Genome-Wide Identification of the 1-Aminocyclopropane-1-carboxylic Acid Synthase (ACS) Genes and Their Possible Role in Sand Pear (Pyrus pyrifolia) Fruit Ripening
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Treatments
2.2. Measurements of Ethylene Production
2.3. RNA Isolation and RNA-Seq
2.4. Genome-Wide Identification of ACS Genes
2.5. Phylogenetic Analysis
2.6. Quantitative Real-Time PCR (qRT-PCR) Analysis
3. Results
3.1. ACS Gene Sequence Identification
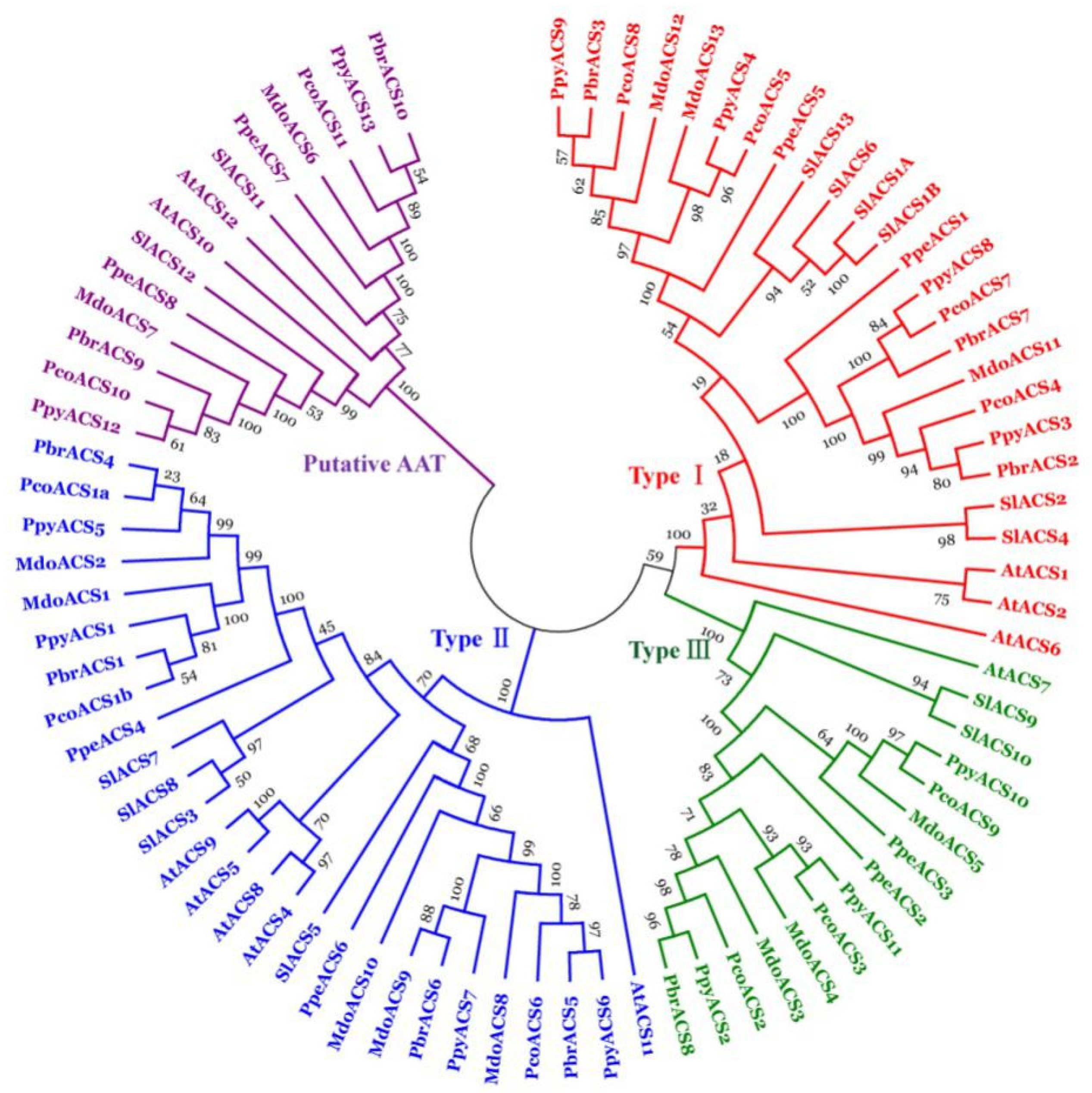
3.2. Phylogenetic Analysis of Putative ACS Genes
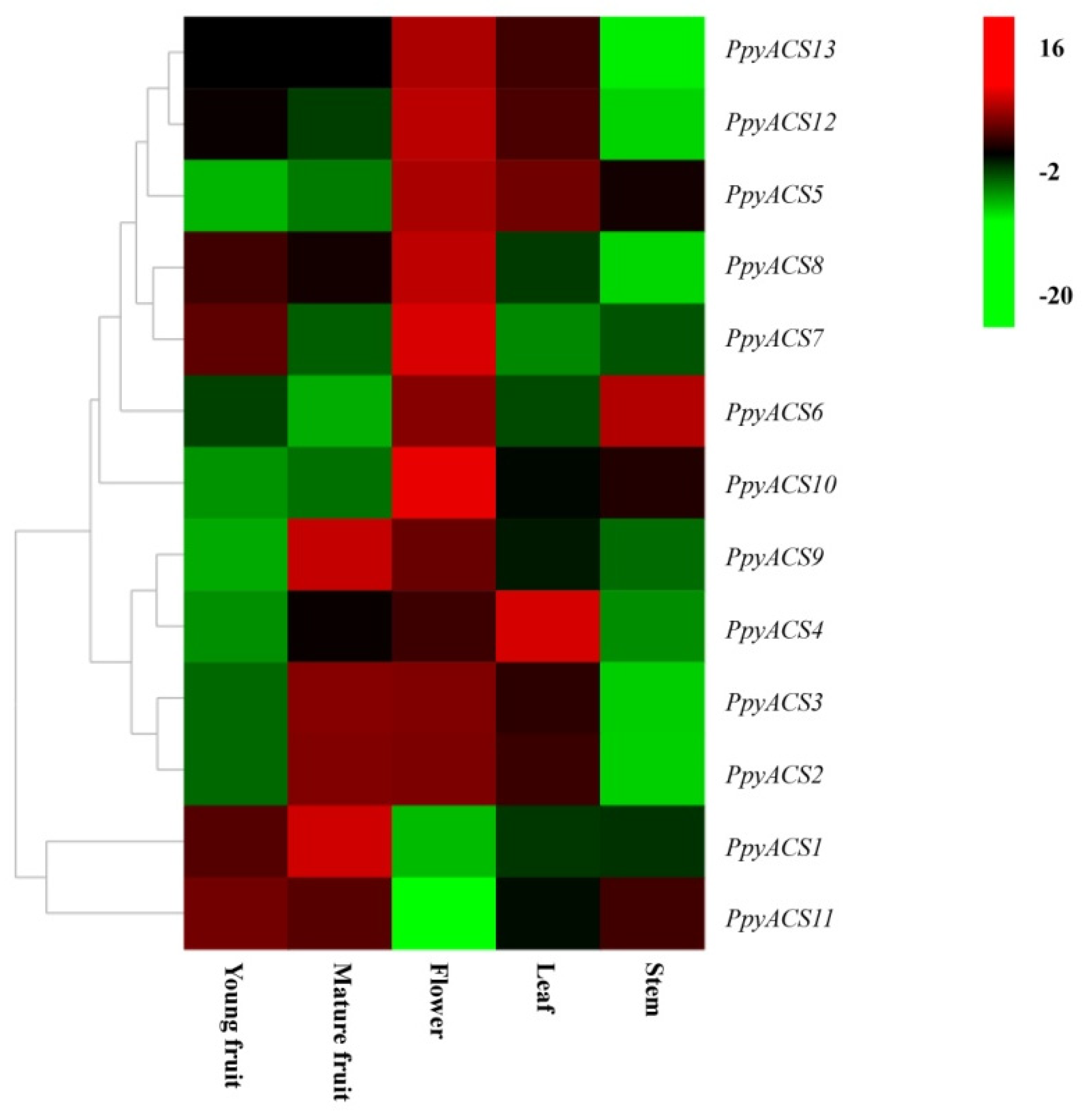
3.3. Expression of PpyACSs in Different Organs
3.4. RNA-Seq of Sand Pear Fruit and Identification of PpyACSs
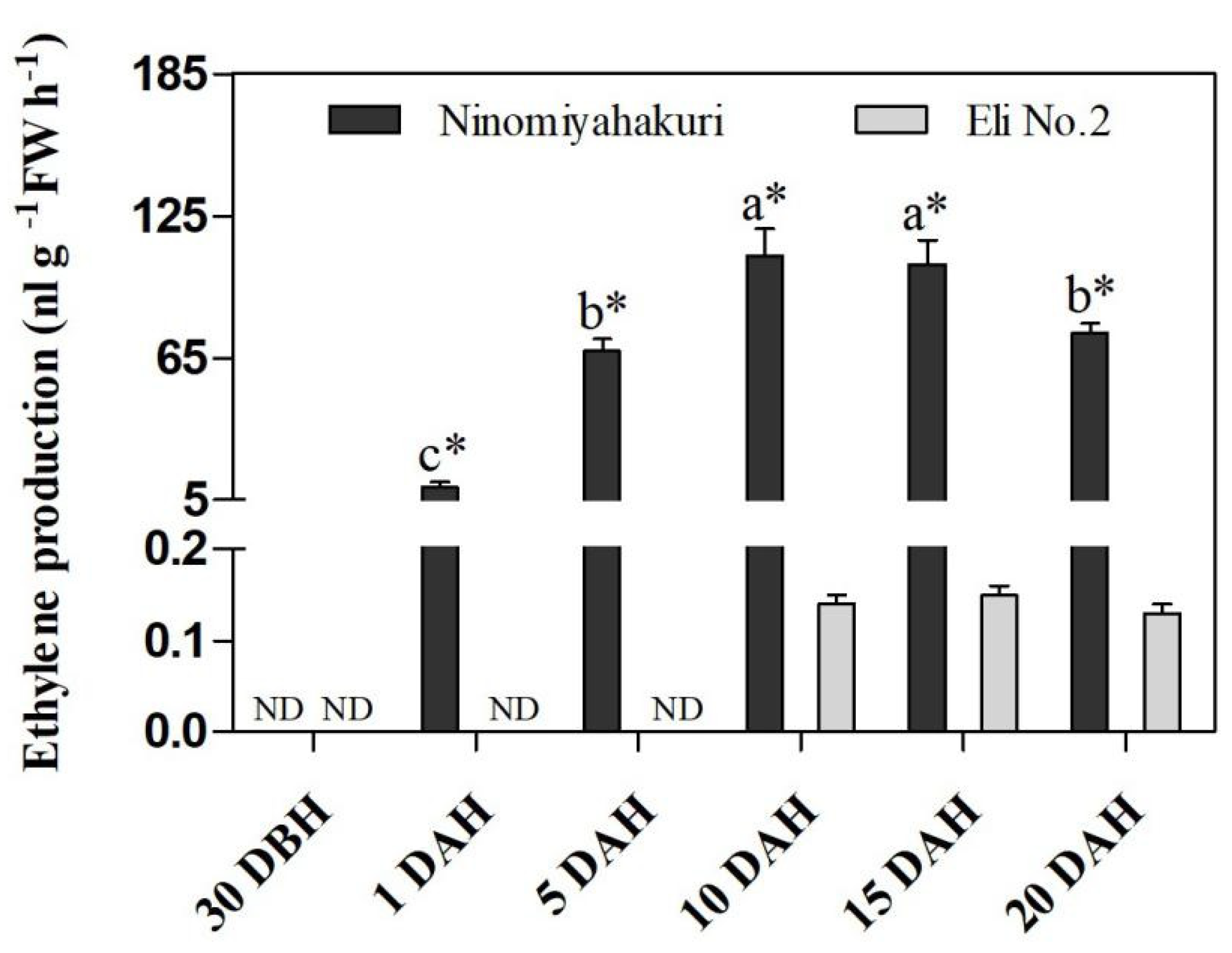
3.5. Changes in Internal Ethylene Concentration in Flesh during Fruit Ripening
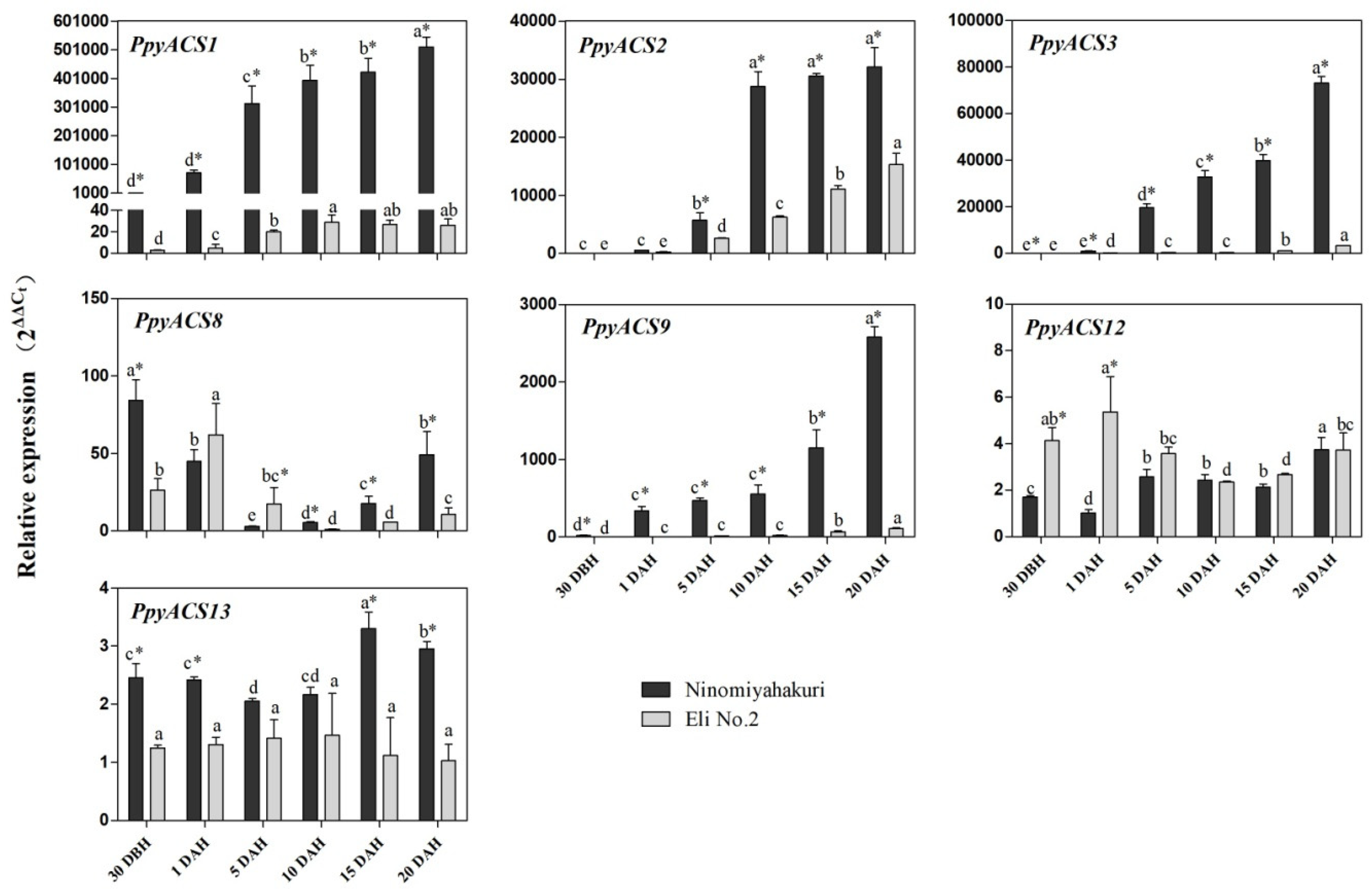
3.6. Expression of PpyACSs during Fruit Ripening
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Li, S.; Chen, K.; Grierson, D. A critical evaluation of the role of ethylene and MADS transcription factors in the network controlling fleshy fruit ripening. New Phytol. 2019, 221, 1724–1741. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dubois, M.; Van den Broeck, L.; Inzé, D. The pivotal role of ethylene in plant growth. Trends Plant Sci. 2018, 23, 311–323. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Tang, M.; Liu, M.; Su, D.; Chen, J.; Gao, Y.; Bouzayen, M.; Li, Z. The molecular regulation of ethylene in fruit ripening. Small Methods 2020, 4, 1900485. [Google Scholar] [CrossRef]
- Itai, A.; Kawata, T.; Tanabe, K.; Tamura, F.; Uchiyama, M.; Tomomitsu, M.; Shiraiwa, N. Identification of 1-aminocyclopropane-1-carboxylic acid synthase genes controlling the ethylene level of ripening fruit in Japanese pear (Pyrus pyrifolia Nakai). Mol. Gen. Genet. 1999, 261, 42–49. [Google Scholar] [CrossRef] [PubMed]
