Genome-Wide Analysis of the ABCB Family and Its Expression in Adventitious Root Development of Paeonia ostii
Abstract
:1. Introduction
2. Materials and Methods
2.1. PoABCB Gene Identification and Physicochemical Properties of the Putative Proteins
2.2. Phylogenetic Analysis
2.3. Analysis of Conserved Motif and Gene Structure
2.4. Cis-Acting Element Analysis
2.5. Chromosome Localization and Collinearity Analysis
2.6. Analysis of Transcript Profiles of the PoABCB Gene Family During Adventitious Root Induction
3. Results
3.1. Identification of PoABCBs and Physicochemical Properties of the Putative Proteins
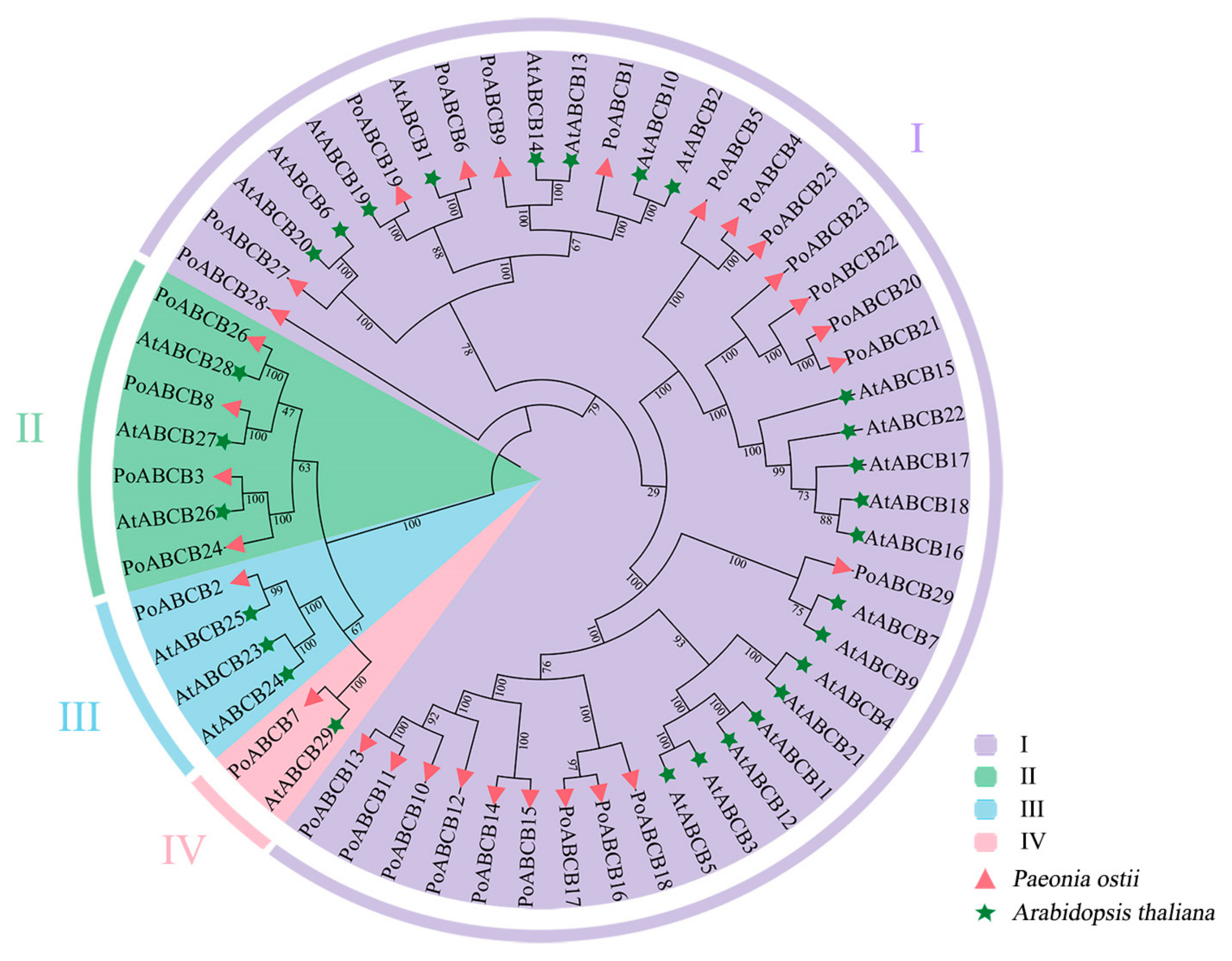
3.2. PoABCB Phylogenetic Analysis
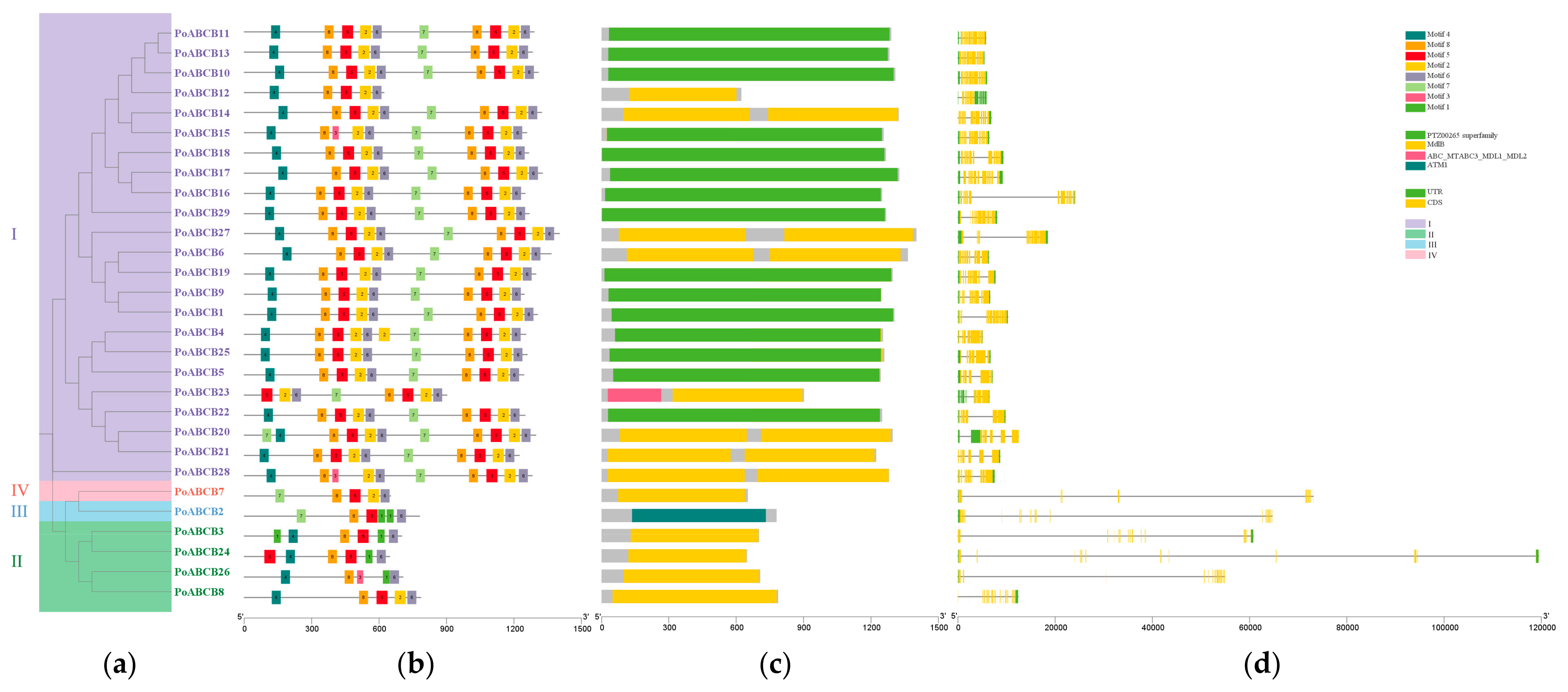
3.3. Analysis of PoABCB Gene Structure and Conserved Motifs
3.4. PoABCB Cis-Acting Element Analysis
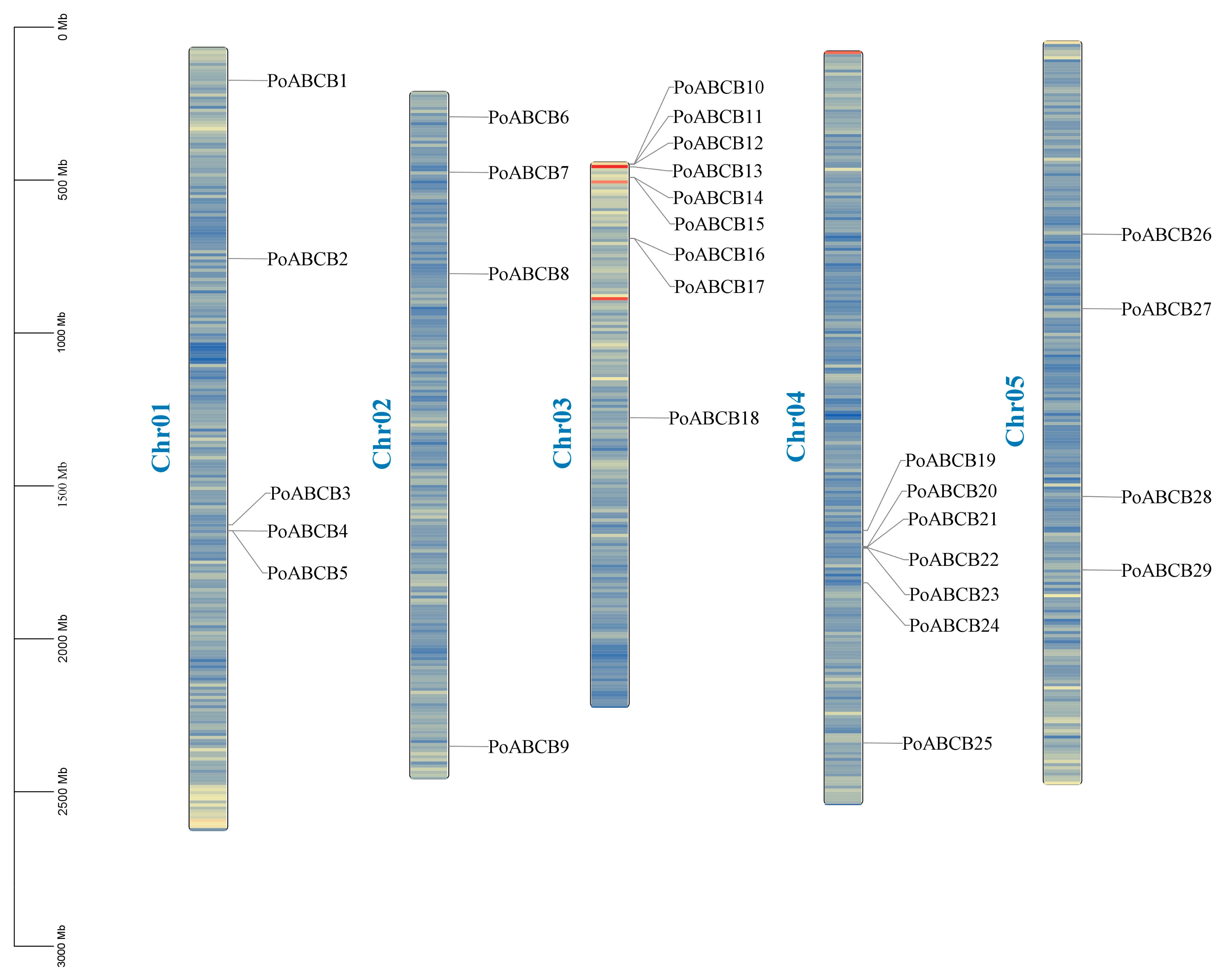
3.5. Locations of PoABCBs on Chromosomes
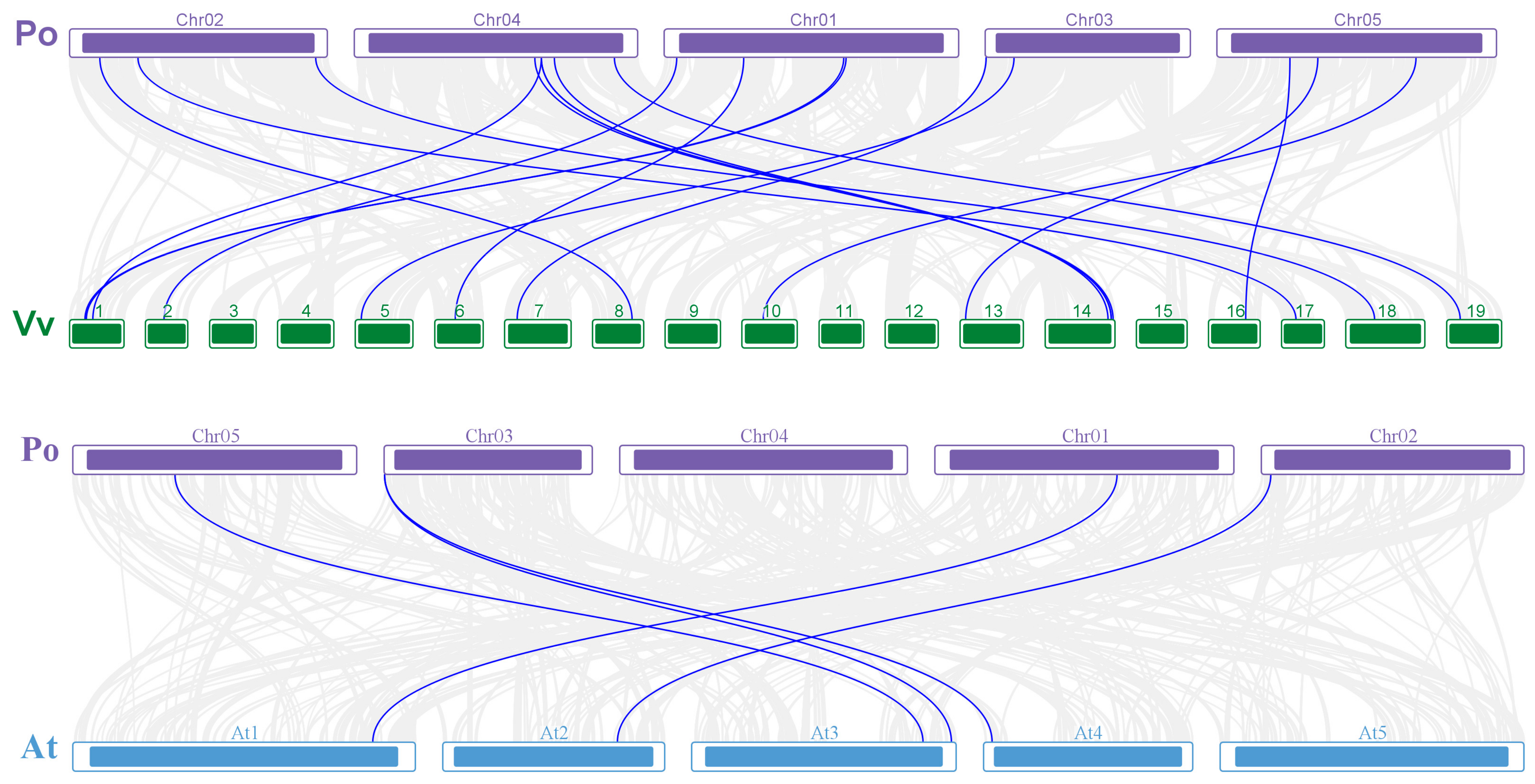
3.6. PoABCB Collinearity Analysis
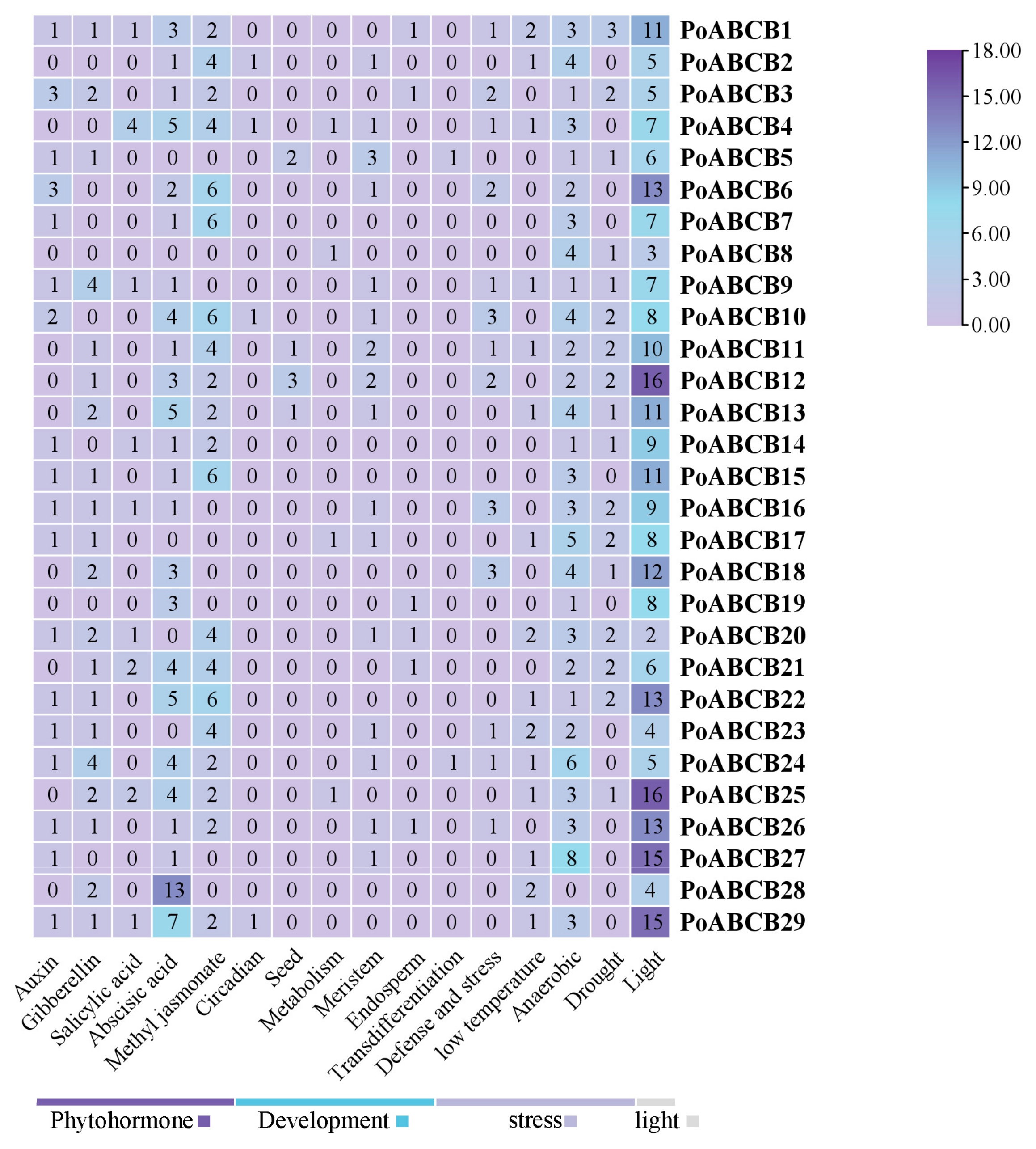
3.7. Analysis of PoABCB Transcript Profiles at the Stem Base During Root Induction
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Appendix A
| qRT-RCP Primer Name | Primer Sequence (5′-3′) |
|---|---|
| Tubulin-F | TGAGCACCAAAGAAGTGGACGAAC |
| Tubulin-R | CACACGCCTGAACATCTCCTGAA |
| PoABCB1-F | AAAACAAAGAAGAGAAGAAA |
| PoABCB1-R | ATAAAGAAGACTGGAACTGA |
| PoABCB2-F | TCAAGTAAGGATGTAACCCC |
| PoABCB2-R | TATCTTCCAACCATAAGTAT |
| PoABCB3-F | ATTTGGAGATTAAGTGGTTG |
| PoABCB3-R | TGATAAGATGAAGTTGTGAG |
| PoABCB4-F | GAGGTGGGGATTACTTCAAA |
| PoABCB4-R | GGAGTCTGTGGAGTCTGGGC |
| PoABCB5-F | GGAGATTCAGAAAAAAAACC |
| PoABCB5-R | GCAACCTAAAGATCCCAGTG |
| PoABCB6-F | AACATCACCTCCTTCAACAA |
| PoABCB6-R | CCATCAAAACATAATCTAAC |
| PoABCB7-F | GCGTTATCTCTGTCTACTCG |
| PoABCB7-R | CTCCCAACACTAAACCCTCA |
| PoABCB8-F | TGGAGGGAGAAAGAGAAACG |
| PoABCB8-R | AGAATGCCAGAAGTGGATGC |
| PoABCB9-F | GGACCAACCATCAAACTCAA |
| PoABCB9-R | ACCAATCCAAGATAAACCAA |
| PoABCB10-F | AAGGGGGCAGAGGAGAAAAC |
| PoABCB10-R | AGCACCTACTGAACCGACAA |
| PoABCB11-F | TGCTCTGGCTGTATGGTATG |
| PoABCB11-R | TGGTGTCCTGTTGATGGTCT |
| PoABCB12-F | CTCATATAAATACGCAGGCA |
| PoABCB12-R | AATCCATTCCCAATAGCACC |
| PoABCB13-F | GGATGGTCACAGGAGAGAGA |
| PoABCB13-R | CCACAAGAAGAGGAATAGAG |
| PoABCB14-F | CGAGGAAAGATGATAGAAAG |
| PoABCB14-R | GACAGATAATGAATCGGAGC |
| PoABCB15-F | CTATTCATTCTGGAAAGACT |
| PoABCB15-R | GACCCATCTGCTGCCTCAAC |
| PoABCB16-F | GGGTGAGAAGGTTGGAAAGT |
| PoABCB16-R | AGGGAAGATAGCATGACGAG |
| PoABCB17-F | GTTCTGTTACCTCTTATGGG |
| PoABCB17-R | GCTACTTGACTTGCTTCCTC |
| PoABCB18-F | TCTTGATACCGAATCTGAAC |
| PoABCB18-R | GACTCCATTTTTAACCACTG |
| PoABCB19-F | GTGGACAGTATTGCAGTAGT |
| PoABCB19-R | TGTGTGATGGTGTTGTAGCT |
| PoABCB20-F | GTTCGCCTCCAACAAACAAA |
