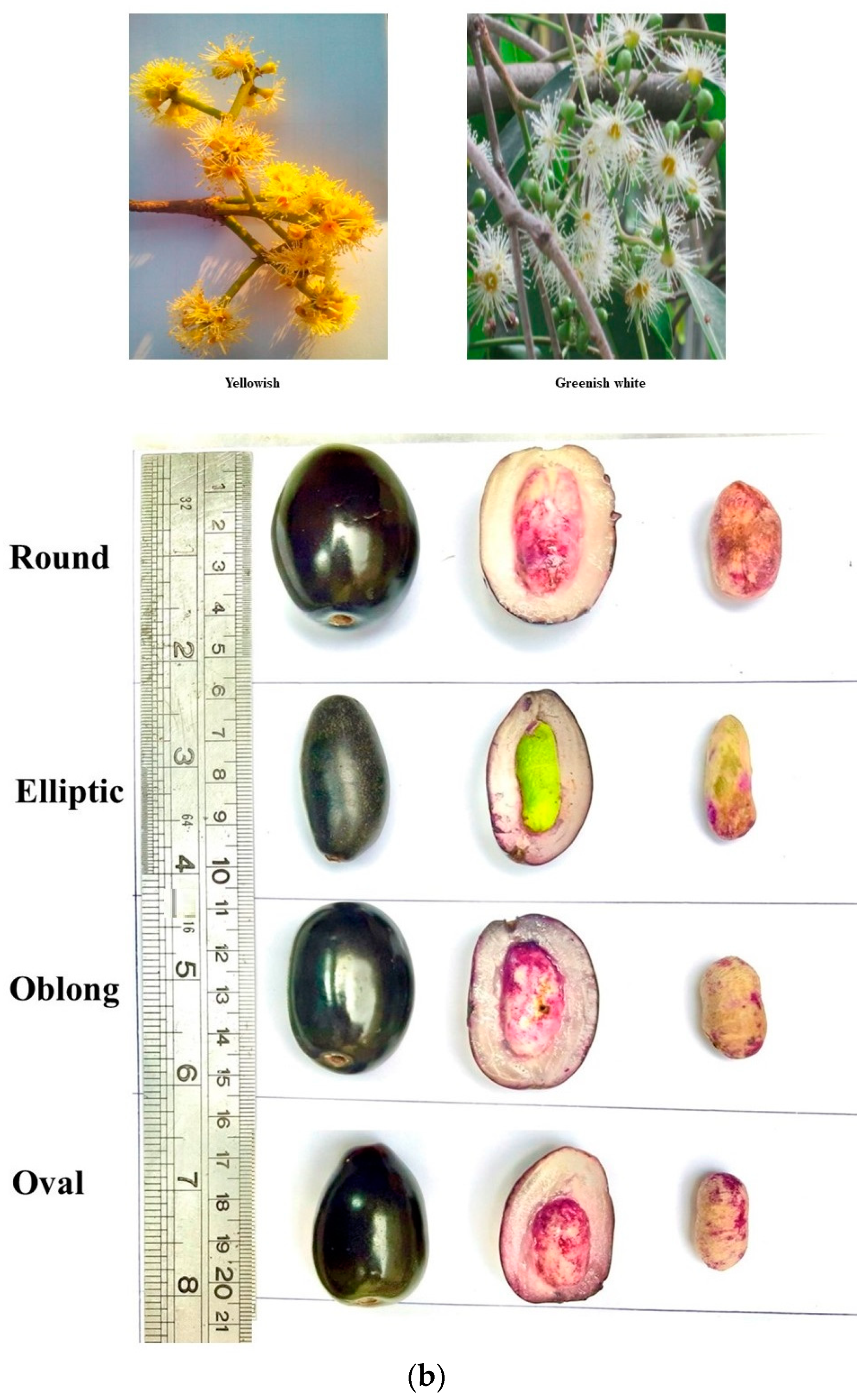
| Sr. No. | Collection Site | Accession ID | Sr. No. | Collection Site | Accession ID |
|---|---|---|---|---|---|
| 1 | Khushab | KDC-01 | 31 | Faisalabad | SFH-04 |
| 2 | KDC-02 | 32 | SPJ-05 | ||
| 3 | KDC-03 | 33 | SFD-06 | ||
| 4 | KDC-04 | 34 | SFG-07 | ||
| 5 | KDC-05 | 35 | SFM-08 | ||
| 6 | Kasur | QFS-01 | 36 | SFD-09 | |
| 7 | QDS-01 | 37 | SFD-10 | ||
| 8 | QDC-01 | 38 | Toba Tek Singh | TTR-01 | |
| 9 | QDC-03 | 39 | TUK-01 | ||
| 10 | Q3DC-02 | 40 | TUK-02 | ||
| 11 | Q3DC-01 | 41 | TUK-03 | ||
| 12 | QFS-02 | 42 | TTD-01 | ||
| 13 | Sahiwal | CSP-25 | 43 | TTD-02 | |
| 14 | CSP-26 | 44 | TTD-03 | ||
| 15 | CSP-27 | 45 | Bhakkar | KKD-01 | |
| 16 | KRC-133-9 | 46 | KKD-02 | ||
| 17 | KRC-2-1339 | 47 | KKF-01 | ||
| 18 | KRC-3-1339 | 48 | KKF-02 | ||
| 19 | KRC-4-1339 | 49 | BKR-03 | ||
| 20 | SHW-01 | 50 | Jhang | JWF-01 | |
| 21 | Sheikhupura | SKS-01 | 51 | JWF-02 | |
| 22 | SKD-01 | 52 | JWD-03 | ||
| 23 | SKD-02 | 53 | Mianwali | MWD-01 | |
| 24 | SKH-03 | 54 | MWD-02 | ||
| 25 | SKUK-02 | 55 | MWF-01 | ||
| 26 | SKD-50 | 56 | MWF-02 | ||
| 27 | SKD-03 | 57 | MSD-01 | ||
| 28 | Faisalabad | SFD-01 | 58 | MSD-02 | |
| 29 | SFS-02 | 59 | Rahim Yar Khan | CUK-01 | |
| 30 | SFS-03 | 60 | MKR-01 |
| Sr. No. | Characters | Categories |
|---|---|---|
| 1 | Tree shape (TS) | Upright (UR), spreading (SP), drooping (Dr) |
| 2 | Bark color (BC) | Light grey (LG), grey (G), brown (B) |
| 3 | Trunk wood color (SwC) | Reddish grey (RG), brownish grey (BG), Dull brown (DB) |
| 4 | Leaf surface (lsurf) | Coriaceous (C), mild coriaceous (MC) |
| 5 | Leaf shape (LSP) | Broadly ovate (BO), elliptic oblong (EO), elliptic (EP), lanceolate (Ln) |
| 6 | Flower color (Fw.C) | Light yellow (LY), greenish white (GW) |
| 7 | Fruit shape (FSp) | Round (Rd), oblong (Ob), oval (Ov), elliptic (EL) |
| 8 | Fruit color (FC) | Pinkish (Pu), deep purple (DP), Bluish black (BB) |
| 9 | Pulp color (PC) | Whitish (W), pinkish (P), purple pink (PP) |
| 10 | Fruit waxiness (FWx) | Low (Lo), medium (Md), high (H) |
| 11 | Fruit base (FB) | Depressed (D), flat (F), projected (Pr) |
| RAPD Primers | Primer Sequence (5′-3′) | ISSR Primers | Primer Sequence (5′-3′) |
|---|---|---|---|
| OPB 08 | GTCCACACGG | UBC 817 | (CA)8A |
| OPA 13 | CAGCACCCAC | UBC 834 | (AG)8YT |
| OPD 18 | GAGAGCCAAC | UBC 835 | (AG)8YC |
| OPG 4 | AGCGTGTGTG | UBC 840 | (GA)8YT |
| OPG 13 | CTCTCCGCCA | UBC 844 | (CT)8RC |