- Bapat, V.A.; Trivedi, P.K.; Ghosh, A.; Sane, V.A.; Ganapathi, T.R.; Nath, P. Ripening of fleshy fruit: Molecular insight and the role of ethylene. Biotechnol. Adv. 2010, 28, 94–107. [Google Scholar] [CrossRef]
- Pattyn, J.; Vaughan-Hirsch, J.; Van de Poel, B. The regulation of ethylene biosynthesis: A complex multilevel control circuitry. New Phytol. 2021, 229, 770–782. [Google Scholar] [CrossRef]
- Kende, H. Ethylene biosynthesis. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1993, 44, 283–307. [Google Scholar] [CrossRef]
- Liu, M.; Pirrello, J.; Chervin, C.; Roustan, J.; Bouzayen, M. Ethylene control of fruit ripening: Revisiting the complex network of transcriptional regulation. Plant Physiol. 2015, 169, 2380–2390. [Google Scholar] [CrossRef] [Green Version]
- Sunako, T.; Sakuraba, W.; Senda, M.; Akada, S.; Ishikawa, R.; Niizeki, M.; Harada, T. An allele of the ripening-specific 1-aminocyclopropane-1-carboxylic acid synthase gene (ACS1) in apple fruit with a long storage life. Plant Physiol. 1999, 119, 1297–1304. [Google Scholar] [CrossRef] [Green Version]
- Wang, A.; Yamakake, J.; Kudo, H.; Wakasa, Y.; Hatsuyama, Y.; Igarashi, M.; Kasai, A.; Li, T.; Harada, T. Null mutation of the MdACS3 gene, coding for a ripening-specific 1-aminocyclopropane-1-carboxylate synthase, leads to long shelf life in apple fruit. Plant Physiol. 2009, 151, 391–399. [Google Scholar] [CrossRef] [Green Version]
- Li, T.; Tan, D.; Liu, Z.; Jiang, Z.; Wei, Y.; Zhang, L.; Li, X.; Yuan, H.; Wang, A. Apple MdACS6 regulates ethylene biosynthesis during fruit development involving ethylene-responsive factor. Plant Cell Physiol. 2015, 56, 1909–1917. [Google Scholar] [CrossRef] [Green Version]