| PoABCB20-R | ATCCCTAACACCTTCACCGC |
| PoABCB21-F | GAAGTGTTGTTATTGGACGA |
| PoABCB21-R | GGCTAAGAGAGACGAGTGGT |
| PoABCB22-F | GGAGCAGAAAACTACCACGA |
| PoABCB22-R | ATACAGCACCACAGGCATAG |
| PoABCB23-F | TAGCGTAGTTGTGGCACATA |
| PoABCB23-R | GCAGGTGTCCTTTGGAGTTT |
| PoABCB24-F | ATTATCCTCAATACCGCCCT |
| PoABCB24-R | GTCTACCCTGAATCCCTCCA |
| PoABCB25-F | ACAGTGCCATTCAATCGGTT |
| PoABCB25-R | GTTCTTCCTGCTTCTATCTT |
| PoABCB26-F | TTTCAATCCCAACCACTGTC |
| PoABCB26-R | TCTTCGGAGGAGCAAACCAG |
| PoABCB27-F | GAAGGAAACAGAGGATACAG |
| PoABCB27-R | GATAACATAAGCAAGGAGAG |
| PoABCB28-F | GTTCCCCTCATTCTTCTCAT |
| PoABCB28-R | CCCGTTCCAACTCCCTTTAT |
| PoABCB29-F | GTAAGTGGGGAAGATAGTTA |
| PoABCB29-R | AGCAGAAGTCCAAATAGAGG |
References
- Wen, S.S.; Chen, L.; Tian, R.N. Micropropagation of tree peony (Paeonia sect. Moutan): A review. Plant Cell Tissue Organ Cult. 2020, 141, 1–14. [Google Scholar] [CrossRef]
- Almeida, M.R.D.; Bastiani, D.D.; Gaeta, M.L.; Mariath, J.E.D.A.; Costa, F.D.; Retallick, J.; Nolan, L.; Tai, H.H.; Strömvik, M.V.; Fett-Neto, A.G. Comparative transcriptional analysis provides new insights into the molecular basis of adventitious rooting recalcitrance in Eucalyptus. Plant Sci. 2015, 239, 155–165. [Google Scholar] [CrossRef] [PubMed]
- Costa, C.T.D.; Gaeta, M.L.; Mariath, J.E.D.A.; Offringa, R.; Fett-Neto, A.G. Comparative adventitious root development in pre-etiolated and flooded Arabidopsis hypocotyls exposed to different auxins. Plant Physiol. Biochem. 2018, 127, 161–168. [Google Scholar] [CrossRef]
- Liu, R.; Xue, Y.Q.; Ci, H.T.; Gao, J.; Wang, S.L.; Zhang, X.X. Establishment of highly efficient plant regeneration of Paeonia ostii ‘Fengdan’ through optimization of callus, adventitious shoot, and rooting induction. Hortic. Plant J. 2022, 8, 777–786. [Google Scholar] [CrossRef]
- Carraro, N.; Tisdale-Orr, T.E.; Clouse, R.M.; Knöller, A.S.; Spicer, R. Diversification and expression of the PIN, AUX/LAX, and ABCB families of putative auxin transporters in Populus. Front. Plant Sci. 2012, 3, 17. [Google Scholar] [CrossRef]
- Mellor, N.L.; Voß, U.; Ware, A.; Janes, G.; Barrack, D.; Bishopp, A.; Bennett, M.J.; Geisler, M.; Wells, D.M.; Band, L.R. Systems approaches reveal that ABCB and PIN proteins mediate co-dependent auxin efflux. Plant Cell 2022, 34, 2309–2327. [Google Scholar] [CrossRef]
- Kaneda, M.; Schuetz, M.; Lin, B.S.P.; Chanis, C.; Hamberger, B.; Western, T.L.; Ehlting, J.; Samuels, A.L. ABC transporters coordinately expressed during lignification of Arabidopsis stems include a set of ABCBs associated with auxin transport. J. Exp. Bot. 2011, 62, 2063–2077. [Google Scholar] [CrossRef]
- Wu, G.S.; Cameron, J.N.; Ljung, K.; Spalding, E.P. A role for ABCB19-mediated polar auxin transport in seedling photomorphogenesis mediated by cryptochrome 1 and phytochrome B. Plant J. 2010, 62, 179–191. [Google Scholar] [CrossRef]