| OPX 4 | CCGCTACCGA | UBC 864 | (ATGA)3 (TG)3 |
| OPF 2 | GAGGATCCCT | UBC 807 | (AG)8T |
| 85SP10A16 | AGCCAGCGAA | ||
| OPQ 4 | AGTGCGCTGA | ||
| 70SP10C1 | CAGGCCCTTC |
| Areas | Tree Shape | Bark Color | Trunk Wood Color | Leaf Surface | Leaf Shape | Flower Color | Fruit Shape | Fruit Color | Pulp Color | Fruit Waxiness | Fruit Base | ||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Upright (UR) | Spreading (SP) | Drooping (Dr) | Light Grey (LG) | Grey (G) | Brown (B) | Reddish Grey (RG) | Brownish Grey (BG) | Dull Brown (DB) | Coriaceous (C) | Mild Coriaceous (MC) | Broadly Ovate (BO) | Elliptic Oblong (EO) | Elliptic (EP) | Lanceolate (Ln) | Light Yellow (LY) | Greenish White (GW) | Round (Rd) | Oblong (Ob) | Oval (Ov) | Elliptic (EL) | Pinkish (Pu) | Deep Purple (DP) | Bluish Black (BB) | Whitish (W) | Pinkish (Pu) | Purple Pink (PP) | Low (Lo) | Medium (Md) | High (H) | Depressed (D) | Flat (F) | Projected (Pr) | |
| KHUSHAB | 60 | 0 | 40 | 0 | 40 | 60 | 40 | 60 | 0 | 20 | 80 | 0 | 20 | 80 | 0 | 100 | 0 | 20 | 60 | 20 | 0 | 0 | 20 | 80 | 0 | 40 | 60 | 20 | 20 | 60 | 40 | 20 | 40 |
| KASUR | 29 | 57 | 14 | 14 | 29 | 57 | 43 | 57 | 0 | 57 | 43 | 29 | 14 | 43 | 14 | 14 | 86 | 0 | 71 | 29 | 0 | 14 | 29 | 57 | 14 | 43 | 43 | 29 | 57 | 14 | 43 | 14 | 43 |
| SAHIWAL | 75 | 13 | 13 | 75 | 25 | 0 | 13 | 50 | 38 | 13 | 88 | 0 | 50 | 50 | 0 | 100 | 0 | 25 | 50 | 25 | 0 | 0 | 63 | 38 | 38 | 25 | 38 | 25 | 63 | 13 | 13 | 13 | 75 |
| SHEIKHUPURA | 43 | 29 | 29 | 57 | 0 | 43 | 14 | 43 | 43 | 71 | 29 | 14 | 0 | 86 | 0 | 71 | 29 | 29 | 14 | 29 | 29 | 0 | 43 | 57 | 14 | 43 | 43 | 0 | 100 | 0 | 0 | 43 | 57 |
| FAISALABAD | 60 | 20 | 20 | 40 | 20 | 40 | 40 | 40 | 20 | 40 | 60 | 30 | 30 | 40 | 0 | 80 | 20 | 50 | 30 | 20 | 0 | 10 | 20 | 70 | 30 | 40 | 30 | 30 | 60 | 10 | 0 | 40 | 60 |
| TOBA TEK SINGH | 0 | 57 | 43 | 86 | 14 | 0 | 14 | 86 | 0 | 71 | 29 | 0 | 29 | 71 | 0 | 100 | 0 | 0 | 71 | 14 | 14 | 0 | 29 | 71 | 29 | 29 | 43 | 0 | 100 | 0 | 14 | 71 | 14 |
| BHAKKAR | 40 | 60 | 0 | 0 | 60 | 40 | 20 | 20 | 60 | 40 | 60 | 20 | 20 | 60 | 0 | 100 | 0 | 20 | 20 | 60 | 0 | 0 | 60 | 40 | 40 | 20 | 40 | 40 | 20 | 40 | 40 | 20 | 40 |
| JHANG | 33 | 0 | 67 | 67 | 0 | 33 | 0 | 33 | 67 | 33 | 67 | 0 | 0 | 100 | 0 | 100 | 0 | 0 | 33 | 67 | 0 | 0 | 33 | 67 | 33 | 33 | 33 | 33 | 33 | 33 | 33 | 33 | 33 |
| MIANWALI | 50 | 17 | 33 | 0 | 67 | 33 | 50 | 0 | 50 | 50 | 50 | 0 | 33 | 67 | 0 | 83 | 17 | 17 | 33 | 50 | 0 | 17 | 33 | 50 | 33 | 33 | 33 | 33 | 67 | 0 | 33 | 17 | 50 |