- Zeng, W.; Pan, L.; Liu, H.; Niu, L.; Lu, Z.; Cui, G.; Wang, Z. Characterization of 1-aminocyclopropane-1-carboxylic acid synthase (ACS) genes during nectarine fruit development and ripening. Tree Genet. 2015, 11, 18. [Google Scholar] [CrossRef]
- Li, X.; Liu, L.; Ming, M.; Hu, H.; Zhang, M.; Fan, J.; Song, B.; Zhang, S.; Wu, J. Comparative transcriptomic analysis provides insight into the domestication and improvement of pear (P. pyrifolia) fruit. Plant Physiol. 2019, 180, 435–452. [Google Scholar] [CrossRef] [Green Version]
- Mudassar, A.; Li, J.Z.; Yang, Q.S.; Jamil, W.; Teng, Y.W.; Bai, S.L. Phylogenetic, molecular, and functional characterization of PpyCBF proteins in Asian pears (Pyrus pyrifolia). Int. J. Mol. Sci. 2019, 20, 2074. [Google Scholar]
- Guo, L.Q.; Zhang, J.G.; Liu, X.X.; Rao, G.D. Polyploidy-related differential gene expression between diploid and synthesized allotriploid and allotetraploid hybrids of Populus. Mol. Breed. 2019, 39, 69. [Google Scholar]
- Wang, L.; Feng, Z.; Wang, X.; Wang, X.; Zhang, X. DEGseq: An R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 2010, 26, 136–138. [Google Scholar] [CrossRef]
- Qi, L.Y.; Chen, L.; Wang, C.S.; Zhang, S.L.; Yang, Y.J.; Liu, J.L.; Li, D.L.; Song, J.K.; Wang, R. Characterization of the auxin efflux transporter PIN proteins in pear. Plants 2020, 9, 349. [Google Scholar] [CrossRef] [Green Version]
- Jung, S.; Lee, T.; Cheng, C.; Buble, K.; Zheng, P.; Yu, J.; Humann, J.; Ficklin, S.; Gasic, K.; Scott, K.; et al. 15 years of GDR: New data and functionality in the genome database for Rosaceae. Nucleic Acids Res. 2019, 47, D1137–D1145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jung, S.; Bassett, C.; Bielenberg, D.G.; Cheng, C.