- Hilleary, R. The sum is greater than the parts: Co-dependent auxin efflux is mediated by ABCBs and PINs. Plant Cell 2022, 34, 2114–2115. [Google Scholar] [CrossRef]
- Chen, J.; Hu, Y.J.; Hao, P.C.; Tsering, T.; Xia, J.; Zhang, Y.Q.; Roth, O.; Njo, M.F.; Sterck, L.; Hu, Y.; et al. ABCB-mediated shootward auxin transport feeds into the root clock. EMBO Rep. 2023, 24, e56271. [Google Scholar] [CrossRef]
- Jasinski, M.; Ducos, E.; Martinoia, E.; Boutry, M. The ATP-binding cassette transporters: Structure, function, and gene family comparison between rice and Arabidopsis. Plant Physiol. 2003, 131, 1169–1177. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, V.N.T.; Moon, S.; Jung, K.-H. Genome-wide expression analysis of rice ABC transporter family across spatio-temporal samples and in response to abiotic stresses. J. Plant Physiol. 2014, 171, 1276–1288. [Google Scholar] [CrossRef] [PubMed]
- Theodoulou, F.L. Plant ABC transporters. Biochim. Biophys. Acta. 2000, 1465, 79–103. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Fernández, R.; Davies, T.G.; Coleman, J.O.; Rea, P.A. The Arabidopsis thaliana ABC protein superfamily, a complete inventory. J. Biol. Chem. 2001, 276, 30231–30244. [Google Scholar] [CrossRef]
- Kamimoto, Y.; Terasaka, K.; Hamamoto, M.; Takanashi, K.; Fukuda, S.; Shitan, N.; Sugiyama, A.; Suzuki, H.; Shibata, D.; Wang, B.; et al. Arabidopsis ABCB21 is a facultative auxin importer/exporter regulated by cytoplasmic auxin concentration. Plant Cell Physiol. 2012, 53, 2090–2100. [Google Scholar] [CrossRef] [PubMed]
- Hwang, J.-U.; Song, W.-Y.; Hong, D.; Ko, D.; Yamaoka, Y.; Jang, S.; Yim, S.; Lee, E.; Khare, D.; Kim, K.; et al. Plant ABC Transporters Enable Many Unique Aspects of a Terrestrial Plant’s Lifestyle. Mol. Plant 2016, 9, 338–355. [Google Scholar] [CrossRef]
- Dahuja, A.; Kumar, R.R.; Sakhare, A.; Watts, A.; Singh, B.; Goswami, S.; Sachdev, A.; Praveen, S. Role of ATP-binding cassette transporters in maintaining plant homeostasis under abiotic and biotic stresses. Physiol. Plant. 2021, 171, 785–801. [Google Scholar] [CrossRef]
- Geisler, M.M. Embracing substrate multispecificity in plant ABC transporters. Mol. Plant 2024, 17, 990–992. [Google Scholar] [CrossRef]
- Jenness, M.K.; Carraro, N.; Pritchard, C.A.; Murphy, A.S. Corrigendum: The Arabidopsis ATP-BINDING CASSETTE Transporter ABCB21 Regulates Auxin Levels in Cotyledons, the Root Pericycle, and Leaves. Front. Plant Sci. 2020, 11, 351. [Google Scholar] [CrossRef]
- Tamizhselvan, P.; Madhavan, S.; Constan-Aguilar, C.; Elrefaay, E.R.; Liu, J.; Pěnčík, A.; Novák, O.; Cairó, A.; Hrtyan, M.; Geisler, M.; et al. Chloroplast Auxin Efflux Mediated by ABCB28 and ABCB29 Fine-Tunes Salt and Drought Stress Responses in Arabidopsis. Plants 2023, 13, 7. [Google Scholar] [CrossRef]