| RAHIM YAR KHAN | 50 | 50 | 0 | 0 | 50 | 50 | 50 | 50 | 0 | 100 | 0 | 0 | 50 | 50 | 0 | 100 | 0 | 100 | 0 | 0 | 0 | 0 | 0 | 100 | 50 | 0 | 50 | 50 | 50 | 0 | 0 | 50 | 50 |
| TOTAL | 45 | 30 | 25 | 38 | 28 | 33 | 28 | 45 | 27 | 47 | 53 | 12 | 25 | 62 | 2 | 82 | 18 | 23 | 42 | 30 | 5 | 5 | 35 | 60 | 27 | 33 | 40 | 23 | 62 | 15 | 20 | 32 | 48 |
| CV% | 36.552 | 41.108 | 41.045 | 31.140 | 28.206 | 32.101 | 41.315 | 34.445 | 41.508 | 25.761 | 36.243 | ||||||||||||||||||||||
| Accession | Tree Shape | Bark Color | Stem Wood Color | Fruit Color | Fruit Waxiness | Fruit Shape | Fruit Base | Pulp Color | Leaf Surface | Leaf Shape | Flower Color |
|---|---|---|---|---|---|---|---|---|---|---|---|
| KDC-01 | Drooping | Brown | Reddish grey | Bluish Black | Medium | Oblong | Flat | Pinkish | Mild coriaceous | Elliptic | Light yellow |
| KDC-02 | Upright | Grey | Reddish grey | Bluish Black | High | Oblong | Depressed | Purple pink | Mild coriaceous | Elliptic | Light yellow |
| KDC-03 | Upright | Brown | Brownish grey | Bluish Black | High | Round | Flat | Purple pink | Mild coriaceous | Elliptic | Light yellow |
| KDC-04 | Upright | Brown | Brownish grey | Bluish Black | High | Oval | Depressed | Purple pink | Mild coriaceous | Elliptic | Light yellow |
| KDC-05 | Drooping | Grey | Brownish grey | Deep purple | Low | Oval | Projected | Pinkish | Coriaceous | Elliptic oblong | Light yellow |
| QFS-01 | Spreading | Grey | Reddish grey | Pinkish | Medium | Oval | Projected | Whitish | Coriaceous | Elliptic oblong | Greenish white |
| QDS-01 | Spreading | Brown | Brownish grey | Deep purple | Medium | Oblong | Depressed | Pinkish | Coriaceous | Elliptic | Greenish white |
| QDC-01 | Spreading | Brown | Brownish grey | Bluish Black | Medium | Oblong | Depressed | Purple pink | Mild coriaceous | Lanceolate | Greenish white |
| QDC-03 | Drooping | Grey | Reddish grey | Deep purple | Medium | Oval | Flat | Pinkish | Coriaceous | Elliptic | Greenish white |
| Q3DC-02 | Spreading | Brown | Reddish grey | Bluish Black | Low | Oblong | Flat | Pinkish | Coriaceous | Broadly ovate | Greenish white |
| Q3DC-01 | Upright | Light grey | Brownish grey | Bluish Black | Low | Oblong | Projected | Purple pink | Mild coriaceous | Broadly ovate | Greenish white |
| QFS-02 | Upright | Brown | Brownish grey | Bluish Black | High | Oval | Depressed | Purple pink | Mild coriaceous | Elliptic | Light yellow |