-H.; Dardick, C.; Main, D.; Meisel, L.; Slovin, J.; Troggio, M.; Schaffer, R.J. A standard nomenclature for gene designation in the Rosaceae. Tree Genet. 2015, 11, 108. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.; Frank, M.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Xu, L.; Dong, Z.; Fang, L.; Luo, Y.; Wei, Z.; Guo, H.; Zhang, G.; Gu, Y.; Coleman-Derr, D.; Xia, Q.; et al. OrthoVenn2: A web server for whole-genome comparison and annotation of orthologous clusters across multiple species. Nucleic Acids Res. 2019, 47, 52–58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamagami, T.; Tsuchisaka, A.; Yamada, K.; Haddon, W.; Harden, L.; Theologis, A. Biochemical diversity among the 1-amino-cyclopropane-1-carboxylate synthase isozymes encoded by the Arabidopsis gene family. J. Biol. Chem. 2003, 278, 49102–49112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Dai, M.; Cai, D.; Shi, Z. Screening for quantitative real-time PCR reference genes with high stable expression using the mRNA-sequencing data for pear. Tree Genet. 2019, 15, 54. [Google Scholar] [CrossRef]
- Li, H.; Huang, C.-H.; Ma, H. Whole-Genome Duplications in Pear and Apple. In The Pear Genome; Korban, S.S., Ed.; Springer International Publishing: Cham, Switzerland, 2019; pp. 279–299. [Google Scholar]
- Yoshida, H.; Nagata, M.; Saito, K.; Wang, K.; Ecker, J. Arabidopsis ETO1 specifically interacts with and negatively regulates type 2 1-aminocyclopropane-1-carboxylate synthases. BMC Plant Biol. 2005, 5, 14. [Google Scholar] [CrossRef] [Green Version]
- Zhu, J.-H.; Xu, J.; Chang, W.-J.; Zhang, Z.-L. Isolation and molecular characterization of 1-aminocyclopropane-1-carboxylic acid synthase genes in Heveabrasiliensis. Int. J. Mol. Sci. 2015, 16, 4136–4149. [Google Scholar] [CrossRef] [Green Version]