- Sukumar, P.; Maloney, G.S.; Muday, G.K. Localized induction of the ATP-binding cassette B19 auxin transporter enhances adventitious root formation in Arabidopsis. Plant Physiol. 2013, 162, 1392–1405. [Google Scholar] [CrossRef] [PubMed]
- Yuan, J.H.; Jiang, S.J.; Jian, J.B.; Liu, M.Y.; Yue, Z.; Xu, J.B.; Li, J.; Xu, C.Y.; Lin, L.H.; Jing, Y.; et al. Genomic basis of the giga-chromosomes and giga-genome of tree peony Paeonia ostii. Nat. Commun. 2022, 13, 7328. [Google Scholar] [CrossRef] [PubMed]
- Su, A.; Ge, S.Y.; Zhou, B.Y.; Wang, Z.Y.; Zhou, L.P.; Zhang, Z.W.; Yan, X.Y.; Wang, Y.; Li, D.L.; Zhang, H.; et al. Analysis of the Tomato mTERF Gene Family and Study of the Stress Resistance Function of SLmTERF-13. Plants 2023, 12, 2862. [Google Scholar] [CrossRef]
- Zhang, D.D.; Yu, Z.F.; Zeng, B.; Liu, X.Y. Genome-wide analysis of the ABC gene family in almond and functional predictions during flower development, freezing stress, and salt stress. BMC Plant Biol. 2024, 24, 12. [Google Scholar] [CrossRef]
- Zhao, M.M.; Liu, Z.W.; Gan, J.T.; Yang, C.; Lu, A.; Han, Q.Q.; Yang, H.T.; Xu, Y.H.; Sun, G.L.; Wu, D.C. Identification and expression analysis of XIP gene family members in rice. Genetica 2024, 152, 83–100. [Google Scholar] [CrossRef]
- Zhang, S.; Hu, X.W.; Dong, J.J.; Du, M.X.; Song, J.Q.; Xu, S.Y.; Zhao, C.J. Identification, evolution, and expression analysis of OsBSK gene family in Oryza sativa Japonica. BMC Plant Biol. 2022, 22, 565. [Google Scholar] [CrossRef]
- Han, S.; Jiao, Z.Y.; Niu, M.X.; Yu, X.; Huang, M.B.; Liu, C.; Wang, H.L.; Zhou, Y.Y.; Mao, W.; Wang, X.F.; et al. Genome-Wide Comprehensive Analysis of the GASA Gene Family in Populus. Int. J. Mol. Sci. 2021, 22, 12336. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Que, F.; Zhu, Y.Q.; Liu, Q.N.; Wei, Q.; Ramakrishnan, M. Genome-Wide Identification, Expansion, Evolution, and Expression Analysis Reveals ABCB Genes Important for Secondary Cell Wall Development in Moso Bamboo (Phyllostachys edulis). Agronomy 2023, 13, 1828. [Google Scholar] [CrossRef]
- Ma, J.J.; Han, M.Y. Genomewide analysis of ABCBs with a focus on ABCB1 and ABCB19 in Malus domestica. J. Genet. 2016, 95, 141–149. [Google Scholar] [CrossRef]
- Tang, R.N.; Zhu, Y.J.; Yang, S.M.; Wang, F.; Chen, G.Z.; Chen, J.L.; Zhao, K.; Liu, Z.J.; Peng, D.H. Genome-Wide Identification and Analysis of WRKY Gene Family in Melastoma dodecandrum. Int. J. Mol. Sci. 2023, 24, 14904. [Google Scholar] [CrossRef] [PubMed]
- Xue, G.X.; Hu, L.F.; Zhu, L.M.; Chen, Y.; Qiu, C.; Fan, R.F.; Ma, X.X.; Cao, Z.J.; Chen, J.H.; Shi, J.S.; et al. Genome-Wide Identification and Expression Analysis of CCO Gene Family in Liriodendron chinense. Plants 2023, 12, 1975. [Google Scholar] [CrossRef]
