Understanding Root Rot Disease in Agricultural Crops
Abstract
:1. Introduction
2. Common Causal Agents of Root Rots
2.1. Fungi
2.2. Oomycetes
2.3. Bacteria and Viruses
3. Molecular Mechanisms of Resistance Against Root Rot Pathogens
4. Common Causal Agents of Root Rots
4.1. Fungal Root Rot
4.1.1. Rhizoctonia Solani
4.1.2. Transgenic Approach to Combat Rhizoctonia solani
4.1.3. Fusarium solani Root Rot: The Case of Pea and Similitudes with Soybean
4.1.4. Transgenic Approach to Counter Fusarium solani Root Rot
4.1.5. Fusarium graminareum Root Rot
4.1.6. Fusarium Root Rot in Cereals
4.1.7. Phoma Root Rot
4.1.8. Thielaviopsis basicola Root Rot
4.1.9. A Transgenic Approach to Counter Thielaviopsis basicola Root Rot
4.2. Oomycete Root Rot
4.2.1. Aphanomyces Root Rot
4.2.2. Role of Nodulation and Mycorrhizas in Aphanomyces Root Rot
4.2.3. Pythium Root Rot
4.2.4. Phytophthora Root Rot
4.2.5. Phytophthora Root Rot in Other Crops
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Kumari, N.; Katoch, S. Wilt and Root Rot Complex of Important Pulse Crops: Their Detection and Integrated Management. In Management of Fungal Pathogens in Pulses; Springer: Berlin/Heidelberg, Germany, 2020; pp. 93–119. [Google Scholar]
- Erwin, D.C.; Ribeiro, O.K. Phytophthora Diseases Worldwide; American Phytopathological Society (APS Press): St. Paul, MN, USA, 1996; ISBN 0890542120. [Google Scholar]
- Gaulin, E.; Jacquet, C.; Bottin, A.; Dumas, B. Root rot disease of legumes caused by Aphanomyces euteiches. Mol. Plant Pathol. 2007, 8, 539–548. [Google Scholar] [CrossRef]
- Bodah, E.T. Root rot diseases in plants: A review of common causal agents and management strategies. Agric. Res. Technol. Open Access J. 2017, 5, 555661. [Google Scholar]
- Kraft, J.M.; Haware, M.P.; Jimenez-Diaz, R.M.; Bayaa, B.; Harrabi, M. Screening techniques and sources of resistance to root rots and wilts in cool season food legumes. Euphytica 1993, 73, 27–39. [Google Scholar] [CrossRef] [Green Version]
- Hughes, T.J.; Grau, C.R. Aphanomyces root rot (common root rot) of legumes. Plant Heal. Instr. 2007. [Google Scholar] [CrossRef]
- Bhat, K.A.; Masood, S.D.; Bhat, N.A.; Bhat, M.A.; Razvi, S.M.; Mir, M.R.; Sabina, A.; Wani, N.; Habib, M. Current status of post harvest soft rot in vegetables: A review. Asian J. Plant Sci. 2010, 9, 200–208. [Google Scholar] [CrossRef] [Green Version]
- Kikumoto, T. Ecology and biocontrol of soft rot of Chinese cabbage. Jpn. J. Phytopathol. 2000, 66, 60–62. [Google Scholar] [CrossRef]
- Liao, C.-H. Control of foodborne pathogens and soft-rot bacteria on bell pepper by three strains of bacterial antagonists. J. Food Prot. 2009, 72, 85–92. [Google Scholar] [CrossRef] [Green Version]
- Perombelon, M.C.M.; Kelman, A. Ecology of the soft rot erwinias. Annu. Rev. Phytopathol. 1980, 18, 361–387. [Google Scholar] [CrossRef]
- Bock, K.R. Studies on cassava brown streak virus disease in Kenya. Trop. Sci. 1994, 34, 134–145. [Google Scholar]
- Hillocks, R.J.; Raya, M.D.; Mtunda, K.; Kiozia, H. Effects of brown streak virus disease on yield and quality of cassava in Tanzania. J. Phytopathol. 2001, 149, 389–394. [Google Scholar] [CrossRef]
- Louws, F.; Sun, J.; Whittington, H.; Driver, J.; Peeden, K.; Liu, B. Evaluation of fungicides and mustard meal to manage black root rot of strawberry and analysis of Pythium, Fusarium, and Rhizoctonia on strawberry roots. Phytopathology 2012, 102, 72. [Google Scholar]
- Manici, L.M.; Caputo, F.; Baruzzi, G. Additional experiences to elucidate the microbial component of soil suppressiveness towards strawberry black root rot complex. Ann. Appl. Biol. 2005, 146, 421–431. [Google Scholar] [CrossRef]
- Particka, C.A.; Hancock, J.F. Field evaluation of strawberry genotypes for tolerance to black root rot on fumigated and nonfumigated soil. J. Am. Soc. Hortic. Sci. 2005, 130, 688–693. [Google Scholar] [CrossRef] [Green Version]
- Xue, A.G. Biological control of pathogens causing root rot complex in field pea using Clonostachys rosea strain ACM941. Phytopathology 2003, 93, 329–335. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hosseini, S.; Elfstrand, M.; Heyman, F.; Jensen, D.F.; Karlsson, M. Deciphering common and specific transcriptional immune responses in pea towards the oomycete pathogens Aphanomyces euteiches and Phytophthora pisi. BMC Genom. 2015, 16, 627. [Google Scholar] [CrossRef] [Green Version]
- Tu, J.C. Effects of soil compaction, temperature, and moisture on the development of the Fusarium root rot complex of pea in southwestern Ontario. Phytoprotection 1994, 75, 125–131. [Google Scholar] [CrossRef] [Green Version]
- Zitnick-Anderson, K.; Simons, K.; Pasche, J.S. Detection and qPCR quantification of seven Fusarium species associated with the root rot complex in field pea. Can. J. Plant Pathol. 2018, 40, 261–271. [Google Scholar] [CrossRef]
- Hamon, C.; Baranger, A.; Coyne, C.J.; McGee, R.J.; Le Goff, I.; L’Anthoëne, V.; Esnault, R.; Riviere, J.-P.; Klein, A.; Mangin, P. New consistent QTL in pea associated with partial resistance to Aphanomyces euteiches in multiple French and American environments. Theor. Appl. Genet. 2011, 123, 261–281. [Google Scholar] [CrossRef] [PubMed]
- Carling, D.E.; Baird, R.E.; Gitaitis, R.D.; Brainard, K.A.; Kuninaga, S. Characterization of AG-13, a newly reported anastomosis group of Rhizoctonia solani. Phytopathology 2002, 92, 893–899. [Google Scholar] [CrossRef] [Green Version]
- Tewoldemedhin, Y.T.; Lamprecht, S.C.; McLeod, A.; Mazzola, M. Characterization of Rhizoctonia spp. recovered from crop plants used in rotational cropping systems in the Western Cape province of South Africa. Plant Dis. 2006, 90, 1399–1406. [Google Scholar] [CrossRef] [Green Version]
- Allmaras, R.R.; Fritz, V.A.; Pfleger, F.L.; Copeland, S.M. Impaired internal drainage and Aphanomyces euteiches root rot of pea caused by soil compaction in a fine-textured soil. Soil Tillage Res. 2003, 70, 41–52. [Google Scholar] [CrossRef]
- Falcon, M.F.; Fox, R.L.; Trujillo, E.E. Interactions of soil pH, nutrients and moisture on Phytophthora root rot of avocado. Plant Soil 1984, 81, 165–176. [Google Scholar] [CrossRef]
- Rhoades, C.C.; Brosi, S.L.; Dattilo, A.J.; Vincelli, P. Effect of soil compaction and moisture on incidence of phytophthora root rot on American chestnut (Castanea dentata) seedlings. For. Ecol. Manag. 2003, 184, 47–54. [Google Scholar] [CrossRef]
- La Porta, N.; Capretti, P.; Thomsen, I.M.; Kasanen, R.; Hietala, A.M.; Von Weissenberg, K. Forest pathogens with higher damage potential due to climate change in Europe. Can. J. Plant Pathol. 2008, 30, 177–195. [Google Scholar] [CrossRef]
- Kubiak, K.; Żółciak, A.; Damszel, M.; Lech, P.; Sierota, Z. Armillaria pathogenesis under climate changes. Forests 2017, 8, 100. [Google Scholar] [CrossRef] [Green Version]
- Klopfenstein, N.B. Approaches to Predicting Potential Impacts of Climate Change on Forest Disease: An Example with Armillaria Root Disease; US Department of Agriculture, Forest Service, Rocky Mountain Research Station: Washington, DC, USA, 2009.
- Papavizas, G.C.; Ayers, W.A. Aphanomyces Species and Their Root Diseases in Pea and Sugarbeet; US Department of Agriculture, Forest Service, Rocky Mountain Research Station: Washington, DC, USA, 1974.
- Oelke, L.M.; Bosland, P.W.; Steiner, R. Differentiation of race specific resistance to Phytophthora root rot and foliar blight in Capsicum annuum. J. Am. Soc. Hortic. Sci. 2003, 128, 213–218. [Google Scholar] [CrossRef] [Green Version]
- Tu, J.C.; Papadopoulos, A.P.; Hao, X.; Zheng, J. The relationship of Pythium root rot and rhizosphere microorganisms in a closed circulating and an open system in rockwool culture of tomato. In Proceedings of the International Symposium on Growing Media and Hydroponics, Windsor, ON, Canada, 19 May 1997; Volume 481, pp. 577–586. [Google Scholar]
- Zhu, Y.; Saltzgiver, M. A systematic analysis of apple root resistance traits to Pythium ultimum infection and the underpinned molecular regulations of defense activation. Hortic. Res. 2020, 7, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Anderson, N.A. The genetics and pathology of Rhizoctonia solani. Annu. Rev. Phytopathol. 1982, 20, 329–347. [Google Scholar] [CrossRef]
- Paulitz, T.C.; Adams, K. Composition and distribution of Pythium communities in wheat fields in eastern Washington State. Phytopathology 2003, 93, 867–873. [Google Scholar] [CrossRef] [Green Version]
- Sneh, B.; Jabaji-Hare, S.; Neate, S.M.; Dijst, G. Rhizoctonia Species: Taxonomy, Molecular Biology, Ecology, Pathology and Disease Control; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2013; ISBN 9401729018. [Google Scholar]
- Burton, R.J.; Coley-Smith, J.R.; Wareing, P.W.; Gladders, P. Rhizoctonia oryzae and R. solani associated with barley stunt disease in the United Kingdom. Trans. Br. Mycol. Soc. 1988, 91, 409–417. [Google Scholar] [CrossRef]
- McKinley, A. Evaluation of progeny of genetically engineered barley plants for resistance to rhizoctonia oryzae and Rhizoctonia solani AG-8. Honor’s Thesis, Washington State University, Pullman, WA, USA, 2003. [Google Scholar]
- Neate, S.M. A comparison of controlled environment and field trials for detection of resistance in cereal cultivars to root rot caused by Rhizoctonia solani. Plant Pathol. 1989, 38, 494–501. [Google Scholar] [CrossRef]
- Rush, C.M.; Carling, D.E.; Harveson, R.M.; Mathieson, J.T. Prevalence and pathogenicity of anastomosis groups of Rhizoctonia solani from wheat and sugar beet in Texas. Plant Dis. 1994, 78, 349–352. [Google Scholar] [CrossRef]
- Karaca, G.H.; Ozkoc, I.; Erper, I. Determination of the anastomosis grouping and virulence of Rhizoctonia solani Kühn isolates associated with bean plants grown in Samsun/Turkey. Pak. J. Biol. Sci. 2002, 5, 434–437. [Google Scholar] [CrossRef]
- Muyolo, N.G.; Lipps, P.E.; Schmitthenner, A.F. Reactions of dry bean, lima bean, and soybean cultivars to Rhizoctonia root and hypocotyl rot and web blight. Plant Dis. 1993, 77, 234–238. [Google Scholar] [CrossRef]
- Oladzad, A.; Zitnick-Anderson, K.; Jain, S.; Simons, K.; Osorno, J.M.; McClean, P.E.; Pasche, J. Genotypes and genomic regions associated with Rhizoctonia solani resistance in common bean. Front. Plant Sci. 2019, 10, 956. [Google Scholar] [CrossRef] [Green Version]
- Peña, P.A.; Steadman, J.R.; Eskridge, K.M.; Urrea, C.A. Identification of sources of resistance to damping-off and early root/hypocotyl damage from Rhizoctonia solani in common bean (Phaseolus vulgaris L.). Crop Prot. 2013, 54, 92–99. [Google Scholar] [CrossRef] [Green Version]
- Keinath, A.P.; Farnham, M.W. Differential cultivars and criteria for evaluating resistance to Rhizoctonia solani in seedling Brassica oleracea. Plant Dis. 1997, 81, 946–952. [Google Scholar] [CrossRef] [Green Version]
- Rollins, P.A.; Keinath, A.P.; Farnham, M.W. Effect of inoculum type and anastomosis group of Rhizoctonia solani causing wirestem of cabbage seedlings in a controlled environment. Can. J. Plant Pathol. 1999, 21, 119–124. [Google Scholar] [CrossRef]
- Yanga, G.H.; Chenb, J.Y.; Pua, W.Q. New disease report. Plant Pathol. 2007, 56, 351. [Google Scholar]
- Grisham, M.P.; Anderson, N.A. Pathogenicity and host specificity of Rhizoctonia solani isolated from carrots. Phytopathology 1983, 73, 1564–1569. [Google Scholar] [CrossRef]
- Naito, S.; Kanematsu, S. Characterization and pathogenicity of a new anastomosis subgroup AG-2-3 of Rhizoctonia solani Kühn isolated from leaves of soybean. Jpn. J. Phytopathol. 1994, 60, 681–690. [Google Scholar] [CrossRef] [Green Version]
- Punja, Z.K. Transgenic carrots expressing a thaumatin-like protein display enhanced resistance to several fungal pathogens. Can. J. Plant Pathol. 2005, 27, 291–296. [Google Scholar] [CrossRef]
- Assunção, I.P.; Nascimento, L.D.; Ferreira, M.F.; Oliveira, F.J.; Michereff, S.J.; Lima, G.S.A. Reaction of faba bean genotypes to Rhizoctonia solani and resistance stability. Hortic. Bras. 2011, 29, 492–497. [Google Scholar] [CrossRef] [Green Version]
- Rashid, K.Y.; Bernier, C.C. Genetic diversity among isolates of Rhizoctonia solani and sources of resistance in Vicia faba. Can. J. Plant Pathol. 1993, 15, 23–28. [Google Scholar] [CrossRef]
- Valkonen, J.P.T.; Von Heiroth, W.; Savela, M. Fungi and Gram-negative bacteria as soilborne minor pathogens of goat’s rue (Galega orientalis Lam.). Ann. Appl. Biol. 1993, 123, 257–269. [Google Scholar] [CrossRef]
- Dijst, G.; Schneider, J.H.M. Flowerbulbs diseases incited by Rhizoctonia species. In Rhizoctonia Species: Taxonomy, Molecular Biology, Ecology, Pathology and Disease Control; Springer: Berlin/Heidelberg, Germany, 1996; pp. 279–288. [Google Scholar]
- Fähler, B.; Petersen, P. Rapid greenhouse screening of maize for resistance to Rhizoctonia solani AG2-2IIIB/Ein schnelles Screening von Mais auf Resistenz gegenüber Rhizoctonia solani AG2-2IIIB im Gewächshaus. J. Plant Dis. Prot. 2004, 111, 292–301. [Google Scholar]
- Garg, A.; Prasanna, B.M.; Sharma, R.C.; Rathore, R.S.; Saxena, S.C.; Chauhan, S.V.S. Identification of resistance sources to banded leaf and sheath blight (Rhizoctonia solani f. sp. sasakil) in maize. Indian Phytopathol. 2011, 60, 162–166. [Google Scholar]