- Shirasawa, K.; Itai, A.; Isobe, S. Chromosome-scale genome assembly of Japanese pear (Pyrus pyrifolia) variety ‘Nijisseiki’. DNA Res. 2021, 28, dsab001. [Google Scholar] [CrossRef]
- Yamane, M.; Abe, D.; Yasui, S.; Yokotani, N.; Kimata, W.; Ushijima, K.; Nakano, R.; Kubo, Y.; Inaba, A. Differential expression of ethylene biosynthetic genes in climacteric and non-climacteric Chinese pear fruit. Postharvest Biol. Technol. 2007, 44, 220–227. [Google Scholar] [CrossRef]
- Lee, J.-H.; Kim, Y.-C.; Choi, D.; Han, J.-H.; Jung, Y.; Lee, S. RNA expression, protein activity, and interactions in the ACC synthase gene family in cucumber (Cucumis sativus L.). Hortic. Environ. Biotechnol. 2018, 59, 81–91. [Google Scholar] [CrossRef]
- Li, T.; Tan, D.; Yang, X.; Wang, A. Exploring the apple genome reveals six ACC synthase genes expressed during fruit ripening. Sci. Hortic. 2013, 157, 119–123. [Google Scholar] [CrossRef] [Green Version]
- Yuan, H.; Yue, P.; Bu, H.; Han, D.; Wang, A. Genome-wide analysis of ACO and ACS genes in pear (Pyrus ussuriensis). In Vitro Cell. Dev. Biol. Plant 2020, 56, 193–199. [Google Scholar] [CrossRef]
- Paul, V.; Pandey, R.; Srivastava, G.C. The fading distinctions between classical patterns of ripening in climacteric and non-climacteric fruit and the ubiquity of ethylene-an overview. J. Food Sci. Technol. 2012, 49, 1–21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Itai, A.; Kotaki, T.; Tanabe, K.; Tamura, F.; Kawaguchi, D.; Fukuda, M. Rapid identification of 1-aminocyclopropane-1-carboxylate (ACC) synthase genotypes in cultivars of Japanese pear (Pyrus pyrifolia Nakai) using CAPS markers. Theor. Appl. Genet. 2003, 106, 1266–1272. [Google Scholar] [CrossRef]
- Tan, D.; Li, T.; Wang, A. Apple 1-aminocyclopropane-1-carboxylic acid synthase genes, MdACS1 and MdACS3a, are expressed in different systems of ethylene biosynthesis. Plant Mol. Biol. Report 2012, 31, 204–209. [Google Scholar] [CrossRef]