- Chen, X.L.; Mo, F.L.; Shen, C.Y.; Meng, L.J.; Zou, Y.X.; Xue, X.P.; Cheng, M.Z.; Meng, F.Y.; Qi, H.N.; Wang, A.X. Genome-wide identification and expression analysis of the SlNAC gene family in tomato based on a high-quality genome. Hortic. Environ. Biotechnol. 2022, 63, 887–901. [Google Scholar] [CrossRef]
- Akram, J.; Siddique, R.; Shafiq, M.; Tabassum, B.; Manzoor, M.T.; Javed, M.A.; Anwar, S.; Nisa, B.U.; Saleem, M.H.; Javed, B.; et al. Genome-wide identification of CCO gene family in cucumber (Cucumis sativus) and its comparative analysis with A. thaliana. BMC Plant Biol. 2023, 23, 640. [Google Scholar] [CrossRef]
- Elsanosi, H.A.; Zhang, J.H.; Mostafa, S.; Geng, X.Y.; Zhou, G.S.; Awdelseid, A.H.M.; Song, L. Genome-wide identification, structural and gene expression analysis of BTB gene family in soybean. BMC Plant Biol. 2024, 24, 663. [Google Scholar] [CrossRef]
- Shang, W.Q.; Wang, Z.; He, S.L.; He, D.; Dong, N.L.; Guo, Y. Changes of endogenous IAA and related activities during rooting of Paeonia suffruticosa in vitro. J. Northwest Univ. (Nat. Sci. Ed.) 2021, 49, 129–136. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Itoh, J.; Nonomura, K.; Ikeda, K.; Yamaki, S.; Inukai, Y.; Yamagishi, H.; Kitano, H.; Nagato, Y. Rice plant development: From zygote to spikelet. Plant Cell Physiol. 2005, 46, 23–47. [Google Scholar] [CrossRef]
- Guan, L.; Murphy, A.S.; Peer, W.A.; Gan, L.J.; Li, Y.; Cheng, Z.M. Physiological and Molecular Regulation of Adventitious Root Formation. Crit. Rev. Plant Sci. 2015, 34, 506–521. [Google Scholar] [CrossRef]
- Frick, E.M.; Strader, L.C. Roles for IBA-derived auxin in plant development. J. Exp. Bot. 2018, 69, 169–177. [Google Scholar] [CrossRef]
- Song, X.B.; Huang, R.M.; Liu, H.; Zhang, J.; Chang, Y.Y.; Pei, D. Transcriptome profiling of indole-3-butyric acid–induced adventitious root formation in softwood cuttings of walnut. Hortic. Plant J. 2024, 10, 1336–1348. [Google Scholar] [CrossRef]
- Yang, C.H.; Wang, D.D.; Zhang, C.; Ye, M.H.; Kong, N.N.; Ma, H.L.; Chen, Q. Comprehensive Analysis and Expression Profiling of PIN, AUX/LAX, and ABCB Auxin Transporter Gene Families in Solanum tuberosum under Phytohormone Stimuli and Abiotic Stresses. Biology 2021, 10, 127. [Google Scholar] [CrossRef] [PubMed]
- Jenness, M.K.; Tayengwa, R.; Murphy, A.S. An ATP-binding cassette transporter, ABCB19, regulates leaf position and morphology during phototropin1-mediated blue light responses. Plant Physiol. 2020, 184, 1601–1612. [Google Scholar] [CrossRef]
- Nguyen, V.N.T.; Usman, B.; Kim, E.J.; Shim, S.H.; Jeon, J.S.; Jung, K.H. An ATP-binding cassette transporter, OsABCB24, is involved in female gametophyte development and early seed growth in rice. Physiol. Plant. 2024, 176, e14354. [Google Scholar] [CrossRef]