- Sharma, R.C.; Rai, S.N.; Batsa, B.K. Identifying resistance to banded leaf and sheath blight of maize. Indian Phytopathol. 2012, 58, 121–122. [Google Scholar]
- Zhao, M.; Zhang, Z.; Zhang, S.; Li, W.; Jeffers, D.P.; Rong, T.; Pan, G. Quantitative trait loci for resistance to banded leaf and sheath blight in maize. Crop Sci. 2006, 46, 1039–1045. [Google Scholar] [CrossRef]
- Kataria, H.; Verma, P.R.; Gisi, U. Variability in the sensitivity of Rhizoctonia solani anastomosis groups to fungicides. J. Phytopathol. 1991, 133, 121–133. [Google Scholar] [CrossRef]
- Yang, J.; Verma, P.R. Screening genotypes for resistance to pre-emergence damping-off and postemergence seedling root rot of oilseed rape and canola caused by Rhizoctonia solani AG-2-1. Crop Prot. 1992, 11, 443–448. [Google Scholar] [CrossRef]
- Verma, P.R. Biology and control of Rhizoctonia solani on rapeseed: A review. Phytoprotection 1996, 77, 99–111. [Google Scholar] [CrossRef] [Green Version]
- Erper, I.; Karaca, G.H.; Turkkan, M.; Ozkoc, I. Characterization and pathogenicity of Rhizoctonia spp. from onion in Amasya, Turkey. J. Phytopathol. 2006, 154, 75–79. [Google Scholar] [CrossRef]
- Sharma-Poudyal, D.; Paulitz, T.C.; du Toit, L.J. Evaluation of onion genotypes for resistance to stunting caused by Rhizoctonia solani AG 8. HortScience 2015, 50, 551–554. [Google Scholar] [CrossRef] [Green Version]
- Hwang, S.F.; Gossen, B.D.; Conner, R.L.; Chang, K.F.; Turnbull, G.D.; Lopetinsky, K.; Howard, R.J. Management strategies to reduce losses caused by Rhizoctonia seedling blight of field pea. Can. J. Plant Sci. 2007, 87, 145–155. [Google Scholar] [CrossRef]
- McCoy, R.J.; Kraft, J.M. Comparison of techniques and inoculum in evaluating peas (Pisum sativum) for resistance to stem rot caused by Rhizobium solani. Plant Dis. 1984, 68, 53–55. [Google Scholar] [CrossRef]
- Leach, S.S.; Webb, R.E. Evaluation of potato cultivars, clones and a true seed population for resistance to Rhizoctonia solani. Am. Potato J. 1993, 70, 317–328. [Google Scholar] [CrossRef]
- Naz, F.; Rauf, C.A.; Abbasi, N.A.; Haque, I.; Ahmad, I. Influence of inoculum levels of Rhizoctonia solani and susceptibility on new potato germplasm. Pak. J. Bot. 2008, 40, 2199–2209. [Google Scholar]
- Zhang, X.-Y.; Yu, X.-X.; Yu, Z.; Xue, Y.-F.; Qi, L.-P. A simple method based on laboratory inoculum and field inoculum for evaluating potato resistance to black scurf caused by Rhizoctonia solani. Breed. Sci. 2014, 64, 156–163. [Google Scholar] [CrossRef] [Green Version]
- Yanar, Y.; Yılmaz, G.; Cesmeli, I.; Coskun, S. Characterization of Rhizoctonia solani isolates from potatoes in turkey and screening potato cultivars for resistance to AG-3 isolates. Phytoparasitica 2005, 33, 370–376. [Google Scholar] [CrossRef]
- Datta, K.; Velazhahan, R.; Oliva, N.; Ona, I.; Mew, T.; Khush, G.S.; Muthukrishnan, S.; Datta, S.K. Over-expression of the cloned rice thaumatin-like protein (PR-5) gene in transgenic rice plants enhances environmental friendly resistance to Rhizoctonia solani causing sheath blight disease. Theor. Appl. Genet. 1999, 98, 1138–1145. [Google Scholar] [CrossRef]
- Eizenga, G.C.; Lee, F.N.; Rutger, J.N. Screening Oryza species plants for rice sheath blight resistance. Plant Dis. 2002, 86, 808–812. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jha, S.; Tank, H.G.; Prasad, B.D.; Chattoo, B.B. Expression of Dm-AMP1 in rice confers resistance to Magnaporthe oryzae and Rhizoctonia solani. Transgenic Res. 2009, 18, 59–69. [Google Scholar] [CrossRef]
- Li, Z.; Pinson, S.R.M.; Marchetti, M.A.; Stansel, J.W.; Park, W.D. Characterization of quantitative trait loci (QTLs) in cultivated rice contributing to field resistance to sheath blight (Rhizoctonia solani). Theor. Appl. Genet. 1995, 91, 382–388. [Google Scholar] [CrossRef] [PubMed]
- Mazzola, M.; Wong, O.T.; Cook, R.J. Virulence of Rhizoctonia oryzae and R. solani AG-8 on wheat and detection of R. oryzae in plant tissue by PCR. Phytopathology 1996, 86, 354. [Google Scholar] [CrossRef]
- Bradley, C.A.; Hartman, G.L.; Nelson, R.L.; Mueller, D.S.; Pederson, W.L. Response of ancestral soybean lines and commercial cultivars to Rhizoctonia root and hypocotyl rot. Plant Dis. 2001, 85, 1091–1095. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muyolo, N.G.; Lipps, P.E.; Schmitthenner, A.F. Anastomosis grouping and variation in virulence among isolates of Rhizoctonia solani associated with dry bean and soybean in Ohio and Zaire. Phytopathology 1993, 83, 438–444. [Google Scholar] [CrossRef]
- Rahman, M.T.; Bhuiyan, M.K.A.; Karim, M.A.; Rubayet, M.T. Screening of Soybean Resistance Genotypes Against Fusarium oxysporum, Macrophomina phaseolina, Rhizoctonia solani and Sclerotium rolfsii. Res. Agric. Vet. Sci. 2018, 2, 139–156. [Google Scholar]
- Yang, X.B.; Berggren, G.T.; Snow, J.P. Types of Rhizoctonia foliar blight on soybean in Louisiana. Plant Dis. 1990, 74, 501–504. [Google Scholar] [CrossRef]
- Nagendran, S.; Hammerschmidt, R.; McGrath, J.M. Identification of sugar beet germplasm EL51 as a source of resistance to post-emergence Rhizoctonia damping-off. Eur. J. Plant Pathol. 2009, 123, 461–471. [Google Scholar] [CrossRef]
- Panella, L.; Frese, L.; Srivastava, H.M.; Lange, W. Screening and utilizing Beta genetic resources with resistance to Rhizoctonia root rot and Cercospora leaf spot in a sugar beet breeding program. In Proceedings of the 4th International Beta Genetic Resources Workshop and World Beta Network Conference, Izmir, Turkey, 28 February–3 March 1996; International Plant Genetic Resources Institute: Rome, Italy, 1998; pp. 62–72. [Google Scholar]
- Scholten, O.E.; Panella, L.W.; De Bock, T.S.M.; Lange, W. A greenhouse test for screening sugar beet (Beta vulgaris) for resistance to Rhizoctonia solani. Eur. J. Plant Pathol. 2001, 107, 161–166. [Google Scholar] [CrossRef]
- Date, H.; Yagi, S.; Okamoto, Y.; Oniki, M. On the leaf blight of tomatoes by Thanatephorus cucumeris (Frank) Donk (Rhizoctonia solani). Ann. Phytopathol. Soc. Jpn. 1984, 50, 1375–1381. [Google Scholar]
- Nikraftar, F.; Taheri, P.; Rastegar, M.F.; Tarighi, S. Tomato partial resistance to Rhizoctonia solani involves antioxidative defense mechanisms. Physiol. Mol. Plant Pathol. 2013, 81, 74–83. [Google Scholar] [CrossRef]
- Taheri, P.; Tarighi, S. The role of pathogenesis-related proteins in the tomato-Rhizoctonia solani interaction. J. Bot. 2012. [Google Scholar] [CrossRef] [Green Version]
- Mahoney, A.K.; Babiker, E.M.; Paulitz, T.C.; See, D.; Okubara, P.A.; Hulbert, S.H. Characterizing and mapping resistance in synthetic-derived wheat to Rhizoctonia root rot in a green bridge environment. Phytopathology 2016, 106, 1170–1176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Okubara, P.A.; Leston, N.; Micknass, U.; Kogel, K.-H.; Imani, J. Rapid Quantitative Assessment of Rhizoctonia Resistance in Roots of Selected Wheat and Barley Genotypes. Plant Dis. 2016, 100, 640–644. [Google Scholar] [CrossRef] [Green Version]
- Coleman, J.J. The F usarium solani species complex: Ubiquitous pathogens of agricultural importance. Mol. Plant Pathol. 2016, 17, 146–158. [Google Scholar] [CrossRef] [PubMed]
- Foroud, N.A.; Chatterton, S.; Reid, L.M.; Turkington, T.K.; Tittlemier, S.A.; Gräfenhan, T. Fusarium diseases of Canadian grain crops: Impact and disease management strategies. In Future Challenges in Crop Protection Against Fungal Pathogens; Springer: Berlin/Heidelberg, Germany, 2014; pp. 267–316. [Google Scholar]
- Zhang, N.; O’Donnell, K.; Sutton, D.A.; Nalim, F.A.; Summerbell, R.C.; Padhye, A.A.; Geiser, D.M. Members of the Fusarium solani species complex that cause infections in both humans and plants are common in the environment. J. Clin. Microbiol. 2006, 44, 2186–2190. [Google Scholar] [CrossRef] [Green Version]
- Šišić, A.; Baćanović-Šišić, J.; Al-Hatmi, A.M.S.; Karlovsky, P.; Ahmed, S.A.; Maier, W.; De Hoog, G.S.; Finckh, M.R. The ‘forma specialis’ issue in Fusarium: A case study in Fusarium solani f. sp. pisi. Sci. Rep. 2018, 8, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.; Soni, S.K.; Awasthi, A.; Kalra, A. Evaluation of vermicompost doses for management of root-rot disease complex in Coleus forskohlii under organic field conditions. Australas. Plant Pathol. 2012, 41, 397–403. [Google Scholar] [CrossRef]
- Singh, R.; Kalra, A.; Ravish, B.S.; Divya, S.; Parameswaran, T.N.; Srinivas, K.; Bagyaraj, D.J. Effect of potential bioinoculants and organic manures on root-rot and wilt, growth, yield and quality of organically grown Coleus forskohlii in a semiarid tropical region of Bangalore (India). Plant Pathol. 2012, 61, 700–708. [Google Scholar] [CrossRef]
- Lops, F.; Cibelli, F.; Raimondo, M.L.; Carlucci, A. First report of stem wilt and root rot of Schlumbergera truncata caused by Fusarium oxysporum f. sp. Opuntiarum in Southern Italy. Plant Dis. 2013, 97, 846. [Google Scholar] [CrossRef] [PubMed]
- Cohen, R.; Orgil, G.; Burger, Y.; Saar, U.; Elkabetz, M.; Tadmor, Y.; Edelstein, M.; Belausov, E.; Maymon, M.; Freeman, S. Differences in the responses of melon accessions to fusarium root and stem rot and their colonization by Fusarium oxysporum f. sp. radicis-cucumerinum. Plant Pathol. 2015, 64, 655–663. [Google Scholar] [CrossRef]
- Miller-Garvin, J.E.; Viands, D.R. Selection for resistance to Fusarium root rot, and associations among resistances to six diseases in alfalfa. Crop Sci. 1994, 34, 1461–1465. [Google Scholar] [CrossRef]
- Linkmeyer, A.; Götz, M.; Hu, L.; Asam, S.; Rychlik, M.; Hausladen, H.; Hess, M.; Hückelhoven, R. Assessment and introduction of quantitative resistance to Fusarium head blight in elite spring barley. Phytopathology 2013, 103, 1252–1259. [Google Scholar] [CrossRef] [PubMed]
- Xue, A.; Ho, K.; Butler, G.; Vigier, B.; Babcock, C. Pathogenicity of Fusarium species causing head blight in barley. Phytoprotection 2006, 87, 55–61. [Google Scholar] [CrossRef] [Green Version]
- You, M.P.; Barbett, M.J.; Nichols, P.G.H. New sources of resistance identified in Trifolium subterraneum breeding lines and cultivars to root rot caused by Fusarium avenaceum and Pythium irregulare and their relationship to seedling survival. Australas. Plant Pathol. 2005, 34, 237–244. [Google Scholar] [CrossRef]
- Chittem, K.; Porter, L.; McPhee, K.; Khan, M.; Goswami, R.S. Fusarium avenaceum as causal agent of root rot in field peas and its control. Phytopathology 2010, 100, S25. [Google Scholar]
- Eranthodi, A.; Schneiderman, D.; Harris, L.J.; Witte, T.E.; Sproule, A.; Hermans, A.; Overy, D.P.; Chatterton, S.; Liu, J.; Li, T. Enniatin Production Influences Fusarium avenaceum Virulence on Potato Tubers, but not on Durum Wheat or Peas. Pathogens 2020, 9, 75. [Google Scholar] [CrossRef] [Green Version]
- Li, W.J.; Feng, J.; Chang, K.F.; Conner, R.L.; Hwang, S.F.; Strelkov, S.E.; Gossen, B.D.; McLaren, D.L. Microsatellite DNA markers indicate quantitative trait loci controlling resistance to pea root rot caused by Fusarium avenaceum (Corda ex Fries) Sacc. Plant Pathol. J. 2012, 11, 114–119. [Google Scholar] [CrossRef]
- Golinski, P.; Kaczmarek, Z.; Kiecana, I.; Wisniewska, H.; Kaptur, P.; Kostecki, M.; Chelkowski, J. Fusarium head blight of common Polish winter wheat cultivars–comparison of effects of Fusarium avenaceum and Fusarium culmorum on yield components. J. Phytopathol. 2002, 150, 135–141. [Google Scholar] [CrossRef]
- Wojciechowski, S.; Chelkowski, J.; Ponitka, A.; ŠLusarkiewicz-Jarzina, A. Evaluation of spring and winter wheat reaction to Fusarium culmorum and Fusarium avenaceum. J. Phytopathol. 1997, 145, 99–103. [Google Scholar] [CrossRef]
- Mesterhazy, A.; Bartók, T.; Kászonyi, G.; Varga, M.; Tóth, B.; Varga, J. Common resistance to different Fusarium spp. causing Fusarium head blight in wheat. Eur. J. Plant Pathol. 2005, 112, 267–281. [Google Scholar] [CrossRef]
- Bai, G.; Shaner, G. Management and resistance in wheat and barley to Fusarium head blight. Annu. Rev. Phytopathol. 2004, 42, 135–161. [Google Scholar] [CrossRef]
- Ūsele, G.; Beinarovica, I.; Mezaka, I.; Legzdina, L. Comparison of spring barley (Hordeum vulgare L.) screening methods for Fusarium head blight resistance breeding. Zemdirbyste-Agriculture 2013, 100, 317. [Google Scholar] [CrossRef] [Green Version]
- Gavrilova, O.P.; Gagkaeva, T.Y.; Loskutov, I.G. Screening of parent material for breeding oat varieties resistant to Fusarium disease and accumulation of mycotoxins in grain. Russ. Agric. Sci. 2012, 38, 33–35. [Google Scholar] [CrossRef]
- Browne, R.A.; Cooke, B.M. Resistance of wheat to Fusarium spp. in an in vitro seed germination assay and preliminary investigations into the relationship with Fusarium head blight resistance. Euphytica 2005, 141, 23–32. [Google Scholar] [CrossRef]
- Erginbas-Orakci, G.; Morgounov, A.; Dababat, A.A. Determination of resistance in winter wheat genotypes to the dryland root rots caused by Fusarium culmorum in Turkey. Int. J. Agric. Wildl. Sci. 2018, 4, 193–202. [Google Scholar] [CrossRef]