| CSP-25 | Upright | Grey | Dull Brown | Bluish Black | Medium | Round | Depressed | Purple pink | Mild coriaceous | Elliptic oblong | Light yellow |
| CSP-26 | Spreading | Light grey | Dull Brown | Deep purple | Medium | Oblong | Projected | Whitish | Mild coriaceous | Elliptic | Light yellow |
| CSP-27 | Upright | Light grey | Brownish grey | Deep purple | Low | Oblong | Projected | Whitish | Mild coriaceous | Elliptic | Light yellow |
| KRC-133-9 | Upright | Light grey | Brownish grey | Bluish Black | Medium | Oval | Projected | Whitish | Mild coriaceous | Elliptic oblong | Light yellow |
| KRC-2-1339 | Upright | Grey | Reddish grey | Deep purple | High | Round | Flat | Purple pink | Coriaceous | Elliptic | Light yellow |
| KRC-3-1339 | Upright | Light grey | Brownish grey | Bluish Black | Medium | Round | Depressed | Pinkish | Mild coriaceous | Elliptic oblong | Light yellow |
| KRC-4-1339 | Upright | Light grey | Brownish grey | Deep purple | Medium | Oval | Depressed | Purple pink | Mild coriaceous | Elliptic oblong | Light yellow |
| SHW-01 | Drooping | Light grey | Dull Brown | Deep purple | Low | Oval | Projected | Pinkish | Mild coriaceous | Elliptic | Light yellow |
| SKS-01 | Drooping | Light grey | Brownish grey | Deep purple | Medium | Oval | Projected | Whitish | Coriaceous | Broadly ovate | Greenish white |
| SKD-01 | Upright | Light grey | Dull Brown | Bluish Black | Medium | Oval | Projected | Purple pink | Mild coriaceous | Elliptic | Light yellow |
| SKD-02 | Upright | Light grey | Dull Brown | Bluish Black | Medium | Round | Flat | Pinkish | Coriaceous | Elliptic | Light yellow |
| SKH-03 | Spreading | Brown | Dull Brown | Deep purple | Medium | Oblong | Projected | Pinkish | Coriaceous | Elliptic | Light yellow |
| SKUK-02 | Spreading | Brown | Brownish grey | Deep purple | Medium | Elliptic | Flat | Pinkish | Coriaceous | Elliptic | Light yellow |
| SKD-50 | Upright | Light grey | Brownish grey | Bluish Black | Medium | Elliptic | Projected | Purple pink | Coriaceous | Elliptic | Greenish white |
| SKD-03 | Drooping | Brown | Reddish grey | Bluish Black | Medium | Round | Projected | Pinkish | Mild coriaceous | Elliptic | Light yellow |
| SFD-01 | Upright | Brown | Brownish grey | Bluish Black | Medium | Round | Projected | Purple pink | Mild coriaceous | Elliptic | Light yellow |
| SFS-02 | Drooping | Light grey | Dull Brown | Deep purple | Low | Oval | Projected | Pinkish | Mild coriaceous | Elliptic oblong | Light yellow |