- Huang, G.; Li, T.; Li, X.; Tan, D.; Jiang, Z.; Wei, Y.; Li, J.; Wang, A. Comparative transcriptome analysis of climacteric fruit of Chinese pear (Pyrus ussuriensis) reveals new insights into fruit ripening. PLoS ONE 2014, 9, e107562. [Google Scholar] [CrossRef] [Green Version]
- Zhang, T.; Qiao, Q.; Zhong, Y. Detecting adaptive evolution and functional divergence in aminocyclopropane-1-carboxylate synthase (ACS) gene family. Comput. Biol. Chem. 2012, 38, 10–16. [Google Scholar] [CrossRef]
- Barry, C.S.; Llop-Tous, M.I.; Grierson, D. The regulation of 1-aminocyclopropane-1-carboxylic acid synthase gene expression during the transition from system-1 to system-2 ethylene synthesis in tomato. Plant Physiol. 2000, 123, 979–986. [Google Scholar] [CrossRef] [Green Version]
- Booker, M.A.; DeLong, A. Producing the ethylene signal: Regulation and diversification of ethylene biosynthetic enzymes. Plant Physiol. 2015, 169, 42–50. [Google Scholar] [CrossRef] [Green Version]
- Gao, Y.; Ma, J.; Zheng, J.C.; Chen, J.; Chen, M.; Zhou, Y.B.; Fu, J.D.; Xu, Z.S.; Ma, Y.Z. The elongation factor GmEF4 is involved in the response to drought and salt tolerance in soybean. Int. J. Mol. Sci. 2019, 20, 3001. [Google Scholar] [CrossRef] [Green Version]
- Wang, W.; Wu, H.; Liu, J.H. Genome-wide identification and expression profiling of copper-containing amine oxidase genes in sweet orange (Citrus sinensis). Tree Genet. Genomes 2017, 13, 31. [Google Scholar] [CrossRef]
- Jakubowicz, M.; Sadowski, J. 1-aminocyclopropane-1-carboxylate synthase genes and expression. Acta Physiol. Plant. 2002, 24, 459–478. [Google Scholar] [CrossRef]