- Scholten, O.E.; Steenhuis-Broers, G.; Osman, A.; Bremer, E. Screening for resistance to Fusarium head blight in spring wheat cultivars. In Proceedings of the Joint Organic Congress, Odense, Denmark, 30–31 May 2006. [Google Scholar]
- Hori, K.; Kobayashi, T.; Sato, K.; Takeda, K. QTL analysis of Fusarium head blight resistance using a high-density linkage map in barley. Theor. Appl. Genet. 2005, 111, 1661–1672. [Google Scholar] [CrossRef]
- Mamo, B.E.; Steffenson, B.J. Genome-wide association mapping of Fusarium head blight resistance and agromorphological traits in barley landraces from Ethiopia and Eritrea. Crop Sci. 2015, 55, 1494–1512. [Google Scholar] [CrossRef]
- Massman, J.; Cooper, B.; Horsley, R.; Neate, S.; Dill-Macky, R.; Chao, S.; Dong, Y.; Schwarz, P.; Muehlbauer, G.J.; Smith, K.P. Genome-wide association mapping of Fusarium head blight resistance in contemporary barley breeding germplasm. Mol. Breed. 2011, 27, 439–454. [Google Scholar] [CrossRef]
- Ali, M.L.; Taylor, J.H.; Jie, L.; Sun, G.; William, M.; Kasha, K.J.; Reid, L.M.; Pauls, K.P. Molecular mapping of QTLs for resistance to Gibberella ear rot, in corn, caused by Fusarium graminearum. Genome 2005, 48, 521–533. [Google Scholar] [CrossRef]
- Asran, M.R.; Buchenauer, H. Pathogenicity of Fusarium graminearum isolates on maize (Zea mays L.) cultivars and relation with deoxynivalenol and ergosterol contents/Pathogenität von Fusarium graminearum Isolaten an Mais-(Zea mays L.) Sorten und Beziehung zu Deoxynivalenol-und Ergost. J. Plant Dis. Prot. 2003, 110, 209–219. [Google Scholar]
- du Toit, L.J.; Kirby, H.W.; Pedersen, W.L. Evaluation of an aeroponics system to screen maize genotypes for resistance to Fusarium graminearum seedling blight. Plant Dis. 1997, 81, 175–179. [Google Scholar] [CrossRef] [Green Version]
- Löffler, M.; Kessel, B.; Ouzunova, M.; Miedaner, T. Population parameters for resistance to Fusarium graminearum and Fusarium verticillioides ear rot among large sets of early, mid-late and late maturing European maize (Zea mays L.) inbred lines. Theor. Appl. Genet. 2010, 120, 1053–1062. [Google Scholar] [CrossRef] [PubMed]
- Silva, E.; Mora, E.A.; Medina, A.; Vásquez, J.; Valdez, D.; Danial, D.L.; Parlevliet, J.E. Fusarium ear rot and how to screen for resistance in open pollinated maize in the Andean regions. Euphytica 2007, 153, 329–337. [Google Scholar] [CrossRef]
- Acharya, B.; Lee, S.; Mian, M.A.R.; Jun, T.-H.; McHale, L.K.; Michel, A.P.; Dorrance, A.E. Identification and mapping of quantitative trait loci (QTL) conferring resistance to Fusarium graminearum from soybean PI 567301B. Theor. Appl. Genet. 2015, 128, 827–838. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ellis, M.L.; Wang, H.; Paul, P.A.; St. Martin, S.K.; McHale, L.K.; Dorrance, A.E. Identification of soybean genotypes resistant to Fusarium graminearum and genetic mapping of resistance quantitative trait loci in the cultivar Conrad. Crop Sci. 2012, 52, 2224–2233. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.X.; Xue, A.G.; Zhang, H.J.; Nagasawa, A.E.; Tambong, J.T. Response of soybean cultivars to root rot caused by Fusarium species. Can. J. Plant Sci. 2010, 90, 767–776. [Google Scholar] [CrossRef]
- Anderson, J.A.; Stack, R.W.; Liu, S.; Waldron, B.L.; Fjeld, A.D.; Coyne, C.; Moreno-Sevilla, B.; Fetch, J.M.; Song, Q.J.; Cregan, P.B.; et al. DNA markers for Fusarium head blight resistance QTLs in two wheat populations. Theor. Appl. Genet. 2001, 102, 1164–1168. [Google Scholar] [CrossRef]
- Campbell, K.A.G.; Lipps, P.E. Allocation of resources: Sources of variation in Fusarium head blight screening nurseries. Phytopathology 1998, 88, 1078–1086. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Del Blanco, I.; Frohberg, R.; Stack, R.; Berzonsky, W.; Kianian, S. Detection of QTL linked to Fusarium head blight resistance in Sumai 3-derived North Dakota bread wheat lines. Theor. Appl. Genet. 2003, 106, 1027–1031. [Google Scholar] [CrossRef]
- Fuentes, R.G.; Mickelson, H.R.; Busch, R.H.; Dill-Macky, R.; Evans, C.K.; Thompson, W.G.; Wiersma, J.V.; Xie, W.; Dong, Y.; Anderson, J.A. Resource allocation and cultivar stability in breeding for Fusarium head blight resistance in spring wheat. Crop Sci. 2005, 45, 1965–1972. [Google Scholar] [CrossRef]
- He, X.; Singh, P.K.; Duveiller, E.; Schlang, N.; Dreisigacker, S.; Singh, R.P. Identification and characterization of international Fusarium head blight screening nurseries of wheat at CIMMYT, Mexico. Eur. J. Plant Pathol. 2013, 136, 123–134. [Google Scholar] [CrossRef]
- Liu, Y.; Zheng, Y.; Wei, Y.; Zhou, M.; Liu, C. Genotypic differences to crown rot caused by Fusarium pseudograminearum in barley (Hordeum vulgare L.). Plant Breed. 2012, 131, 728–732. [Google Scholar] [CrossRef]
- Liu, Y.; Ma, J.; Yan, W.; Yan, G.; Zhou, M.; Wei, Y.; Zheng, Y.; Liu, C. Different tolerance in bread wheat, durum wheat and barley to Fusarium crown rot disease caused by Fusarium pseudograminearum. J. Phytopathol. 2012, 160, 412–417. [Google Scholar] [CrossRef]
- Poole, G.J.; Smiley, R.W.; Paulitz, T.C.; Walker, C.A.; Carter, A.H.; See, D.R.; Garland-Campbell, K. Identification of quantitative trait loci (QTL) for resistance to Fusarium crown rot (Fusarium pseudograminearum) in multiple assay environments in the Pacific Northwestern US. Theor. Appl. Genet. 2012, 125, 91–107. [Google Scholar] [CrossRef] [Green Version]
- Wildermuth, G.B.; Morgan, J.M. Genotypic differences in partial resistance to crown rot caused by Fusarium pseudograminearum in relation to an osmoregulation gene in wheat. Australas. Plant Pathol. 2004, 33, 121–123. [Google Scholar] [CrossRef] [Green Version]
- da Silva, W.L.; Clark, C.A. Infection of sweetpotato by Fusarium solani and Macrophomina phaseolina prior to harvest. Plant Dis. 2013, 97, 1636–1644. [Google Scholar] [CrossRef] [Green Version]
- Klingelfuss, L.H.; Yorinori, J.T.; Arias, C.A.A.; Destro, D. Reaction of soybean cultivars to sudden death syndrome and disease scoring methods for screening resistance. Embrapa Soja-Artigo em Periódico Indexado 2002, 2, 257–264. [Google Scholar] [CrossRef]
- Mueller, D.S.; Hartman, G.L.; Nelson, R.L.; Pedersen, W.L. Evaluation of Glycine max germ plasm for resistance to Fusarium solani f. sp. glycines. Plant Dis. 2002, 86, 741–746. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mueller, D.S.; Nelson, R.L.; Hartman, G.L.; Pedersen, W.L. Response of commercially developed soybean cultivars and the ancestral soybean lines to Fusarium solani f. sp. glycines. Plant Dis. 2003, 87, 827–831. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bilgi, V.N.; Bradley, C.A.; Khot, S.D.; Grafton, K.F.; Rasmussen, J.B. Response of dry bean genotypes to Fusarium root rot, caused by Fusarium solani f. sp. phaseoli, under field and controlled conditions. Plant Dis. 2008, 92, 1197–1200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hagerty, C.H.; Cuesta-Marcos, A.; Cregan, P.B.; Song, Q.; McClean, P.; Noffsinger, S.; Myers, J.R. Mapping Fusarium solani and Aphanomyces euteiches root rot resistance and root architecture quantitative trait loci in common bean. Crop Sci. 2015, 55, 1969–1977. [Google Scholar] [CrossRef] [Green Version]
- Schneider, K.A.; Grafton, K.F.; Kelly, J.D. QTL analysis of resistance to Fusarium root rot in bean. Crop Sci. 2001, 41, 535–542. [Google Scholar] [CrossRef]
- Hagedorn, D.J. Testing commercial Pea varieties for reaction to Fusarium root rot, Fusarium solani f. pisi. Phytopathology 1960, 50, 637. [Google Scholar]
- Porter, L.D.; Kraft, J.M.; Grünwald, N.J. Release of pea germplasm with Fusarium resistance combined with desirable yield and anti-lodging traits. J. Plant Regist. 2014, 8, 191–194. [Google Scholar] [CrossRef]
- Miedaner, T.; Bolduan, C.; Melchinger, A.E. Aggressiveness and mycotoxin production of eight isolates each of Fusarium graminearum and Fusarium verticillioides for ear rot on susceptible and resistant early maize inbred lines. Eur. J. Plant Pathol. 2010, 127, 113–123. [Google Scholar] [CrossRef]
- De Gruyter, J.; Aveskamp, M.M.; Woudenberg, J.H.C.; Verkley, G.J.M.; Groenewald, J.Z.; Crous, P.W. Molecular phylogeny of Phoma and allied anamorph genera: Towards a reclassification of the Phoma complex. Mycol. Res. 2009, 113, 508–519. [Google Scholar] [CrossRef] [PubMed]
- Kirkpatrick, T.L.; Rockroth, C.S. Compendium of Cotton Diseases; American Phytopathological Society (APS Press): St. Paul, MN, USA, 2001; ISBN 0890542791. [Google Scholar]
- Geldenhuis, M.M.; Roux, J.; Cilliers, A.J.; Wingfield, B.D.; Wingfield, M.J. Clonality in South African isolates and evidence for a European origin of the root pathogen Thielaviopsis basicola. Mycol. Res. 2006, 110, 306–311. [Google Scholar] [CrossRef] [PubMed]
- Coumans, J.V.F.; Poljak, A.; Raftery, M.J.; Backhouse, D.; Pereg-Gerk, L. Analysis of cotton (Gossypium hirsutum) root proteomes during a compatible interaction with the black root rot fungus Thielaviopsis basicola. Proteomics 2009, 9, 335–349. [Google Scholar] [CrossRef]
- Zaman, N.; Ahmed, S. Survey of root rot of groundnut in rainfed areas of Punjab, Pakistan. Afr. J. Biotechnol. 2012, 11, 4791–4794. [Google Scholar]
- Nandris, D.; Chadoeuf, J.; Pierrat, J.C.; Joannes, H.; Geiger, J.-P.; Nicole, M. Modelling rubber-tree root diseases, simulations of various inoculum rates and methods of control. Eur. J. For. Pathol. 1996, 26, 25–44. [Google Scholar] [CrossRef]
- Manjunatha, S.V.; Naik, M.K.; Khan, M.F.R.; Goswami, R.S. Evaluation of bio-control agents for management of dry root rot of chickpea caused by Macrophomina phaseolina. Crop Prot. 2013, 45, 147–150. [Google Scholar] [CrossRef]
- Thomidis, T.; Exadaktylou, E. Effectiveness of cyproconazole to control Armillaria root rot of apple, walnut and kiwifruit. Crop Prot. 2012, 36, 49–51. [Google Scholar] [CrossRef]
- Aguín-Casal, O.; Sáinz-Osés, M.J.; Mansilla-Vázquez, J.P. Armillaria species infesting vineyards in northwestern Spain. Eur. J. Plant Pathol. 2004, 110, 683–687. [Google Scholar] [CrossRef]
- Bhat, Z.A.; Sheikh, F.A.; Mubarak, T.; Bhat, J.A.; Zargar, M.A.; Wani, A.A.; Rather, G.H.; Itoo, H.U. On Farm Testing and Popularization of Integrated Management Module of Apple Root Rot Under High Altitude Temperate Conditions. J. Krishi Vigyan 2012, 1, 54–57. [Google Scholar]
- Sánchez, M.A.G.; Cazorla, F.M.; Cayo, R.; de Vicente, A.; Jiménez, R.M.P. Studies of soil and rhizosphera bacteria to improve biocontrol of avocado white root rot caused by Rosellinia necatrix. S. Michele all’Adige Italy 2004, 27, 169–172. [Google Scholar]
- Bugbee, W.M.; Campbell, L.G. Combined resistance in sugar beet to Rhizoctonia solani, Phoma betae, and Botrytis cinerea. Plant Dis. 1990, 74, 353. [Google Scholar] [CrossRef]
- Mao, W.; Carroll, R.B.; Whittington, D.P. Association of Phoma terrestris, Pythium irregulare, and Fusarium acuminatum in causing red root rot of corn. Plant Dis. 1998, 82, 337–342. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coleman, P.M.; Ellerbrock, L.A.; Lorbeer, J.W. Reaction of selected onion cultigens to pink root under field conditions in New York. Plant Dis. 1997, 81, 138–142. [Google Scholar] [CrossRef] [PubMed]
- Wiriyajitsomboon, P. Characterization of Setophoma terrestris Causing Pink Root in Onion, Disease Management, and Age-Related Resistance; Michigan State University: East Lansing, MI, USA, 2015; ISBN 1339303280. [Google Scholar]
- Hollingsworth, C.R.; Gray, F.A.; Groose, R.W. Evidence for the heritability of resistance to brown root rot of alfalfa, caused by Phoma sclerotioides. Can. J. Plant Pathol. 2005, 27, 64–70. [Google Scholar] [CrossRef]
- Berkenkamp, B.; McCartney, D.; Bittman, S. Resistance of alfalfa cultivars to brown root rot. Can. J. Plant Sci. 1991, 71, 211–213. [Google Scholar] [CrossRef] [Green Version]
- Kumar, V.; Joshi, S.G.; Bell, A.A.; Rathore, K.S. Enhanced resistance against Thielaviopsis basicola in transgenic cotton plants expressing Arabidopsis NPR1 gene. Transgenic Res. 2013, 22, 359–368. [Google Scholar] [CrossRef] [PubMed]
- Rajasekaran, K.; Cary, J.W.; Jaynes, J.M.; Cleveland, T.E. Disease resistance conferred by the expression of a gene encoding a synthetic peptide in transgenic cotton (Gossypium hirsutum L.) plants. Plant Biotechnol. J. 2005, 3, 545–554. [Google Scholar] [CrossRef] [PubMed]
- Wheeler, T.A.; Gannaway, J.R. Identification of germplasm resistant to Thielaviopsis basicola in the USDA cotton germplasm collection. In Proceedings of the World Cotton Conference-4, Lubbock, TX, USA, 10–14 September 2007; pp. 10–14. [Google Scholar]
- Wheeler, T.A.; Gannaway, J.R.; Keating, K. Identification of resistance to Thielaviopsis basicola in diploid cotton. Plant Dis. 1999, 83, 831–833. [Google Scholar] [CrossRef] [Green Version]
- Bai, D.; Reeleder, R.; Brandie, J.E. Identification of two RAPD markers tightly linked with the Nicotiana debneyi gene for resistance to black root rot of tobacco. Theor. Appl. Genet. 1995, 91, 1184–1189. [Google Scholar] [CrossRef]