| SFS-03 | Drooping | Grey | Brownish grey | Deep purple | Low | Oval | Projected | Pinkish | Coriaceous | Elliptic oblong | Light yellow |
| SFH-04 | Upright | Light grey | Dull Brown | Bluish Black | High | Oblong | Projected | Whitish | Coriaceous | Elliptic | Light yellow |
| SPJ-05 | Spreading | Grey | Reddish grey | Bluish Black | Medium | Oblong | Flat | Pinkish | Mild coriaceous | Broadly ovate | Greenish white |
| SFD-06 | Upright | Light grey | Reddish grey | Bluish Black | Medium | Round | Flat | Purple pink | Mild coriaceous | Broadly ovate | Light yellow |
| SFG-07 | Upright | Brown | Brownish grey | Bluish Black | Medium | Round | Flat | Pinkish | Coriaceous | Elliptic | Light yellow |
| SFM-08 | Spreading | Brown | Reddish grey | Pinkish | Low | Round | Flat | Whitish | Coriaceous | Elliptic oblong | Light yellow |
| SFD-09 | Upright | Light grey | Reddish grey | Bluish Black | Medium | Oval | Flat | Whitish | Mild coriaceous | Broadly ovate | Greenish white |
| SFD-10 | Upright | Brown | Brownish grey | Bluish Black | Medium | Round | Projected | Purple pink | Mild coriaceous | Elliptic | Light yellow |
| TTR-01 | Spreading | Light grey | Brownish grey | Bluish Black | Medium | Oblong | Projected | Pinkish | Coriaceous | Elliptic | Light yellow |
| TUK-01 | Spreading | Grey | Brownish grey | Bluish Black | Medium | Oblong | Flat | Whitish | Coriaceous | Elliptic oblong | Light yellow |
| TUK-02 | Spreading | Light grey | Brownish grey | Deep purple | Medium | Oblong | Depressed | Whitish | Coriaceous | Elliptic | Light yellow |
| TUK-03 | Spreading | Light grey | Brownish grey | Deep purple | Medium | Oval | Flat | Pinkish | Mild coriaceous | Elliptic | Light yellow |
| TTD-01 | Drooping | Light grey | Brownish grey | Bluish Black | Medium | Oblong | Flat | Purple pink | Coriaceous | Elliptic | Light yellow |
| TTD-02 | Drooping | Light grey | Reddish grey | Bluish Black | Medium | Elliptic | Flat | Purple pink | Mild coriaceous | Elliptic oblong | Light yellow |
| TTD-03 | Drooping | Light grey | Brownish grey | Bluish Black | Medium | Oblong | Flat | Purple pink | Coriaceous | Elliptic | Light yellow |
| KKD-01 | Upright | Brown | Dull Brown | Bluish Black | High | Round | Depressed | Purple pink | Mild coriaceous | Elliptic oblong | Light yellow |
| KKD-02 | Drooping | Brown | Dull Brown | Bluish Black | High | Oblong | Depressed | Purple pink | Mild coriaceous | Elliptic | Light yellow |