- Van de Poel, B.; Van Der Straeten, D. 1-aminocyclopropane-1-carboxylic acid (ACC) in plants: More than just the precursor of ethylene! Front. Plant. Sci. 2014, 5, 640. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matarasso, N.; Schuster, S.; Avni, A. A novel plant cysteine protease has a dual function as a regulator of 1-aminocyclopropane-1-carboxylic acid synthase gene expression. Plant. Cell 2005, 17, 1205–1216. [Google Scholar] [CrossRef] [PubMed] [Green Version]




| Gene Name | Gene ID | Chromosome | Protein Length (aa) | No. of Exons | Molecular Weight (kDa) | Isoelectric Point (pI) | Type | Subcellular Prediction |
|---|---|---|---|---|---|---|---|---|
| PpyACS1 | Ppy15g2510.1 | Chr15:20022979..20024784+ | 473 | 4 | 53.24 | 6.47 | Type II | Cytoplasm |
| PpyACS5 | Ppy02g1684.1 | Chr02:14156236..14158531+ | 473 | 4 | 55.54 | 6.9 | Type II | Cytoplasm |
| PpyACS6 | Ppy08g0607.1 | Chr08:4476046..4478458- | 488 | 5 | 59.57 | 6.69 | Type II | Cytoplasm |
| PpyACS7 | Ppy15g0540.1 | Chr15:3580798..3589288- | 502 | 5 | 54.67 | 7.06 | Type II | Cytoplasm |
| PpyACS3 | Ppy01g0738.1 | Chr01:12651750..12654529- | 495 | 4 | 54.86 | 7.96 | Type I | Chloroplast |
| PpyACS8 | Ppy07g1563.1 | Chr07:23041756..23045681+ | 529 | 5 | 53.18 | 8.53 | Type I | Chloroplast |
| PpyACS9 | Ppy06g0809.1 | Chr06:12079561..12081861- | 487 | 4 | 56.82 | 7.56 | Type I | Chloroplast |
| PpyACS4 | Ppy14g0933.1 | Chr14:11862299..11864545- | 487 | 4 | 55.15 | 6.64 | Type I | Chloroplast |
| PpyACS2 | Ppy15g1745.1 | Chr15:12377889..12379594- | 446 | 3 | 50.1 | 5.65 | Type III | Chloroplast |
| PpyACS10 | Ppy02g0565.1 | Chr02:3901240..3902919- | 447 | 3 | 50.31 | 5.63 | Type III | Chloroplast |
| PpyACS11 | Ppy02g0566.1 | Chr02:3918274..3920013- | 445 | 3 | 49.84 | 5.84 | Type III | Chloroplast |
| PpyACS12 | Ppy04g0675.1 | Chr04:6740597..6743142+ | 538 | 4 | 58.89 | 8.76 | Putative AAT | Chloroplast |