- Trojak-Goluch, A.; Berbeć, A. Potential of Nicotiana glauca (Grah.) as a source of resistance to black root rot Thielaviopsis basicola (Berk. and Broome) Ferr. in tobacco improvement. Plant Breed. 2005, 124, 507–510. [Google Scholar] [CrossRef]
- Harper, J.T.; Waanders, E.; Keeling, P.J. On the monophyly of chromalveolates using a six-protein phylogeny of eukaryotes. Int. J. Syst. Evol. Microbiol. 2005, 55, 487–496. [Google Scholar] [CrossRef] [Green Version]
- van West, P.; Appiah, A.A.; Gow, N.A.R. Advances in research on oomycete root pathogens. Physiol. Mol. Plant Pathol. 2003, 62, 99–113. [Google Scholar] [CrossRef]
- Chamnanpunt, J.; Shan, W.; Tyler, B.M. High frequency mitotic gene conversion in genetic hybrids of the oomycete Phytophthora sojae. Proc. Natl. Acad. Sci. USA 2001, 98, 14530–14535. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fry, W.E.; Goodwin, S.B. Re-emergence of potato and tomato late blight in the United States. Plant Dis. 1997, 81, 1349–1357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schettini, T.M.; Legg, E.J.; Michelmore, R.W. Insensitivity to metalaxyl in California populations of Bremia lactucae and resistance of California lettuce cultivars to downy mildew. Phytopathology 1991, 81, 64–70. [Google Scholar] [CrossRef]
- Harveson, R.M.; Rush, C.M. The influence of irrigation frequency and cultivar blends on the severity of multiple root diseases in sugar beets. Plant Dis. 2002, 86, 901–908. [Google Scholar] [CrossRef]
- Akamatsu, H.O.; Grünwald, N.J.; Chilvers, M.I.; Porter, L.D.; Peever, T.L. Development of codominant simple sequence repeat, single nucleotide polymorphism and sequence characterized amplified region markers for the pea root rot pathogen, Aphanomyces euteiches. J. Microbiol. Methods 2007, 71, 82–86. [Google Scholar] [CrossRef] [PubMed]
- Taguchi, K.; Okazaki, K.; Takahashi, H.; Kubo, T.; Mikami, T. Molecular mapping of a gene conferring resistance to Aphanomyces root rot (black root) in sugar beet (Beta vulgaris L.). Euphytica 2010, 173, 409–418. [Google Scholar] [CrossRef]
- Campbell, L.G.; Klotz, K.L. Postharvest storage losses associated with Aphanomyces root rot in sugarbeet. J. Sugarbeet Res. 2006, 43, 113–128. [Google Scholar] [CrossRef] [Green Version]
- Taguchi, K.; Ogata, N.; Kubo, T.; Kawasaki, S.; Mikami, T. Quantitative trait locus responsible for resistance to Aphanomyces root rot (black root) caused by Aphanomyces cochlioides Drechs. in sugar beet. Theor. Appl. Genet. 2009, 118, 227–234. [Google Scholar] [CrossRef]
- Saunders, J.W.; Mcgrath, J.M.; Theurer, J.C.; Halloin, J.M. Registration ofSR87′Sugarbeet Germplasm with Low Tare and Resistances to Cereospora and Aphanomyces. Crop Sci. 2000, 40, 1833. [Google Scholar]
- Windels, C.E. Aphanomyces root rot on sugar beet. Plant Health Prog. 2000, 1, 8. [Google Scholar] [CrossRef]
- Fitzpatrick, S.; Brummer, J.; Hudelson, B.; Malvick, D.; Grau, C. Aphanomyces root rot resistance (Races 1 and 2). In Proceedings of the North American Alfalfa Improvement Conference, Bozeman, MT, USA, 2–6 August 1998. [Google Scholar]
- Grau, C.R.; Muehlchen, A.M.; Tofte, J.E.; Smith, R.R. Variability in virulence of Aphanomyces euteiches. Plant Dis. 1991, 75, 1153–1156. [Google Scholar] [CrossRef]
- McGee, R.J.; Coyne, C.J.; Pilet-Nayel, M.-L.; Moussart, A.; Tivoli, B.; Baranger, A.; Hamon, C.; Vandemark, G.; McPhee, K. Registration of pea germplasm lines partially resistant to aphanomyces root rot for breeding fresh or freezer pea and dry pea types. J. Plant Regist. 2012, 6, 203–207. [Google Scholar] [CrossRef]
- Pfender, W.F.; Hagedorn, D.J. Aphanomyces euteiches f. sp. phaseoli, a causal agent of bean root and hypocotyl rot. Phytopathology 1982, 72, 306–310. [Google Scholar]
- Fagoaga, C.; Rodrigo, I.; Conejero, V.; Hinarejos, C.; Tuset, J.J.; Arnau, J.; Pina, J.A.; Navarro, L.; Peña, L. Increased tolerance to Phytophthora citrophthora in transgenic orange plants constitutively expressing a tomato pathogenesis related protein PR-5. Mol. Breed. 2001, 7, 175–185. [Google Scholar] [CrossRef]
- Matheron, M.E.; Wright, G.C.; Porchas, M. Resistance to Phytophthora citrophthora and P. parasitica and nursery characteristics of several citrus rootstocks. Plant Dis. 1998, 82, 1217–1225. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Park, B.; Veeraraghavan, N.; Jung, K.; Lee, Y.-H.; Blair, J.E.; Geiser, D.M.; Isard, S.; Mansfield, M.A.; Nikolaeva, E.; et al. Phytophthora database: A forensic database supporting the identification and monitoring of Phytophthora. Plant Dis. 2008, 92, 966–972. [Google Scholar] [CrossRef] [Green Version]
- Thomidis, T.; Exadaktylou, E.; Sotiropoulos, T. Susceptibility of three citrus rootstocks towards Phytophthora cactorum, P. citrophthora, P. parasitica and P. citricola/Anfälligkeit dreier Citrus-Veredelungsunterlagen gegenüber Phytophthora cactorum, P. citrophthora, P. parasitica und P. citricola. J. Plant Dis. Prot. 2005, 112, 204–207. [Google Scholar]
- Tuzcu, Ö.; Cinar, A.; Göksedef, M.O.; Özsan, M.; Biçici, M. Resistance of citrus rootstocks to Phytophthora citrophthora during winter dormancy. Plant Dis. 1984, 68, 502–505. [Google Scholar] [CrossRef]
- Yildirim, B.; Yeşiloğlu, T.; Incesu, M. Fruit yield and quality of Santa Teresa lemon on seven rootstocks in Adana (Turkey). Afr. J. Agric. Res. 2010, 5, 1077–1081. [Google Scholar]
- Azevedo, F.A.; Mourão Filho, F.A.A.; Mendes, B.M.J.; Almeida, W.A.B.; Schinor, E.H.; Pio, R.; Barbosa, J.M.; Guidetti-Gonzalez, S.; Carrer, H.; Lam, E. Genetic transformation of Rangpur lime (Citrus limonia osbeck) with thebO (bacterio-opsin) genen and its initial evaluation for Phytophthora nicotianae resistance. Plant Mol. Biol. Rep. 2006, 24, 185–196. [Google Scholar] [CrossRef]
- Benfradj, N.; Metoui, N.; Boughalleb-M’Hamdi, N. Screening for tolerance of different citrus rootstocks against zoospores of Phytophthora nicotianae in infested soil. J. Phytopathol. Pest. Manag. 2016, 3, 63–75. [Google Scholar]
- Graham, J.H. Root regeneration and tolerance of citrus rootstocks to root rot caused by Phytophthora nicotianae. Phytopathology 1995, 85, 111–117. [Google Scholar] [CrossRef] [Green Version]
- Sakupwanya, M.N.; Labuschagne, N.; Loots, T.; Apostolides, Z. Towards developing a metabolic-marker based predictive model for Phytophthora nicotianae tolerance in citrus rootstocks. J. Plant Pathol. 2018, 100, 269–277. [Google Scholar] [CrossRef]
- Washington, W.S.; McGee, P.; Flett, S.P.; Jerie, P.H.; Ashcroft, W.J. Cultivars and fungicides affect Phytophthora root rot in processing tomatoes. Australas. Plant Pathol. 2001, 30, 309–315. [Google Scholar] [CrossRef]
- Carisse, O.; Khanizadeh, S. Relative resistance of newly released apple rootstocks to Phytophthora cactorum. Can. J. Plant Sci. 2006, 86, 199–204. [Google Scholar] [CrossRef]
- Eikemo, H.; Brurberg, M.B.; Davik, J. Resistance to Phytophthora cactorum in diploid Fragaria species. HortScience 2010, 45, 193–197. [Google Scholar] [CrossRef] [Green Version]
- Mclntosh, E.D.L. Proceedings of the 1974 APDW workshop on crown rot of apple trees. Can. Plant Dis. Surv. 1975, 55, 109–116. [Google Scholar]
- Mangandi, J.; Verma, S.; Osorio, L.; Peres, N.A.; van de Weg, E.; Whitaker, V.M. Pedigree-based analysis in a multiparental population of octoploid strawberry reveals QTL alleles conferring resistance to Phytophthora cactorum. G3 Genes Genomes Genet. 2017, 7, 1707–1719. [Google Scholar] [CrossRef] [Green Version]
- Sewell, G.W.F.; Wilson, J.F. Resistance Trials of Some Apple Rootstock Varieties to Phytophthora cactorum (L. & C.) Schroet. J. Hortic. Sci. 1959, 34, 51–58. [Google Scholar]
- Utkhede, R.S.; Quamme, H.A. Use of the excised shoot assay to evaluate resistance to Phytophthora cactorum of apple rootstock cultivars. Can. J. Plant Sci. 1988, 68, 851–857. [Google Scholar] [CrossRef]
- Douhan, G.W.; Fuller, E.; McKee, B.; Pond, E. Genetic diversity analysis of avocado (Persea americana Miller) rootstocks selected under greenhouse conditions for tolerance to phytophthora root rot caused by Phytophthora cinnamomi. Euphytica 2011, 182, 209. [Google Scholar] [CrossRef]
- Haymes, K.M.; Van de Weg, W.E.; Arens, P.; Maas, J.L.; Vosman, B.; Den Nijs, A.P.M. Development of SCAR markers linked to a Phytophthora fragariae resistance gene and their assessment in European and North American strawberry genotypes. J. Am. Soc. Hortic. Sci. 2000, 125, 330–339. [Google Scholar] [CrossRef] [Green Version]
- Rugienius, R.; Siksnianas, T.; Stanys, V.; Gelvonauskiene, D.; Bendokas, V. Use of RAPD and SCAR markers for identification of strawberry genotypes carrying red stele (Phytophtora fragariae) resistance gene Rpf1. Agron. Res. 2006, 4, 335–339. [Google Scholar]
- Pattison, J.A.; Wilcox, W.F.; Weber, C.A. Assessing the resistance of red raspberry (Rubus idaeus L.) genotypes to Phytophthora fragariae var. rubi in hydroponic culture. HortScience 2004, 39, 1553–1556. [Google Scholar] [CrossRef]
- Weber, C.A.; Pattison, J.; Samuelian, S. Marker assisted selection for resistance to root rot in red raspberry caused by Phytophthora fragariae var. rubi. In Proceedings of the IX International Rubus and Ribes Symposium, Pucón, Chile, 1–7 December 2005; Volume 777, pp. 311–316. [Google Scholar]
- Burnham, K.D.; Dorrance, A.E.; VanToai, T.T.; St. Martin, S.K. Quantitative trait loci for partial resistance to Phytophthora sojae in soybean. Crop Sci. 2003, 43, 1610–1617. [Google Scholar] [CrossRef] [Green Version]
- Dorrance, A.E.; Schmitthenner, A.F. New sources of resistance to Phytophthora sojae in the soybean plant introductions. Plant Dis. 2000, 84, 1303–1308. [Google Scholar] [CrossRef] [Green Version]
- Bosland, P.W.; Lindsey, D.L. A seedling screen for Phytophthora root rot of pepper, Capsicum annuum. Plant Dis. 1991, 75, 1048–1050. [Google Scholar] [CrossRef]
- Kim, H.-J.; Nahm, S.-H.; Lee, H.-R.; Yoon, G.-B.; Kim, K.-T.; Kang, B.-C.; Choi, D.; Kweon, O.Y.; Cho, M.-C.; Kwon, J.-K.; et al. BAC-derived markers converted from RFLP linked to Phytophthora capsici resistance in pepper (Capsicum annuum L.). Theor. Appl. Genet. 2008, 118, 15. [Google Scholar] [CrossRef]
- Ogundiwin, E.A.; Berke, T.F.; Massoudi, M.; Black, L.L.; Huestis, G.; Choi, D.; Lee, S.; Prince, J.P. Construction of 2 intraspecific linkage maps and identification of resistance QTLs for Phytophthora capsici root-rot and foliar-blight diseases of pepper (Capsicum annuum L.). Genome 2005, 48, 698–711. [Google Scholar] [CrossRef]
- Cerra, S.M. Phytophthora root and stem rot of soybean in Iowa: Minimizing losses through an improved understanding of population structure and implementation of novel management strategies. Master’s Thesis, Iowa State University, Ames, IA, USA, 2007. [Google Scholar]
- Dale, M.L.; Irwin, J.A.G. Glasshouse and field screening of chickpea cultivars for resistance to Phytophthora megasperma f.sp. medicaginis. Aust. J. Exp. Agric. 1991, 31, 663–667. [Google Scholar] [CrossRef]
- Knights, E.J.; Southwell, R.J.; Schwinghamer, M.W.; Harden, S. Resistance to Phytophthora medicaginis Hansen and Maxwell in wild Cicer species and its use in breeding root rot resistant chickpea (Cicer arietinum L.). Aust. J. Agric. Res. 2008, 59, 383–387. [Google Scholar] [CrossRef]
- Vandemark, G.J.; Barker, B.M. Quantifying Phytophthora medicaginis in susceptible and resistant alfalfa with a real-time fluorescent PCR assay. J. Phytopathol. 2003, 151, 577–583. [Google Scholar] [CrossRef] [Green Version]
- Wiersma, D.W.; Grau, C.R.; Undersander, D.J. Alfalfa cultivar performance with differing levels of resistance to Phytophthora and Aphanomyces root rots. J. Prod. Agric. 1995, 8, 259–264. [Google Scholar] [CrossRef]
- Abrinbana, M.; Babai-Ahary, A.; Heravan, I.M. Assessment of resistance in sugarbeet lines to damping-off caused by Pythium ultimum Trow var. ultimum under greenhouse conditions. Plant Pathol. J. 2007, 6, 266–270. [Google Scholar]
- Bates, G.D.; Rothrock, C.S.; Rupe, J.C. Resistance of the Soybean Cultivar Archer to Pythium Damping-Off and Root Rot Caused by Several Pythium spp. Plant Dis. 2008, 92, 763–766. [Google Scholar] [CrossRef] [Green Version]
- Balk, C.S. Assessment of resistance in soybean to Pythium ultimum and sensitivity of Ohio’s diverse Pythium species towards metalaxyl 2014. Master’s Thesis, Ohio State University, Columbus, OH, USA, 2014. [Google Scholar]
- Cheng, L. Pythium Ultimum; NC State University Depepartment of Plant Pathology: Raleigh, NC, USA, 2007. [Google Scholar]
- Lucas, B.; Griffiths, P.D. Evaluation of common bean accessions for resistance to Pythium ultimum. HortScience 2004, 39, 1193–1195. [Google Scholar] [CrossRef] [Green Version]
- Ellis, M.L.; McHale, L.K.; Paul, P.A.; St Martin, S.K.; Dorrance, A.E. Soybean germplasm resistant to Pythium irregulare and molecular mapping of resistance quantitative trait loci derived from the soybean accession PI 424354. Crop Sci. 2013, 53, 1008–1021. [Google Scholar] [CrossRef]
- Farr, D.F.; Rossman, A.Y.; Palm, M.E.; McCray, E.B. Fungal Databases, Systematic Botany and Mycology Laboratory; Agricultural Research Service, US Department of Agriculture: Washington, DC, USA, 2007.