- Liu, W.X.; Feng, X.; Cao, F.B.; Wu, D.Z.; Zhang, G.P.; Vincze, E.; Wang, Y.Z.; Chen, Z.-H.; Wu, F.B. An ATP binding cassette transporter HvABCB25 confers aluminum detoxification in wild barley. J. Hazard. Mater. 2021, 401, 123371. [Google Scholar] [CrossRef]
- Khan, N.; You, F.M.; Datla, R.; Ravichandran, S.; Jia, B.; Cloutier, S. Genome-wide identification of ATP binding cassette (ABC) transporter and heavy metal associated (HMA) gene families in flax (Linum usitatissimum L.). BMC Genom. 2020, 21, 722. [Google Scholar] [CrossRef] [PubMed]
- Pang, K.Y.; Li, Y.J.; Liu, M.H.; Meng, Z.D.; Yu, Y.L. Inventory and general analysis of the ATP-binding cassette (ABC) gene superfamily in maize (Zea mays L.). Gene 2013, 526, 411–428. [Google Scholar] [CrossRef]
- Chen, J.; Hou, S.J.; Zhang, Q.Q.; Meng, J.Q.; Zhang, Y.Y.; Du, J.H.; Wang, C.; Liang, D.; Guo, Y.Q. Genome-Wide Identification and Analysis of the WRKY Gene Family in Asparagus officinalis. Genes 2023, 14, 1704. [Google Scholar] [CrossRef]
- Airoldi, C.A.; Davies, B. Gene Duplication and the Evolution of Plant MADS-box Transcription Factors. J. Genet. Genom. 2012, 39, 157–165. [Google Scholar] [CrossRef]
- Moons, A. Transcriptional profiling of the PDR gene family in rice roots in response to plant growth regulators, redox perturbations and weak organic acid stresses. Planta 2008, 229, 53–71. [Google Scholar] [CrossRef]
- Aslam, M.; Fakher, B.; Jakada, B.H.; Zhao, L.h.; Cao, S.J.; Cheng, Y.; Qin, Y. Genome-Wide Identification and Expression Profiling of CBL-CIPK Gene Family in Pineapple (Ananas comosus) and the Role of AcCBL1 in Abiotic and Biotic Stress Response. Biomolecules 2019, 9, 293. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.Y.; Bovet, L.; Kushnir, S.; Noh, E.W.; Martinoia, E.; Lee, Y. AtATM3 is involved in heavy metal resistance in Arabidopsis. Plant Physiol. 2006, 140, 922–932. [Google Scholar] [CrossRef] [PubMed]
- Shen, C.; Li, X. Genome-wide identification and expression pattern profiling of the ATP-binding cassette gene family in tea plant (Camellia sinensis). Plant Physiol. Biochem. 2023, 202, 107930. [Google Scholar] [CrossRef]
- Tran, L.S.P.; Nakashima, K.; Sakuma, Y.; Osakabe, Y.; Qin, F.; Simpson, S.D.; Maruyama, K.; Fujita, Y.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Co-expression of the stress-inducible zinc finger homeodomain ZFHD1 and NAC transcription factors enhances expression of the ERD1 gene in Arabidopsis: ZFHD and NAC in drought stress response. Plant J. 2006, 49, 46–63. [Google Scholar] [CrossRef]
- Noh, B.; Murphy, A.S.; Spalding, E.P. Multidrug resistance–like genes of Arabidopsis required for auxin transport and auxin-mediated development. Plant Cell 2001, 13, 2441–2454. [Google Scholar] [CrossRef]