| KKF-01 | Drooping | Grey | Dull Brown | Deep purple | Medium | Oval | Projected | Whitish | Coriaceous | Broadly ovate | Light yellow |
| KKF-02 | Drooping | Grey | Brownish grey | Deep purple | Low | Oval | Projected | Whitish | Mild coriaceous | Elliptic | Light yellow |
| BKR-03 | Drooping | Grey | Reddish grey | Deep purple | Low | Oval | Flat | Pinkish | Coriaceous | Elliptic | Light yellow |
| JWF-01 | Drooping | Light grey | Dull Brown | Deep purple | Low | Oval | Projected | Pinkish | Mild coriaceous | Elliptic | Light yellow |
| JWF-02 | Drooping | Light grey | Brownish grey | Bluish Black | Medium | Oval | Flat | Whitish | Coriaceous | Elliptic | Light yellow |
| JWD-03 | Upright | Brown | Dull Brown | Bluish Black | High | Oblong | Depressed | Purple pink | Mild coriaceous | Elliptic | Light yellow |
| MWD-01 | Upright | Brown | Dull Brown | Bluish Black | Medium | Oblong | Depressed | Purple pink | Mild coriaceous | Elliptic | Light yellow |
| MWD-02 | Upright | Brown | Dull Brown | Bluish Black | Medium | Round | Projected | Pinkish | Coriaceous | Elliptic | Light yellow |
| MWF-01 | Drooping | Grey | Reddish grey | Deep purple | Low | Oval | Flat | Pinkish | Coriaceous | Elliptic | Light yellow |
| MWF-02 | Drooping | Grey | Reddish grey | Deep purple | Low | Oblong | Depressed | Whitish | Mild coriaceous | Elliptic | Light yellow |
| MSD-01 | Upright | Grey | Dull Brown | Bluish Black | Medium | Round | Projected | Purple pink | Mild coriaceous | Elliptic oblong | Light yellow |
| MSD-02 | Spreading | Grey | Reddish grey | Pinkish | Medium | Oblong | Flat | Whitish | Coriaceous | Elliptic oblong | Greenish white |
| CUK-01 | Upright | Brown | Reddish grey | Bluish Black | Low | Round | Projected | Whitish | Coriaceous | Elliptic oblong | Light yellow |
| MKR-01 | Spreading | Grey | Brownish grey | Bluish Black | Medium | Round | Flat | Purple pink | Coriaceous | Elliptic | Light yellow |
| Primer | AFS | TNB | NPB | MBF | PIC | RP | %Pol | |
|---|---|---|---|---|---|---|---|---|
| For RAPD markers | ||||||||
| OPA 13 | 50–1050 | 8 | 8 | 0.683 | 0.424 | 3.433 | 100 | |
| OPD 18 | 480–1460 | 7 | 6 | 0.700 | 0.406 | 2.861 | 85.714 | |
| OPG 13 | 600–1500 | 6 | 4 | 0.667 | 0.353 | 1.556 | 66.667 | |
| OPG 4 | 260–1500 | 7 | 5 | 0.524 | 0.352 | 1.931 | 71.428 | |
| OPX 4 | 100–1340 | 8 | 7 | 0.602 | 0.419 | 2.330 | 87.500 | |
| OPF 2 | 670–1600 | 5 | 2 | 0.697 | 0.347 | 0.784 | 40.000 | |
| 85SP10A16 | 100–1300 | 10 | 7 | 0.635 | 0.405 | 2.106 | 70.000 | |