- Katawczik, M. Pythium Irregulare; NC State University Depepartment of Plant Pathology: Raleigh, NC, USA, 2008. [Google Scholar]
- Aliyu, T.H.; Balogun, O.S.; Adesina, O.M. Effect of Pythium Aphanidermatum on Two Cultivars of Pepper (Capsicum spp.). Preprints 2012. [Google Scholar] [CrossRef]
- Fattahi, S.H.; Zafari, D.; Mahmoudi, B. Evaluation of superior sugar beet genotypes for resistance to important root rot pathogens in the greenhouse. J. Sugar Beet. 2011, 27, 25–38. [Google Scholar]
- Mahmoudi, S.B.; Koulaei, H.E.; Hasani, M.; Alaghebandzade, N.; Soltani, J.; Kakueinezhad, M. Developement of sugar beet S1 pollinator lines resistant to Pythium root rot. In Proceedings of the 1st International and 13th Iranian Crop Science Congress 3rd Iranian Seed Science and Technology Conference, Karaj, Iran, 26–28 August 2014. [Google Scholar]
- Parker, K.C. Pythium Aphanidermatum. Soilborne Plant Pathology; NC State University: Raleigh, NC, USA, 2009; Volume 20. [Google Scholar]
- Rosso, M.L.; Rupe, J.C.; Chen, P.; Mozzoni, L.A. Inheritance and genetic mapping of resistance to Pythium damping-off caused by Pythium aphanidermatum in ‘Archer’soybean. Crop Sci. 2008, 48, 2215–2222. [Google Scholar] [CrossRef]
- Richard, C.; Beghdadi, A.; Martin, J.G. Aphanomyces euteiches, a novel root pathogen to alfalfa in Québec. Plant Dis. 1991, 75. [Google Scholar] [CrossRef]
- Tofte, J.E.; Smith, R.R.; Grau, C.R. Reaction of red clover to Aphanomyces euteiches. Plant Dis. 1992, 76, 39–42. [Google Scholar] [CrossRef]
- Van Leur, J.A.G.; Southwell, R.J.; Mackie, J.M. Aphanomyces root rot on faba bean in northern NSW. Australas. Plant Dis. Notes 2008, 3, 8–9. [Google Scholar] [CrossRef]
- Chen, W.; Sharma, H.C.; Muehlbauer, F.J. Compendium of Chickpea and Lentil Diseases and Pests; American Phytopathological Society (APS Press): St. Paul, MN, USA, 2011; ISBN 0890543836. [Google Scholar]
- Beckerman, J. Disease Management Strategies for Horticultural Crops: Phytium Root rot of Herbaceous Plants; Purdue University: West Lafayette, Indiana, 2011. [Google Scholar]
- Nzungize, J.; Gepts, P.; Buruchara, R.; Male, A.; Ragama, P.; Busogoro, J.P.; Baudoin, J.-P. Introgression of Pythium root rot resistance gene into Rwandan susceptible common bean cultivars. Afr. J. Plant Sci. 2011, 5, 193–200. [Google Scholar]
- Broders, K.D.; Wallhead, M.W.; Austin, G.D.; Lipps, P.E.; Paul, P.A.; Mullen, R.W.; Dorrance, A.E. Association of soil chemical and physical properties with Pythium species diversity, community composition, and disease incidence. Phytopathology 2009, 99, 957–967. [Google Scholar] [CrossRef] [Green Version]
- Broders, K.D.; Lipps, P.E.; Paul, P.A.; Dorrance, A.E. Characterization of Pythium spp. associated with corn and soybean seed and seedling disease in Ohio. Plant Dis. 2007, 91, 727–735. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Menzies, J.G.; Ehret, D.L.; Stan, S. Effect of inoculum density of Pythium aphanidermatum on the growth and yield of cucumber plants grown in recirculating nutrient film culture. Can. J. Plant Pathol. 1996, 18, 50–54. [Google Scholar] [CrossRef]
- Utkhede, R.S.; Koch, C.A. Rhizobacterial growth and yield promotion of cucumber plants inoculated with Pythium aphanidermatum. Can. J. Plant Pathol. 1999, 21, 265–271. [Google Scholar] [CrossRef]
- Utkhede, R.S.; Levesque, C.A.; Dinh, D. Pythium aphanidermatum root rot in hydroponically grown lettuce and the effect of chemical and biological agents on its control. Can. J. Plant Pathol. 2000, 22, 138–144. [Google Scholar] [CrossRef]
- Kakueinezhad, M.; Taheri, P.; Mahmoudi, S.B.; Tarighi, S. Resistance assessment and biochemical responses of sugar beet lines against Pythium aphanidermatum, causing root rot. Eur. J. Plant Pathol. 2018, 151, 307–319. [Google Scholar] [CrossRef]
- Petkowski, J.E.; de Boer, R.F.; Norng, S.; Thomson, F.; Minchinton, E.J. Pythium species associated with root rot complex in winter-grown parsnip and parsley crops in south eastern Australia. Australas. Plant Pathol. 2013, 42, 403–411. [Google Scholar] [CrossRef]
- Mavrodi, O.V.; Walter, N.; Elateek, S.; Taylor, C.G.; Okubara, P.A. Suppression of Rhizoctonia and Pythium root rot of wheat by new strains of Pseudomonas. Biol. Control. 2012, 62, 93–102. [Google Scholar] [CrossRef]
- Dissanayake, N.; Hoy, J.W.; Griffin, J.L. Herbicide effects on sugarcane growth, Pythium root rot, and Pythium arrhenomanes. Phytopathology 1998, 88, 530–535. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elmer, W.H.; Gent, M.P.N.; McAvoy, R.J. Partial saturation under ebb and flow irrigation suppresses Pythium root rot of ornamentals. Crop Prot. 2012, 33, 29–33. [Google Scholar] [CrossRef]
- Hardham, A.R.; Blackman, L.M. Molecular cytology of Phytophthora-plant interactions. Australas. Plant Pathol. 2010, 39, 29–35. [Google Scholar] [CrossRef]
- Kroon, L.P.N.M.; Brouwer, H.; de Cock, A.W.A.M.; Govers, F. The genus Phytophthora anno 2012. Phytopathology 2012, 102, 348–364. [Google Scholar] [CrossRef] [Green Version]
- Dirac, M.F.; Menge, J.A. High temperatures are not responsible for lack of infection of citrus roots by Phytophthora citrophthora during the summer, but suppressive soil microorganisms may inhibit infection by P. citrophthora. Plant Soil 2002, 241, 243–249. [Google Scholar] [CrossRef]
- Alvarez, L.A.; Vicent, A.; De la Roca, E.; Bascón, J.; Abad-Campos, P.; Armengol, J.; García-Jiménez, J. Branch cankers on citrus trees in Spain caused by Phytophthora citrophthora. Plant Pathol. 2008, 57, 84–91. [Google Scholar] [CrossRef]
- Graham, J.H.; Menge, J.A. Root diseases. Citrus Health Manag. 1999, 126–135. [Google Scholar]
- Bekker, T.F.; Kaiser, C.; Labuschagne, N. Efficacy of water soluble silicon against Phytophthora cinnamomi root rot of avocado: A progress report. S. Afr. Avocado Grow. Assoc. Yearb. 2006, 29, 58–62. [Google Scholar]
- Anderson, J.M.; Pegg, K.G.; Scott, C.; Drenth, A. Phosphonate applied as a pre-plant dip controls Phytophthora cinnamomi root and heart rot in susceptible pineapple hybrids. Australas. Plant Pathol. 2012, 41, 59–68. [Google Scholar] [CrossRef]
- Pérombelon, M.C.M. Potato diseases caused by soft rot erwinias: An overview of pathogenesis. Plant Pathol. 2002, 51, 1–12. [Google Scholar] [CrossRef]
- Zhao, Y.; Li, P.; Huang, K.; Wang, Y.; Hu, H.; Sun, Y. Control of postharvest soft rot caused by Erwinia carotovora of vegetables by a strain of Bacillus amyloliquefaciens and its potential modes of action. World J. Microbiol. Biotechnol. 2013, 29, 411–420. [Google Scholar] [CrossRef] [PubMed]
- Collmer, A.; Keen, N.T. The role of pectic enzymes in plant pathogenesis. Annu. Rev. Phytopathol. 1986, 24, 383–409. [Google Scholar] [CrossRef]
- Charkowski, A.O. The soft rot Erwinia. In Plant-Associated Bacteria; Springer: Berlin/Heidelberg, Germany, 2007; pp. 423–505. [Google Scholar]
- Liao, C.-H. Analysis of pectate lyases produced by soft rot bacteria associated with spoilage of vegetables. Appl. Environ. Microbiol. 1989, 55, 1677–1683. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, L.F.; Fang, B.P.; Luo, Z.X.; Chen, J.Y.; Zhang, X.J.; Wang, Z.Y. First report of bacterial stem and root rot of sweetpotato caused by a Dickeya sp.(Erwinia chrysanthemi) in China. Plant Dis. 2010, 94, 1503. [Google Scholar] [CrossRef]
- Muimba-Kankolongo, A. Food Crop. Production by Smallholder Farmers in Southern Africa: Challenges and Opportunities for Improvement; Academic Press: Cambridge, MA, USA, 2018; ISBN 0128143843. [Google Scholar]
- Bigirimana, S.; Barumbanze, P.; Ndayihanzamaso, P.; Shirima, R.; Legg, J.P. First report of cassava brown streak disease and associated Ugandan cassava brown streak virus in Burundi. New Dis. Rep. 2011, 24, 588–2044. [Google Scholar] [CrossRef] [Green Version]
- Anjanappa, R.B.; Mehta, D.; Maruthi, M.N.; Kanju, E.; Gruissem, W.; Vanderschuren, H. Characterization of brown streak virus–resistant cassava. Mol. Plant-Microbe Interact. 2016, 29, 527–534. [Google Scholar] [CrossRef] [Green Version]
- Kaweesi, T.; Kawuki, R.; Kyaligonza, V.; Baguma, Y.; Tusiime, G.; Ferguson, M.E. Field evaluation of selected cassava genotypes for cassava brown streak disease based on symptom expression and virus load. Virol. J. 2014, 11, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Ogwok, E.; Odipio, J.; Halsey, M.; Gaitán-Solís, E.; Bua, A.; Taylor, N.J.; Fauquet, C.M.; Alicai, T. Transgenic RNA interference (RNA i)-derived field resistance to cassava brown streak disease. Mol. Plant Pathol. 2012, 13, 1019–1031. [Google Scholar] [CrossRef]
- Beyene, G.; Chauhan, R.D.; Ilyas, M.; Wagaba, H.; Fauquet, C.M.; Miano, D.; Alicai, T.; Taylor, N.J. A virus-derived stacked RNAi construct confers robust resistance to cassava brown streak disease. Front. Plant Sci. 2017, 7, 2052. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yadav, J.S.; Ogwok, E.; Wagaba, H.; Patil, B.L.; Bagewadi, B.; Alicai, T.; Gaitan-Solis, E.; Taylor, N.J.; Fauquet, C.M. RNAi-mediated resistance to Cassava brown streak Uganda virus in transgenic cassava. Mol. Plant Pathol. 2011, 12, 677–687. [Google Scholar] [CrossRef]
- Grant, M.; Lamb, C. Systemic immunity. Curr. Opin. Plant Biol. 2006, 9, 414–420. [Google Scholar] [CrossRef] [PubMed]
- Takemoto, D.; Jones, D.A. Particle bombardment-mediated transient expression to identify localization signals in plant disease resistance proteins and target sites for the proteolytic activity of pathogen effectors. In Plant-Pathogen Interactions; Springer: Berlin/Heidelberg, Germany, 2014; pp. 91–101. [Google Scholar]
- Hammond-Kosack, K.E.; Jones, J.D.G. Plant disease resistance genes. Annu. Rev. Plant Biol. 1997, 48, 575–607. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harris, C.J.; Slootweg, E.J.; Goverse, A.; Baulcombe, D.C. Stepwise artificial evolution of a plant disease resistance gene. Proc. Natl. Acad. Sci. USA 2013, 110, 21189–21194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gaskill, J.O. Breeding for Rhizoctonia resistance in sugarbeet. J. Am. Sugar Beet Technol 1968, 15, 107–119. [Google Scholar] [CrossRef]
- Panella, L. Registration of FC709-2 and FC727 sugarbeet germplasms resistant to Rhizoctonia root rot and Cercospora leaf spot. Crop Sci. 1999, 39, 298–299. [Google Scholar] [CrossRef]
- Lein, J.C.; Sagstetter, C.M.; Schulte, D.; Thurau, T.; Varrelmann, M.; Saal, B.; Koch, G.; Borchardt, D.C.; Jung, C. Mapping of rhizoctonia root rot resistance genes in sugar beet using pathogen response-related sequences as molecular markers. Plant Breed. 2008, 127, 602–611. [Google Scholar] [CrossRef]
- Hecker, R.J.; Ruppel, E.G. Inheritance of Resistance to Rhizoctonia Root Rot in Sugarbeet 1. Crop Sci. 1975, 15, 487–490. [Google Scholar] [CrossRef]
- Peng, H.; Chen, Z.; Fang, Z.; Zhou, J.; Xia, Z.; Gao, L.; Chen, L.; Li, L.; Li, T.; Zhai, W.; et al. Rice Xa21 primed genes and pathways that are critical for combating bacterial blight infection. Sci. Rep. 2015, 5, 12165. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seo, Y.-S.; Chern, M.; Bartley, L.E.; Han, M.; Jung, K.-H.; Lee, I.; Walia, H.; Richter, T.; Xu, X.; Cao, P.; et al. Towards establishment of a rice stress response interactome. PLoS Genet. 2011, 7, e1002020. [Google Scholar] [CrossRef]
- Peng, Y.; Bartley, L.E.; Canlas, P.; Ronald, P.C. OsWRKY IIa transcription factors modulate rice innate immunity. Rice 2010, 3, 36–42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, J.; Choi, D.; Lee, S.; Ryu, C.-M.; Hwang, I. Cytokinins and plant immunity: Old foes or new friends? Trends Plant Sci. 2011, 16, 388–394. [Google Scholar] [CrossRef]