- Cho, M.; Henry, E.M.; Lewis, D.R.; Wu, G.; Muday, G.K.; Spalding, E.P. Block of ATP-binding cassette B19 ion channel activity by 5-nitro-2-(3-phenylpropylamino)-benzoic acid impairs polar auxin transport and root gravitropism. Plant Physiol. 2014, 166, 2091–2099. [Google Scholar] [CrossRef]
- Geisler, M.; Kolukisaoglu, H.U.; Bouchard, R.; Billion, K.; Berger, J.; Saal, B.; Frangne, N.; Koncz-Kalman, Z.; Koncz, C.; Dudler, R.; et al. TWISTED DWARF1, a unique plasma membrane-anchored immunophilin-like protein, interacts with Arabidopsis multidrug resistance-like transporters AtPGP1 and AtPGP19. Mol. Biol. Cell 2003, 14, 4238–4249. [Google Scholar] [CrossRef]
- Ganesan, M.; Kanimozhi, G.; Pradhapsingh, B.; Khan, H.A.; Alhomida, A.S.; Ekhzaimy, A.; Brindha, G.R.; Prasad, N.R. Phytochemicals reverse P-glycoprotein mediated multidrug resistance via signal transduction pathways. Biomed. Pharmacother. 2021, 139, 111632. [Google Scholar] [CrossRef]
- Geisler, M.; Dreyer, I. An auxin homeostat allows plant cells to establish and control defined transmembrane auxin gradients. New Phytol. 2024, 244, 1422–1436. [Google Scholar] [CrossRef]



| Gene Name | Gene ID | Chromosome Location | Number of Amino Acids | Molecular Weight | pI | Prediction of Subcellular Location |
|---|---|---|---|---|---|---|
| PoABCB1 | Pos.gene78377.mRNA-1 | Chr01 | 1304 | 142,714.07 | 8.86 | plas: 13, vacu: 1 |
| PoABCB2 | Pos.gene41311.mRNA-1 | Chr01 | 779 | 86,457.51 | 9.55 | chlo: 6, plas: 5, mito: 1, E.R.: 1, pero: 1 |
| PoABCB3 | Pos.gene47973.mRNA-1 | Chr01 | 701 | 78,309.74 | 8.9 | plas: 10, chlo: 2, vacu: 1, E.R.: 1 |
| PoABCB4 | Pos.gene39088.mRNA-1 | Chr01 | 1253 | 136,897.23 | 9.16 | plas: 10, vacu: 2, cyto: 1, mito: 1 |
| PoABCB5 | Pos.gene45213.mRNA-1 | Chr01 | 1243 | 136,380.35 | 8.25 | plas: 13, vacu: 1 |
| PoABCB6 | Pos.gene46182.mRNA-1 | Chr02 | 1364 | 148,998.75 | 6.94 | plas: 13, vacu: 1 |
| PoABCB7 | Pos.gene9826.mRNA-1 | Chr02 | 651 | 72,203.18 | 8.81 | chlo: 6, plas: 6, mito: 1, pero: 1 |
| PoABCB8 | Pos.gene30683.mRNA-1 | Chr02 | 786 | 85,732.3 | 9.3 | plas: 11, E.R.: 2, chlo: 1 |
| PoABCB9 | Pos.gene58376.mRNA-1 | Chr02 | 1245 | 135,635.15 | 6.55 | plas: 13, vacu: 1 |
| PoABCB10 | Pos.gene19693.mRNA-1 | Chr03 | 1308 | 141,504.59 | 8.55 | plas: 7, vacu: 3, golg: 3, E.R.: 1 |
| PoABCB11 | Pos.gene19692.mRNA-1 | Chr03 | 1290 | 138,874.61 | 8.59 | plas: 8, golg: 3, vacu: 2, E.R.: 1 |
| PoABCB12 | Pos.gene29825.mRNA-1 | Chr03 | 622 | 67,221.6 | 8.03 | plas: 8, vacu: 2, E.R.: 2, golg: 2 |
| PoABCB13 | Pos.gene75380.mRNA-1 | Chr03 | 1282 | 137,992.72 | 7.88 | plas: 7, vacu: 3, golg: 3, E.R.: 1 |