| OPQ 4 | 340–2000 | 8 | 5 | 0.704 | 0.336 | 2.021 | 62.500 | |
| 70SP10C1 | 150–950 | 4 | 4 | 0.587 | 0.465 | 3.781 | 100 | |
| OPB 8 | 850–2300 | 6 | 3 | 0.603 | 0.386 | 1.760 | 50.000 | |
| Mean | 6.90 | 5.10 | 0.640 | 0.389 | 2.256 | 73.381 | ||
| Total | 69 | 51 | - | - | - | - | ||
| For ISSR markers | ||||||||
| UBC 807 | 300–1200 | 8 | 8 | 0.612 | 0.460 | 3.800 | 100 | |
| UBC 817 | 380–1200 | 10 | 10 | 0.521 | 0.459 | 4.431 | 100 | |
| UBC 834 | 400–2000 | 10 | 9 | 0.577 | 0.479 | 3.758 | 90.000 | |
| UBC 835 | 750–2100 | 16 | 13 | 0.475 | 0.455 | 4.376 | 81.250 | |
| UBC 840 | 100–1200 | 9 | 7 | 0.608 | 0.437 | 3.932 | 77.778 | |
| UBC 844 | 50–860 | 4 | 4 | 0.512 | 0.500 | 2.221 | 100 | |
| UBC 864 | 250–900 | 6 | 6 | 0.594 | 0.406 | 2.630 | 100 | |
| Mean | 9 | 8.143 | 0.557 | 0.457 | 3.591 | 92.718 | ||
| Total | 63 | 57 | - | - | - | - | ||
| Source | df | SS | MS | Est. Var | % |
|---|---|---|---|---|---|
| Among Population | 9 | 256.608 | 28.512 | 2.334 | 14% |
| Within Population | 50 | 736.158 | 14.723 | 14.723 | 86% |
| Total | 59 | 992.767 | 17.057 | 100% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Uddin, S.; Jaskani, M.J.; Deng, Z.; Maqbool, R.; Naqvi, S.A.; Parajuli, S.; Sharif, N.; Saleem, A.R.; Ledon, S.; Ikram, S.; et al. Phenotypic and Molecular-Markers-Based Assessment of Jamun (Syzygium cumini) Genotypes from Pakistan. Horticulturae 2024, 10, 879. https://doi.org/10.3390/horticulturae10080879
Uddin S, Jaskani MJ, Deng Z, Maqbool R, Naqvi SA, Parajuli S, Sharif N, Saleem AR, Ledon S, Ikram S, et al. Phenotypic and Molecular-Markers-Based Assessment of Jamun (Syzygium cumini) Genotypes from Pakistan. Horticulturae. 2024; 10(8):879. https://doi.org/10.3390/horticulturae10080879
Chicago/Turabian StyleUddin, Safeer, Muhammad Jafar Jaskani, Zhanao Deng, Rizwana Maqbool, Summar Abbas Naqvi, Saroj Parajuli, Naseem Sharif, Abdul Rahman Saleem, Steven Ledon, Sufian Ikram, and et al. 2024. "Phenotypic and Molecular-Markers-Based Assessment of Jamun (Syzygium cumini) Genotypes from Pakistan" Horticulturae 10, no. 8: 879. https://doi.org/10.3390/horticulturae10080879
APA StyleUddin, S., Jaskani, M. J., Deng, Z., Maqbool, R., Naqvi, S. A., Parajuli, S., Sharif, N., Saleem, A. R., Ledon, S., Ikram, S., Khan, I. A., & Shafqat, W. (2024). Phenotypic and Molecular-Markers-Based Assessment of Jamun (Syzygium cumini) Genotypes from Pakistan. Horticulturae, 10(8), 879. https://doi.org/10.3390/horticulturae10080879