- Argueso, C.T.; Ferreira, F.J.; Epple, P.; To, J.P.C.; Hutchison, C.E.; Schaller, G.E.; Dangl, J.L.; Kieber, J.J. Two-component elements mediate interactions between cytokinin and salicylic acid in plant immunity. PLoS Genet. 2012, 8, e1002448. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, Y.J.; Lin, N.-C.; Martin, G.B. Two distinct Pseudomonas effector proteins interact with the Pto kinase and activate plant immunity. Cell 2002, 109, 589–598. [Google Scholar] [CrossRef] [Green Version]
- Tang, X.; Xie, M.; Kim, Y.J.; Zhou, J.; Klessig, D.F.; Martin, G.B. Overexpression of Pto activates defense responses and confers broad resistance. Plant Cell 1999, 11, 15–29. [Google Scholar] [CrossRef] [Green Version]
- Zamora, M.G.M.; Castagnaro, A.P.; Ricci, J.C.D. Genetic diversity of Pto-like serine/threonine kinase disease resistance genes in cultivated and wild strawberries. J. Mol. Evol. 2008, 67, 211–221. [Google Scholar] [CrossRef]
- Webb, K.M.; Freeman, C.; Broeckling, C.D. Metabolome profiling to understand the defense response of sugar beet (Beta vulgaris) to Rhizoctonia solani AG 2-2 IIIB. Physiol. Mol. Plant Pathol. 2016, 94, 108–117. [Google Scholar] [CrossRef] [Green Version]
- Aliferis, K.A.; Jabaji, S. FT-ICR/MS and GC-EI/MS metabolomics networking unravels global potato sprout’s responses to Rhizoctonia solani infection. PLoS ONE 2012, 7, e42576. [Google Scholar] [CrossRef]
- Bednarek, P.; Osbourn, A. Plant-microbe interactions: Chemical diversity in plant defense. Science 2009, 324, 746–748. [Google Scholar] [CrossRef] [PubMed]
- Sparg, S.; Light, M.E.; Van Staden, J. Biological activities and distribution of plant saponins. J. Ethnopharmacol. 2004, 94, 219–243. [Google Scholar] [CrossRef]
- Zhao, C.-J.; Wang, A.-R.; Shi, Y.-J.; Wang, L.-Q.; Liu, W.-D.; Wang, Z.-H.; Lu, G.-D. Identification of defense-related genes in rice responding to challenge by Rhizoctonia solani. Theor. Appl. Genet. 2008, 116, 501–516. [Google Scholar] [CrossRef]
- Bertini, L.; Palazzi, L.; Proietti, S.; Pollastri, S.; Arrigoni, G.; Polverino de Laureto, P.; Caruso, C. Proteomic analysis of MeJa-induced defense responses in rice against wounding. Int. J. Mol. Sci. 2019, 20, 2525. [Google Scholar] [CrossRef] [Green Version]
- Van Loon, L.C.; Bakker, P.; Pieterse, C.M.J. Systemic resistance induced by rhizosphere bacteria. Annu. Rev. Phytopathol. 1998, 36, 453–483. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, S.G.; Kim, S.T.; Wang, Y.; Yu, S.; Choi, I.S.; Kim, Y.C.; Kim, W.T.; Agrawal, G.K.; Rakwal, R.; Kang, K.Y. The RNase activity of rice probenazole-induced protein1 (PBZ1) plays a key role in cell death in plants. Mol. Cells 2011, 31, 25–31. [Google Scholar] [CrossRef] [PubMed]
- Park, C.; Kim, K.; Shin, R.; Park, J.M.; Shin, Y.; Paek, K. Pathogenesis-related protein 10 isolated from hot pepper functions as a ribonuclease in an antiviral pathway. Plant J. 2004, 37, 186–198. [Google Scholar] [CrossRef]
- Chen, S.; Vaghchhipawala, Z.; Li, W.; Asard, H.; Dickman, M.B. Tomato phospholipid hydroperoxide glutathione peroxidase inhibits cell death induced by Bax and oxidative stresses in yeast and plants. Plant Physiol. 2004, 135, 1630–1641. [Google Scholar] [CrossRef] [Green Version]
- Shrestha, C.L.; Ona, I.; Muthukrishnan, S.; Mew, T.W. Chitinase levels in rice cultivars correlate with resistance to the sheath blight pathogen Rhizoctonia solani. Eur. J. Plant Pathol. 2008, 120, 69–77. [Google Scholar] [CrossRef]
- Taheri, P.; Tarighi, S. Riboflavin induces resistance in rice against Rhizoctonia solani via jasmonate-mediated priming of phenylpropanoid pathway. J. Plant Physiol. 2010, 167, 201–208. [Google Scholar] [CrossRef]
- Punja, Z.K. Genetic engineering of plants to enhance resistance to fungal pathogens—A review of progress and future prospects. Can. J. Plant Pathol. 2001, 23, 216–235. [Google Scholar] [CrossRef]
- Lehtonen, M.J.; Somervuo, P.; Valkonen, J.P.T. Infection with Rhizoctonia solani induces defense genes and systemic resistance in potato sprouts grown without light. Phytopathology 2008, 98, 1190–1198. [Google Scholar] [CrossRef] [Green Version]
- Takemoto, D.; Furuse, K.; Doke, N.; Kazuhito, K. Identification of chitinase and osmotin-like protein as actin-binding proteins in suspension-cultured potato cells. Plant Cell Physiol. 1997, 38, 441–448. [Google Scholar] [CrossRef] [Green Version]
- Lorito, M.; Woo, S.L.; Fernandez, I.G.; Colucci, G.; Harman, G.E.; Pintor-Toro, J.A.; Filippone, E.; Muccifora, S.; Lawrence, C.B.; Zoina, A.; et al. Genes from mycoparasitic fungi as a source for improving plant resistance to fungal pathogens. Proc. Natl. Acad. Sci. USA 1998, 95, 7860–7865. [Google Scholar] [CrossRef] [Green Version]
- Chandra, A.; Saxena, R.; Dubey, A.; Saxena, P. Change in phenylalanine ammonia lyase activity and isozyme patterns of polyphenol oxidase and peroxidase by salicylic acid leading to enhance resistance in cowpea against Rhizoctonia solani. Acta Physiol. Plant 2007, 29, 361–367. [Google Scholar] [CrossRef]
- Wen, K.; Seguin, P.; St.-Arnaud, M.; Jabaji-Hare, S. Real-time quantitative RT-PCR of defense-associated gene transcripts of Rhizoctonia solani-infected bean seedlings in response to inoculation with a nonpathogenic binucleate Rhizoctonia isolate. Phytopathology 2005, 95, 345–353. [Google Scholar] [CrossRef] [PubMed]
- Foley, R.C.; Gleason, C.A.; Anderson, J.P.; Hamann, T.; Singh, K.B. Genetic and genomic analysis of Rhizoctonia solani interactions with Arabidopsis; evidence of resistance mediated through NADPH oxidases. PLoS ONE 2013, 8, e56814. [Google Scholar] [CrossRef]
- Li, H.; Smigocki, A.C. Sugar beet polygalacturonase-inhibiting proteins with 11 LRRs confer Rhizoctonia, Fusarium and Botrytis resistance in Nicotiana plants. Physiol. Mol. Plant Pathol. 2018, 102, 200–208. [Google Scholar] [CrossRef]
- Borras-Hidalgo, O.; Caprari, C.; Hernandez-Estevez, I.; De Lorenzo, G.; Cervone, F. A gene for plant protection: Expression of a bean polygalacturonase inhibitor in tobacco confers a strong resistance against Rhizoctonia solani and two oomycetes. Front. Plant Sci. 2012, 3, 268. [Google Scholar] [CrossRef] [Green Version]
- Kalantari, M.; Motallebi, M.; Zamani, M.R. Bean Polygalacturonase-Inhibiting Protein Expressed in Transgenic Sugar Beet Inhibits Polygalacturonase from Rhizoctonia solani. Biosci. Biotechnol. Res. Asia 2016, 8, 19–28. [Google Scholar] [CrossRef]
- Helliwell, E.E.; Wang, Q.; Yang, Y. Transgenic rice with inducible ethylene production exhibits broad-spectrum disease resistance to the fungal pathogens Magnaporthe oryzae and Rhizoctonia solani. Plant Biotechnol. J. 2013, 11, 33–42. [Google Scholar] [CrossRef] [PubMed]
- Maeda, S.; Dubouzet, J.G.; Kondou, Y.; Jikumaru, Y.; Seo, S.; Oda, K.; Matsui, M.; Hirochika, H.; Mori, M. The rice CYP78A gene BSR2 confers resistance to Rhizoctonia solani and affects seed size and growth in Arabidopsis and rice. Sci. Rep. 2019, 9, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kalpana, K.; Maruthasalam, S.; Rajesh, T.; Poovannan, K.; Kumar, K.K.; Kokiladevi, E.; Raja, J.A.J.; Sudhakar, D.; Velazhahan, R.; Samiyappan, R. Engineering sheath blight resistance in elite indica rice cultivars using genes encoding defense proteins. Plant Sci. 2006, 170, 203–215. [Google Scholar] [CrossRef]
- Gill, U.S.; Lee, S.; Mysore, K.S. Host versus nonhost resistance: Distinct wars with similar arsenals. Phytopathology 2015, 105, 580–587. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hadwiger, L.A.; Beckman, J.M. Chitosan as a component of pea-Fusarium solani interactions. Plant Physiol. 1980, 66, 205–211. [Google Scholar] [CrossRef] [Green Version]
- Isaac, J.; Hartney, S.L.; Druffel, K.; Hadwiger, L.A. The non-host disease resistance response in peas; alterations in phosphorylation and ubiquitination of HMG A and histones H2A/H2B. Plant Sci. 2009, 177, 439–449. [Google Scholar] [CrossRef]
- Hadwiger, L.A. Pea–Fusarium solani interactions contributions of a system toward understanding disease resistance. Phytopathology 2008, 98, 372–379. [Google Scholar] [CrossRef] [Green Version]
- Hadwiger, L.A. Anatomy of a nonhost disease resistance response of pea to Fusarium solani: PR gene elicitation via DNase, chitosan and chromatin alterations. Front. Plant Sci. 2015, 6, 373. [Google Scholar] [CrossRef]
- Kendra, D.F.; Hadwiger, L.A. Characterization of the smallest chitosan oligomer that is maximally antifungal to Fusarium solani and elicits pisatin formation in Pisum sativum. Exp. Mycol. 1984. [Google Scholar] [CrossRef]
- Klosterman, S.J.; Chen, J.; Choi, J.J.; Chinn, E.E.; Hadwiger, L.A. Characterization of a 20 kDa DNase elicitor from Fusarium solani f. sp. phaseoli and its expression at the onset of induced resistance in Pisum sativum. Mol. Plant Pathol. 2001, 2, 147–158. [Google Scholar] [CrossRef]
- Williamson-Benavides, B.A.; Sharpe, R.M.; Nelson, G.; Bodah, E.T.; Porter, L.D.; Dhingra, A. Identification of Fusarium solani f. sp. pisi (Fsp) Responsive Genes in Pisum sativum. Front. Genet. 2020, 11, 950. [Google Scholar] [CrossRef] [PubMed]
- Almeida, M.S.; Cabral, K.M.S.; Zingali, R.B.; Kurtenbach, E. Characterization of two novel defense peptides from pea (Pisum sativum) seeds. Arch. Biochem. Biophys. 2000, 378, 278–286. [Google Scholar] [CrossRef] [PubMed]
- Mauch, F.; Hadwiger, L.A.; Boller, T. Ethylene: Symptom, not signal for the induction of chitinase and β-1, 3-glucanase in pea pods by pathogens and elicitors. Plant Physiol. 1984, 76, 607–611. [Google Scholar] [CrossRef] [Green Version]
- Chang, M.-M.; Hadwiger, L.A.; Horovitz, D. Molecular characterization of a pea β-1, 3-glucanase induced by Fusarium solani and chitosan challenge. Plant Mol. Biol. 1992, 20, 609–618. [Google Scholar] [CrossRef] [PubMed]
- Hadwiger, L.A.; Adams, M.J. Nuclear changes associated with the host-parasite interaction between Fusarium solani and peas. Physiol. Plant Pathol. 1978, 12, 63–72. [Google Scholar] [CrossRef]
- Lozovaya, V.V.; Lygin, A.V.; Zernova, O.V.; Li, S.; Hartman, G.L.; Widholm, J.M. Isoflavonoid accumulation in soybean hairy roots upon treatment with Fusarium solani. Plant Physiol. Biochem. 2004, 42, 671–679. [Google Scholar] [CrossRef]
- Iqbal, M.J.; Yaegashi, S.; Ahsan, R.; Shopinski, K.L.; Lightfoot, D.A. Root response to Fusarium solani f. sp. glycines: Temporal accumulation of transcripts in partially resistant and susceptible soybean. Theor. Appl. Genet. 2005, 110, 1429–1438. [Google Scholar] [CrossRef] [PubMed]
- Mackintosh, S.F.; Matthews, D.E.; VanEtten, H.D. Two additional genes for pisatin demethylation and their relationship to the pathogenicity of Nectria haematococca on pea. Mol. Plant-Microbe Interact. 1989, 2, 354–362. [Google Scholar] [CrossRef]
- Coyne, C.J.; Porter, L.D.; Boutet, G.; Ma, Y.; McGee, R.J.; Lesné, A.; Baranger, A.; Pilet-Nayel, M.-L. Confirmation of Fusarium root rot resistance QTL Fsp-Ps 2.1 of pea under controlled conditions. BMC Plant Biol. 2019, 19, 98. [Google Scholar] [CrossRef] [Green Version]
- Coyne, C.J.; Pilet-Nayel, M.; McGee, R.J.; Porter, L.D.; Smýkal, P.; Grünwald, N.J. Identification of QTL controlling high levels of partial resistance to Fusarium solani f. sp. pisi in pea. Plant Breed. 2015, 134, 446–453. [Google Scholar] [CrossRef]