| PpyACS13 | Ppy01g1669.1 | Chr01:20075291..20079590- | 611 | 6 | 67.55 | 8.3 | Putative AAT | Chloroplast |
| Fruit Type | Main ACSs Involved in Ethylene Production | |
|---|---|---|
| Immature Stage | Mature Stage | |
| Climacteric | PpyACS8, PpyACS12, PpyACS13 | PpyACS1, PpyACS2, PpyACS3, PpyACS9 |
| Non-climacteric | PpyACS8, PpyACS12, PpyACS13 | PpyACS2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, J.-G.; Du, W.; Fan, J.; Yang, X.-P.; Chen, Q.-L.; Liu, Y.; Hu, H.-J.; Luo, Z.-R. Genome-Wide Identification of the 1-Aminocyclopropane-1-carboxylic Acid Synthase (ACS) Genes and Their Possible Role in Sand Pear (Pyrus pyrifolia) Fruit Ripening. Horticulturae 2021, 7, 401. https://doi.org/10.3390/horticulturae7100401
Zhang J-G, Du W, Fan J, Yang X-P, Chen Q-L, Liu Y, Hu H-J, Luo Z-R. Genome-Wide Identification of the 1-Aminocyclopropane-1-carboxylic Acid Synthase (ACS) Genes and Their Possible Role in Sand Pear (Pyrus pyrifolia) Fruit Ripening. Horticulturae. 2021; 7(10):401. https://doi.org/10.3390/horticulturae7100401
Chicago/Turabian StyleZhang, Jing-Guo, Wei Du, Jing Fan, Xiao-Ping Yang, Qi-Liang Chen, Ying Liu, Hong-Ju Hu, and Zheng-Rong Luo. 2021. "Genome-Wide Identification of the 1-Aminocyclopropane-1-carboxylic Acid Synthase (ACS) Genes and Their Possible Role in Sand Pear (Pyrus pyrifolia) Fruit Ripening" Horticulturae 7, no. 10: 401. https://doi.org/10.3390/horticulturae7100401
APA StyleZhang, J. -G., Du, W., Fan, J., Yang, X. -P., Chen, Q. -L., Liu, Y., Hu, H. -J., & Luo, Z. -R. (2021). Genome-Wide Identification of the 1-Aminocyclopropane-1-carboxylic Acid Synthase (ACS) Genes and Their Possible Role in Sand Pear (Pyrus pyrifolia) Fruit Ripening. Horticulturae, 7(10), 401. https://doi.org/10.3390/horticulturae7100401