| PoABCB14 | Pos.gene53906.mRNA-1 | Chr03 | 1323 | 143,505.31 | 8.69 | plas: 13, E.R.: 1 |
| PoABCB15 | Pos.gene49335.mRNA-1 | Chr03 | 1256 | 135,594.19 | 6.67 | nucl: 12, extr: 2 |
| PoABCB16 | Pos.gene17081.mRNA-1 | Chr03 | 1249 | 135,727.45 | 8.04 | plas: 12, E.R.: 2 |
| PoABCB17 | Pos.gene17082.mRNA-1 | Chr03 | 1326 | 142,776.5 | 5.94 | plas: 13, vacu: 1 |
| PoABCB18 | Pos.gene64555.mRNA-1 | Chr03 | 1266 | 136,858.57 | 5.89 | plas: 13, vacu: 1 |
| PoABCB19 | Pos.gene37878.mRNA-1 | Chr04 | 1296 | 141,493.81 | 8.56 | plas: 13, vacu: 1 |
| PoABCB20 | Pos.gene50382.mRNA-1 | Chr04 | 1296 | 142,049.58 | 8.82 | plas: 5, E.R.: 5, chlo: 2, vacu: 1, pero: 1 |
| PoABCB21 | Pos.gene70255.mRNA-1 | Chr04 | 1223 | 133,110.2 | 8.42 | plas: 9, E.R.: 3, chlo: 1, mito: 1 |
| PoABCB22 | Pos.gene83054.mRNA-1 | Chr04 | 1249 | 136,140.8 | 7.99 | plas: 9, E.R.: 3, chlo: 1, vacu: 1 |
| PoABCB23 | Pos.gene42756.mRNA-1 | Chr04 | 901 | 98,715.29 | 5.95 | plas: 13, vacu: 1 |
| PoABCB24 | Pos.gene49791.mRNA-1 | Chr04 | 647 | 72,220.68 | 8.01 | plas: 11, mito: 1, E.R.: 1, pero: 1 |
| PoABCB25 | Pos.gene39446.mRNA-1 | Chr04 | 1259 | 137,893.48 | 9.27 | plas: 13, chlo: 1 |
| PoABCB26 | Pos.gene45557.mRNA-1 | Chr05 | 706 | 77,346.09 | 9.2 | plas: 7, chlo: 4, mito: 1, E.R.: 1, pero: 1 |
| PoABCB27 | Pos.gene77030.mRNA-1 | Chr05 | 1402 | 155,516 | 5.95 | plas: 12, nucl: 1, vacu: 1 |
| PoABCB28 | Pos.gene61627.mRNA-1 | Chr05 | 1280 | 140,874.21 | 8.65 | plas: 14 |
| PoABCB29 | Pos.gene70371.mRNA-1 | Chr05 | 1267 | 137,717.23 | 8.94 | plas: 13, vacu: 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shang, W.; Cui, C.; Liu, X.; Meng, W.; Qiu, Y.; Sun, Y.; Shen, Y.; Liu, W.; Wang, Z.; He, S.; et al. Genome-Wide Analysis of the ABCB Family and Its Expression in Adventitious Root Development of Paeonia ostii. Horticulturae 2025, 11, 138. https://doi.org/10.3390/horticulturae11020138
Shang W, Cui C, Liu X, Meng W, Qiu Y, Sun Y, Shen Y, Liu W, Wang Z, He S, et al. Genome-Wide Analysis of the ABCB Family and Its Expression in Adventitious Root Development of Paeonia ostii. Horticulturae. 2025; 11(2):138. https://doi.org/10.3390/horticulturae11020138
Chicago/Turabian StyleShang, Wenqian, Can Cui, Xi Liu, Weihao Meng, Yongjie Qiu, Yuke Sun, Yuxiao Shen, Weichao Liu, Zheng Wang, Songlin He, and et al. 2025. "Genome-Wide Analysis of the ABCB Family and Its Expression in Adventitious Root Development of Paeonia ostii" Horticulturae 11, no. 2: 138. https://doi.org/10.3390/horticulturae11020138
APA StyleShang, W., Cui, C., Liu, X., Meng, W., Qiu, Y., Sun, Y., Shen, Y., Liu, W., Wang, Z., He, S., Song, Y., & Shi, L. (2025). Genome-Wide Analysis of the ABCB Family and Its Expression in Adventitious Root Development of Paeonia ostii. Horticulturae, 11(2), 138. https://doi.org/10.3390/horticulturae11020138