- Feng, J.; Hwang, R.; Chang, K.F.; Conner, R.L.; Hwang, S.F.; Strelkov, S.E.; Gossen, B.D.; McLaren, D.L.; Xue, A.G. Identification of microsatellite markers linked to quantitative trait loci controlling resistance to Fusarium root rot in field pea. Can. J. Plant Sci. 2011, 91, 199–204. [Google Scholar] [CrossRef]
- Williamson-Benavides, B.A.; Sharpe, R.; Nelson, G.; Bodah, E.T.; Porter, L.D.; Dhingra, A. Identification of root rot tolerance QTLs in pea using Fusarium solani f. sp. pisi-responsive differentially expressed genes. bioRxiv 2021. in review. [Google Scholar]
- Badrhadad, A.; Nazarian-Firouzabadi, F.; Ismaili, A. Fusion of a chitin-binding domain to an antibacterial peptide to enhance resistance to Fusarium solani in tobacco (Nicotiana tabacum). 3 Biotech 2018, 8, 391. [Google Scholar] [CrossRef]
- Charfeddine, M.; Samet, M.; Charfeddine, S.; Bouaziz, D.; Bouzid, R.G. Ectopic Expression of StERF94 Transcription Factor in Potato Plants Improved Resistance to Fusarium solani Infection. Plant Mol. Biol. Rep. 2019, 37, 450–463. [Google Scholar] [CrossRef]
- Zhang, C.; Zhao, X.; Qu, Y.; Teng, W.; Qiu, L.; Zheng, H.; Wang, Z.; Han, Y.; Li, W. Loci and candidate genes in soybean that confer resistance to Fusarium graminearum. Theor. Appl. Genet. 2019, 132, 431–441. [Google Scholar] [CrossRef]
- Nelsen, N.S.; Li, Z.; Warner, A.L.; Matthews, B.F.; Knap, H.T. Genomic Polymorphism Identifies a Subtilisin-Like Protease near the Rhg4 Locus in Soybean. Crop Sci. 2004, 44, 265–273. [Google Scholar] [CrossRef]
- Million, C.R.; Wijeratne, S.; Cassone, B.J.; Lee, S.; Rouf Mian, M.A.; McHale, L.K.; Dorrance, A.E. Hybrid genome assembly of a major quantitative disease resistance locus in soybean toward Fusarium graminearum. Plant Genome 2019, 12, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Scherm, B.; Balmas, V.; Spanu, F.; Pani, G.; Delogu, G.; Pasquali, M.; Migheli, Q. F usarium culmorum: Causal agent of foot and root rot and head blight on wheat. Mol. Plant Pathol. 2013, 14, 323–341. [Google Scholar] [CrossRef]
- Osborne, L.E.; Stein, J.M. Epidemiology of Fusarium head blight on small-grain cereals. Int. J. Food Microbiol. 2007, 119, 103–108. [Google Scholar] [CrossRef]
- Aoki, T.; Ward, T.J.; Kistler, H.C.; O’donnell, K. Systematics, phylogeny and trichothecene mycotoxin potential of Fusarium head blight cereal pathogens. JSM Mycotoxins 2012, 62, 91–102. [Google Scholar] [CrossRef] [Green Version]
- Mesterhazy, A.; Lemmens, M.; Reid, L.M. Breeding for resistance to ear rots caused by Fusarium spp. in maize—A review. Plant Breed. 2012, 131, 1–19. [Google Scholar] [CrossRef]
- Saharan, M.S. Current status of resistant source to Fusarium head blight disease of wheat: A review. Indian Phytopathol. 2020, 73, 3–9. [Google Scholar] [CrossRef]
- Gilbert, J.; Haber, S. Overview of some recent research developments in Fusarium head blight of wheat. Can. J. Plant Pathol. 2013, 35, 149–174. [Google Scholar] [CrossRef]
- Lanubile, A.; Muppirala, U.K.; Severin, A.J.; Marocco, A.; Munkvold, G.P. Transcriptome profiling of soybean (Glycine max) roots challenged with pathogenic and non-pathogenic isolates of Fusarium oxysporum. BMC Genom. 2015, 16, 1089. [Google Scholar] [CrossRef] [Green Version]
- Kolb, F.L.; Bai, G.H.; Muehlbauer, G.J.; Anderson, J.A.; Smith, K.P.; Fedak, G. Symposium on genetic solutions to Fusarium head blight in wheat and barley: Challenges, opportunities, and imperatives. Crop Sci. 2001, 41, 611. [Google Scholar] [CrossRef]
- Rudd, J.C.; Horsley, R.D.; McKendry, A.L.; Elias, E.M. Host plant resistance genes for Fusarium head blight: Sources, mechanisms, and utility in conventional breeding systems. Crop Sci. 2001, 41, 620–627. [Google Scholar] [CrossRef] [Green Version]
- Buerstmayr, H.; Ban, T.; Anderson, J.A. QTL mapping and marker-assisted selection for Fusarium head blight resistance in wheat: A review. Plant Breed. 2009, 128, 1–26. [Google Scholar] [CrossRef]
- Ma, Z.; Xie, Q.; Li, G.; Jia, H.; Zhou, J.; Kong, Z.; Li, N.; Yuan, Y. Germplasms, genetics and genomics for better control of disastrous wheat Fusarium head blight. Theor. Appl. Genet. 2020, 133, 1–28. [Google Scholar] [CrossRef] [PubMed]
- Fugate, K.K.; Ferrareze, J.P.; Bolton, M.D.; Deckard, E.L.; Campbell, L.G. Postharvest jasmonic acid treatment of sugarbeet roots reduces rot due to Botrytis cinerea, Penicillium claviforme, and Phoma betae. Postharvest Biol. Technol. 2012, 65, 1–4. [Google Scholar] [CrossRef]
- Bugbee, W.M.; Cole, D.F. Comparison of thiabendazole and genetic resistance for control of sugar beet storage rot. Phytopathology 1979, 69, 1230–1232. [Google Scholar] [CrossRef]
- Wang, H.; Hwang, S.F.; Chang, K.F.; Gossen, B.D.; Turnbull, G.D.; Howard, R.J. Assessing resistance to spring black stem and leaf spot of alfalfa caused by Phoma spp. Can. J. Plant Sci. 2004, 84, 311–317. [Google Scholar] [CrossRef]
- Marzu, J.C.; Straley, E.; Havey, M.J. Genetic Analyses and Mapping of Pink-Root Resistance in Onion. J. Am. Soc. Hortic. Sci. 2018, 143, 503–507. [Google Scholar] [CrossRef] [Green Version]
- Niu, C.; Lister, H.E.; Nguyen, B.; Wheeler, T.A.; Wright, R.J. Resistance to Thielaviopsis basicola in the cultivated A genome cotton. Theor. Appl. Genet. 2008, 117, 1313. [Google Scholar] [CrossRef] [PubMed]
- Callahan, F.E.; Jenkins, J.N.; Creech, R.G.; Lawrence, G.W. Changes in cotton root proteins correlated with resistance to root knot nematode development. J. Cotton Sci. 1997, 1, 38. [Google Scholar]
- Wubben, M.J.; Callahan, F.E.; Velten, J.; Burke, J.J.; Jenkins, J.N. Overexpression of MIC-3 indicates a direct role for the MIC gene family in mediating Upland cotton (Gossypium hirsutum) resistance to root-knot nematode (Meloidogyne incognita). Theor. Appl. Genet. 2015, 128, 199–209. [Google Scholar] [CrossRef] [PubMed]
- Jacquet, C.; Bonhomme, M. Deciphering resistance mechanisms to the root rot disease of legumes caused by Aphanomyces euteiches with Medicago truncatula genetic and genomic resources. Model Legume Medicago Truncatula 2019, 307–316. [Google Scholar] [CrossRef]
- Badis, Y.; Bonhomme, M.; Lafitte, C.; Huguet, S.; Balzergue, S.; Dumas, B.; Jacquet, C. Transcriptome analysis highlights preformed defences and signalling pathways controlled by the pr A e1 quantitative trait locus (QTL), conferring partial resistance to A phanomyces euteiches in M edicago truncatula. Mol. Plant Pathol. 2015, 16, 973–986. [Google Scholar] [CrossRef]
- Colditz, F.; Nyamsuren, O.; Niehaus, K.; Eubel, H.; Braun, H.-P.; Krajinski, F. Proteomic approach: Identification of Medicago truncatula proteins induced in roots after infection with the pathogenic oomycete Aphanomyces euteiches. Plant Mol. Biol. 2004, 55, 109–120. [Google Scholar] [CrossRef]
- Djébali, N.; Jauneau, A.; Ameline-Torregrosa, C.; Chardon, F.; Jaulneau, V.; Mathé, C.; Bottin, A.; Cazaux, M.; Pilet-Nayel, M.-L.; Baranger, A.; et al. Partial resistance of Medicago truncatula to Aphanomyces euteiches is associated with protection of the root stele and is controlled by a major QTL rich in proteasome-related genes. Mol. Plant-Microbe Interact. 2009, 22, 1043–1055. [Google Scholar] [CrossRef] [Green Version]
- Kiirika, L.M.; Schmitz, U.; Colditz, F. The alternative Medicago truncatula defense proteome of ROS—Defective transgenic roots during early microbial infection. Front. Plant Sci. 2014, 5, 341. [Google Scholar] [CrossRef] [Green Version]
- Kiirika, L.M.; Bergmann, H.F.; Schikowsky, C.; Wimmer, D.; Korte, J.; Schmitz, U.; Niehaus, K.; Colditz, F. Silencing of the Rac1 GTPase MtROP9 in Medicago truncatula stimulates early mycorrhizal and oomycete root colonizations but negatively affects rhizobial infection. Plant Physiol. 2012, 159, 501–516. [Google Scholar] [CrossRef] [Green Version]
- Nyamsuren, O.; Colditz, F.; Rosendahl, S.; Tamasloukht, M.; Bekel, T.; Meyer, F.; Kuester, H.; Franken, P.; Krajinski, F. Transcriptional profiling of Medicago truncatula roots after infection with Aphanomyces euteiches (oomycota) identifies novel genes upregulated during this pathogenic interaction. Physiol. Mol. Plant Pathol. 2003. [Google Scholar] [CrossRef]
- Trapphoff, T.; Beutner, C.; Niehaus, K.; Colditz, F. Induction of distinct defense-associated protein patterns in Aphanomyces euteiches (Oomycota)–elicited and–inoculated Medicago truncatula cell-suspension cultures: A proteome and phosphoproteome approach. Mol. Plant-Microbe Interact. 2009, 22, 421–436. [Google Scholar] [CrossRef] [PubMed]
- Pilet-Nayel, M.-L.; Prospéri, J.-M.; Hamon, C.; Lesne, A.; Lecointe, R.; Le Goff, I.; Hervé, M.; Deniot, G.; Delalande, M.; Huguet, T.; et al. AER1, a major gene conferring resistance to Aphanomyces euteiches in Medicago truncatula. Phytopathology 2009, 99, 203–208. [Google Scholar] [CrossRef] [Green Version]
- Colditz, F.; Braun, H.-P.; Jacquet, C.; Niehaus, K.; Krajinski, F. Proteomic profiling unravels insights into the molecular background underlying increased Aphanomyces euteiches-tolerance of Medicago truncatula. Plant Mol. Biol. 2005, 59, 387–406. [Google Scholar] [CrossRef]
- Lechner, E.; Achard, P.; Vansiri, A.; Potuschak, T.; Genschik, P. F-box proteins everywhere. Curr. Opin. Plant Biol. 2006, 9, 631–638. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Ecker, J.R. Plant responses to ethylene gas are mediated by SCFEBF1/EBF2-dependent proteolysis of EIN3 transcription factor. Cell 2003, 115, 667–677. [Google Scholar] [CrossRef] [Green Version]
- del Pozo, J.C.; Diaz-Trivino, S.; Cisneros, N.; Gutierrez, C. The balance between cell division and endoreplication depends on E2FC-DPB, transcription factors regulated by the ubiquitin-SCFSKP2A pathway in Arabidopsis. Plant Cell 2006, 18, 2224–2235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ivanchenko, M.G.; Muday, G.K.; Dubrovsky, J.G. Ethylene–auxin interactions regulate lateral root initiation and emergence in Arabidopsis thaliana. Plant J. 2008, 55, 335–347. [Google Scholar] [CrossRef]
- Cao, F.Y.; Yoshioka, K.; Desveaux, D. The roles of ABA in plant–pathogen interactions. J. Plant Res. 2011, 124, 489–499. [Google Scholar] [CrossRef]
- Adie, B.A.T.; Pérez-Pérez, J.; Pérez-Pérez, M.M.; Godoy, M.; Sánchez-Serrano, J.-J.; Schmelz, E.A.; Solano, R. ABA is an essential signal for plant resistance to pathogens affecting JA biosynthesis and the activation of defenses in Arabidopsis. Plant Cell 2007, 19, 1665–1681. [Google Scholar] [CrossRef] [Green Version]
- Colditz, F.; Niehaus, K.; Krajinski, F. Silencing of PR-10-like proteins in Medicago truncatula results in an antagonistic induction of other PR proteins and in an increased tolerance upon infection with the oomycete Aphanomyces euteiches. Planta 2007, 226, 57–71. [Google Scholar] [CrossRef] [PubMed]
- Rey, T.; Nars, A.; Bonhomme, M.; Bottin, A.; Huguet, S.; Balzergue, S.; Jardinaud, M.; Bono, J.; Cullimore, J.; Dumas, B. NFP, a L ys M protein controlling N od f actor perception, also intervenes in M edicago truncatula resistance to pathogens. New Phytol. 2013, 198, 875–886. [Google Scholar] [CrossRef]
- Schenkluhn, L.; Hohnjec, N.; Niehaus, K.; Schmitz, U.; Colditz, F. Differential gel electrophoresis (DIGE) to quantitatively monitor early symbiosis-and pathogenesis-induced changes of the Medicago truncatula root proteome. J. Proteom. 2010, 73, 753–768. [Google Scholar] [CrossRef] [PubMed]
- Djébali, N.; Mhadhbi, H.; Lafitte, C.; Dumas, B.; Esquerré-Tugayé, M.-T.; Aouani, M.E.; Jacquet, C. Hydrogen peroxide scavenging mechanisms are components of Medicago truncatula partial resistance to Aphanomyces euteiches. Eur. J. Plant Pathol. 2011, 131, 559. [Google Scholar] [CrossRef]
- Rey, T.; Laporte, P.; Bonhomme, M.; Jardinaud, M.-F.; Huguet, S.; Balzergue, S.; Dumas, B.; Niebel, A.; Jacquet, C. MtNF-YA1, a central transcriptional regulator of symbiotic nodule development, is also a determinant of Medicago truncatula susceptibility toward a root pathogen. Front. Plant Sci. 2016, 7, 1837. [Google Scholar] [CrossRef] [Green Version]
- Kjøller, R.; Rosendahl, S. The presence of the arbuscular mycorrhizal fungus Glomus intraradices influences enzymatic activities of the root pathogen Aphanomyces euteiches in pea roots. Mycorrhiza 1997, 6, 487–491. [Google Scholar] [CrossRef]
- Bødker, L.; Kjøller, R.; Rosendahl, S. Effect of phosphate and the arbuscular mycorrhizal fungus Glomus intraradices on disease severity of root rot of peas (Pisum sativum) caused by Aphanomyces euteiches. Mycorrhiza 1998, 8, 169–174. [Google Scholar] [CrossRef]
- Wang, H. Identification and dissection of soybean QTL conferring resistance to Phytophthora sojae. Ph.D. Thesis, Ohio State University, Columbus, OH, USA, 2011. [Google Scholar]
- Li, X.; Han, Y.; Teng, W.; Zhang, S.; Yu, K.; Poysa, V.; Anderson, T.; Ding, J.; Li, W. Pyramided QTL underlying tolerance to Phytophthora root rot in mega-environments from soybean cultivars ‘Conrad’and ‘Hefeng 25’. Theor. Appl. Genet. 2010, 121, 651–658. [Google Scholar] [CrossRef] [PubMed]
- Njiti, V.N.; Meksem, K.; Iqbal, M.J.; Johnson, J.E.; Kassem, M.A.; Zobrist, K.F.; Kilo, V.Y.; Lightfoot, D.A. Common loci underlie field resistance to soybean sudden death syndrome in Forrest, Pyramid, Essex, and Douglas. Theor. Appl. Genet. 2002, 104, 294–300. [Google Scholar] [CrossRef] [PubMed]
- Hashmi, R.Y. Inheritance of Resistance to Soybean Sudden Death Syndrome (SDS) in Ripley × Spencer F (5) Derived Lines; Southern Illinois University at Carbondale: Carbondale, IL, USA, 2004; ISBN 0496969455. [Google Scholar]
- Wang, H.; Waller, L.; Tripathy, S.; St. Martin, S.K.; Zhou, L.; Krampis, K.; Tucker, D.M.; Mao, Y.; Hoeschele, I.; Saghai Maroof, M.A.; et al. Analysis of genes underlying soybean quantitative trait loci conferring partial resistance to Phytophthora sojae. Plant Genome 2010, 3, 23–40. [Google Scholar] [CrossRef] [Green Version]
- Kazi, S.; Shultz, J.; Afzal, J.; Johnson, J.; Njiti, V.N.; Lightfoot, D.A. Separate loci underlie resistance to root infection and leaf scorch during soybean sudden death syndrome. Theor. Appl. Genet. 2008, 116, 967–977. [Google Scholar] [CrossRef] [Green Version]
- Klepadlo, M.; Balk, C.S.; Vuong, T.D.; Dorrance, A.E.; Nguyen, H.T. Molecular characterization of genomic regions for resistance to Pythium ultimum var. ultimum in the soybean cultivar Magellan. Theor. Appl. Genet. 2019, 132, 405–417. [Google Scholar] [CrossRef] [PubMed]
- Campa, A.; Pérez-Vega, E.; Pascual, A.; Ferreira, J.J. Genetic analysis and molecular mapping of quantitative trait loci in common bean against Pythium ultimum. Phytopathology 2010, 100, 1315–1320. [Google Scholar] [CrossRef] [Green Version]
- Shin, S.; Zheng, P.; Fazio, G.; Mazzola, M.; Main, D.; Zhu, Y. Transcriptome changes specifically associated with apple (Malus domestica) root defense response during Pythium ultimum infection. Physiol. Mol. Plant Pathol. 2016, 94, 16–26. [Google Scholar] [CrossRef] [Green Version]
- Zhu, Y.; Shao, J.; Zhou, Z.; Davis, R.E. Genotype-specific suppression of multiple defense pathways in apple root during infection by Pythium ultimum. Hortic. Res. 2019, 6, 1–17. [Google Scholar] [CrossRef] [Green Version]
- Nath, V.S.; Koyyappurath, S.; Alex, T.E.; Geetha, K.A.; Augustine, L.; Nasser, A.; Thomas, G. Transcriptome-based mining and expression profiling of Pythium responsive transcription factors in Zingiber sp. Funct. Integr. Genom. 2019, 19, 249–264. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Zhao, J.; Zhou, Z. Identifying an elite panel of apple rootstock germplasm with contrasting root resistance to Pythium ultimum. J. Plant Pathol. 2018, 9, 1000461. [Google Scholar] [CrossRef]
- Wrather, J.A.; Anderson, T.R.; Arsyad, D.M.; Tan, Y.; Ploper, L.D.; Porta-Puglia, A.; Ram, H.H.; Yorinori, J.T. Soybean disease loss estimates for the top ten soybean-producing counries in 1998. Can. J. Plant Pathol. 2001, 23, 115–121. [Google Scholar] [CrossRef]
- Han, Y.; Teng, W.; Yu, K.; Poysa, V.; Anderson, T.; Qiu, L.; Lightfoot, D.A.; Li, W. Mapping QTL tolerance to Phytophthora root rot in soybean using microsatellite and RAPD/SCAR derived markers. Euphytica 2008, 162, 231–239. [Google Scholar] [CrossRef]
- Dorrance, A.E.; McClure, S.A.; St. Martin, S.K. Effect of partial resistance on Phytophthora stem rot incidence and yield of soybean in Ohio. Plant Dis. 2003, 87, 308–312. [Google Scholar] [CrossRef] [Green Version]
- Weng, C.; Yu, K.; Anderson, T.R.; Poysa, V. Mapping genes conferring resistance to Phytophthora root rot of soybean, Rps1a and Rps7. J. Hered. 2001, 92, 442–446. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sandhu, D.; Schallock, K.G.; Rivera-Velez, N.; Lundeen, P.; Cianzio, S.; Bhattacharyya, M.K. Soybean Phytophthora resistance gene Rps8 maps closely to the Rps3 region. J. Hered. 2005, 96, 536–541. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dorrance, A.E. Management of Phytophthora sojae of soybean: A review and future perspectives. Can. J. Plant Pathol. 2018, 40, 210–219. [Google Scholar] [CrossRef]
- Ranathunge, K.; Thomas, R.H.; Fang, X.; Peterson, C.A.; Gijzen, M.; Bernards, M.A. Soybean root suberin and partial resistance to root rot caused by Phytophthora sojae. Phytopathology 2008, 98, 1179–1189. [Google Scholar] [CrossRef] [Green Version]
- Drenth, A.; Whisson, S.C.; Maclean, D.J.; Irwin, J.A.G.; Obst, N.R.; Ryley, M.J. The evolution of races of Phytophthora sojae in Australia. Phytopathology 1996, 86, 163–169. [Google Scholar] [CrossRef]
- Leitz, R.A.; Hartman, G.L.; Pedersen, W.L.; Nickell, C.D. Races of Phytophthora sojae on soybean in Illinois. Plant Dis. 2000, 84, 487. [Google Scholar] [CrossRef] [PubMed]
- Malvick, D.K.; Grunden, E. Characteristics of Phytophthora sojae populations in Illinois and implications for management of Phytophthora rot of soybean. Phytopathology 2004, 94, S65. [Google Scholar]
- Sugimoto, T.; Kato, M.; Yoshida, S.; Matsumoto, I.; Kobayashi, T.; Kaga, A.; Hajika, M.; Yamamoto, R.; Watanabe, K.; Aino, M.; et al. Pathogenic diversity of Phytophthora sojae and breeding strategies to develop Phytophthora-resistant soybeans. Breed. Sci. 2012, 61, 511–522. [Google Scholar] [CrossRef] [Green Version]
- Gao, H.; Narayanan, N.N.; Ellison, L.; Bhattacharyya, M.K. Two classes of highly similar coiled coil-nucleotide binding-leucine rich repeat genes isolated from the Rps1-k locus encode Phytophthora resistance in soybean. Mol. Plant-Microbe Interact. 2005, 18, 1035–1045. [Google Scholar] [CrossRef] [Green Version]
- Gao, H.; Bhattacharyya, M.K. The soybean-Phytophthora resistance locus Rps1-k encompasses coiled coil-nucleotide binding-leucine rich repeat-like genes and repetitive sequences. BMC Plant Biol. 2008, 8, 29. [Google Scholar] [CrossRef] [Green Version]
- Graham, M.A.; Marek, L.F.; Shoemaker, R.C. Organization, expression and evolution of a disease resistance gene cluster in soybean. Genetics 2002, 162, 1961–1977. [Google Scholar]
- Sun, J.; Li, L.; Zhao, J.; Huang, J.; Yan, Q.; Xing, H.; Guo, N. Genetic analysis and fine mapping of RpsJS, a novel resistance gene to Phytophthora sojae in soybean [Glycine max (L.) Merr.]. Theor. Appl. Genet. 2014, 127, 913–919. [Google Scholar] [CrossRef]
- Zhang, J.; Xia, C.; Wang, X.; Duan, C.; Sun, S.; Wu, X.; Zhu, Z. Genetic characterization and fine mapping of the novel Phytophthora resistance gene in a Chinese soybean cultivar. Theor. Appl. Genet. 2013, 126, 1555–1561. [Google Scholar] [CrossRef]
- Zhang, J.; Xia, C.; Duan, C.; Sun, S.; Wang, X.; Wu, X.; Zhu, Z. Identification and candidate gene analysis of a novel Phytophthora resistance gene Rps10 in a Chinese soybean cultivar. PLoS ONE 2013, 8, e69799. [Google Scholar] [CrossRef] [Green Version]
- Thomas, R.; Fang, X.; Ranathunge, K.; Anderson, T.R.; Peterson, C.A.; Bernards, M.A. Soybean root suberin: Anatomical distribution, chemical composition, and relationship to partial resistance to Phytophthora sojae. Plant Physiol. 2007, 144, 299–311. [Google Scholar] [CrossRef] [Green Version]
- Fang, X. Chemical composition of soybean root epidermal cell walls. Master’s Thesis, University of Waterloo, Waterloo, ON, Canada, 2006. [Google Scholar]
- Guo, N.; Ye, W.-W.; Wu, X.-L.; Shen, D.-Y.; Wang, Y.-C.; Xing, H.; Dou, D.-L. Microarray profiling reveals microRNAs involving soybean resistance to Phytophthora sojae. Genome 2011, 54, 954–958. [Google Scholar] [CrossRef] [PubMed]
- Wong, J.; Gao, L.; Yang, Y.; Zhai, J.; Arikit, S.; Yu, Y.; Duan, S.; Chan, V.; Xiong, Q.; Yan, J. Roles of small RNA s in soybean defense against P hytophthora sojae infection. Plant J. 2014, 79, 928–940. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cui, X.; Yan, Q.; Gan, S.; Xue, D.; Dou, D.; Guo, N.; Xing, H. Overexpression of gma-miR1510a/b suppresses the expression of a NB-LRR domain gene and reduces resistance to Phytophthora sojae. Gene 2017, 621, 32–39. [Google Scholar] [CrossRef]
- Vontimitta, V.; Danehower, D.A.; Steede, T.; Moon, H.S.; Lewis, R.S. Analysis of a Nicotiana tabacum L. genomic region controlling two leaf surface chemistry traits. J. Agric. Food Chem. 2010, 58, 294–300. [Google Scholar] [CrossRef]
- Jackson, D.M.; Danehower, D.A. Integrated case study: Nicotiana leaf-surface components and their effects on insect pests and diseases. In Plant Cuticles: An Integrated Functional Approach; Plant Cuticles BIOS Science: Oxford, UK, 1996. [Google Scholar]
- Severson, R.F.; Johnson, A.W.; Jackson, D.M. Cuticular constituents of tobacco: Factors affecting their production and their role in insect and disease resistance and smoke quality. Recent Adv. Tob. Sci. 1985, 11, 105–174. [Google Scholar]
- Yang, J.-K.; Tong, Z.-J.; Fang, D.-H.; Chen, X.-J.; Zhang, K.-Q.; Xiao, B.-G. Transcriptomic profile of tobacco in response to Phytophthora nicotianae infection. Sci. Rep. 2017, 7, 1–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meng, H.; Sun, M.; Jiang, Z.; Liu, Y.; Sun, Y.; Liu, D.; Jiang, C.; Ren, M.; Yuan, G.; Yu, W.; et al. Comparative transcriptome analysis reveals resistant and susceptible genes in tobacco cultivars in response to infection by Phytophthora nicotianae. Sci. Rep. 2021, 11. [Google Scholar] [CrossRef]
- Valenzuela-Estrada, L.R.; Bryla, D.R.; Hoashi-Erhardt, W.K.; Moore, P.P.; Forge, T.A. Root traits associated with Phytophthora root rot resistance in red raspberry. In Proceedings of the X International Rubus and Ribes Symposium, Zlatibor, Serbia, 22–26 June 2011; Volume 946, pp. 283–287. [Google Scholar]
- Graham, J.; Hackett, C.A.; Smith, K.; Woodhead, M.; MacKenzie, K.; Tierney, I.; Cooke, D.; Bayer, M.; Jennings, N. Towards an understanding of the nature of resistance to Phytophthora root rot in red raspberry. Theor. Appl. Genet. 2011, 123, 585–601. [Google Scholar] [CrossRef] [PubMed]
- Engelbrecht, J.; Van den Berg, N. Expression of defence-related genes against Phytophthora cinnamomi in five avocado rootstocks. S. Afr. J. Sci. 2013, 109, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Van de Weg, W.E.; Henken, B.; Giezen, S. Assessment of the resistance to Phytophthora fragariae var. fragariae of the USA and Canadian differential series of strawberry genotypes. J. Phytopathol. 1997, 145, 1–6. [Google Scholar]
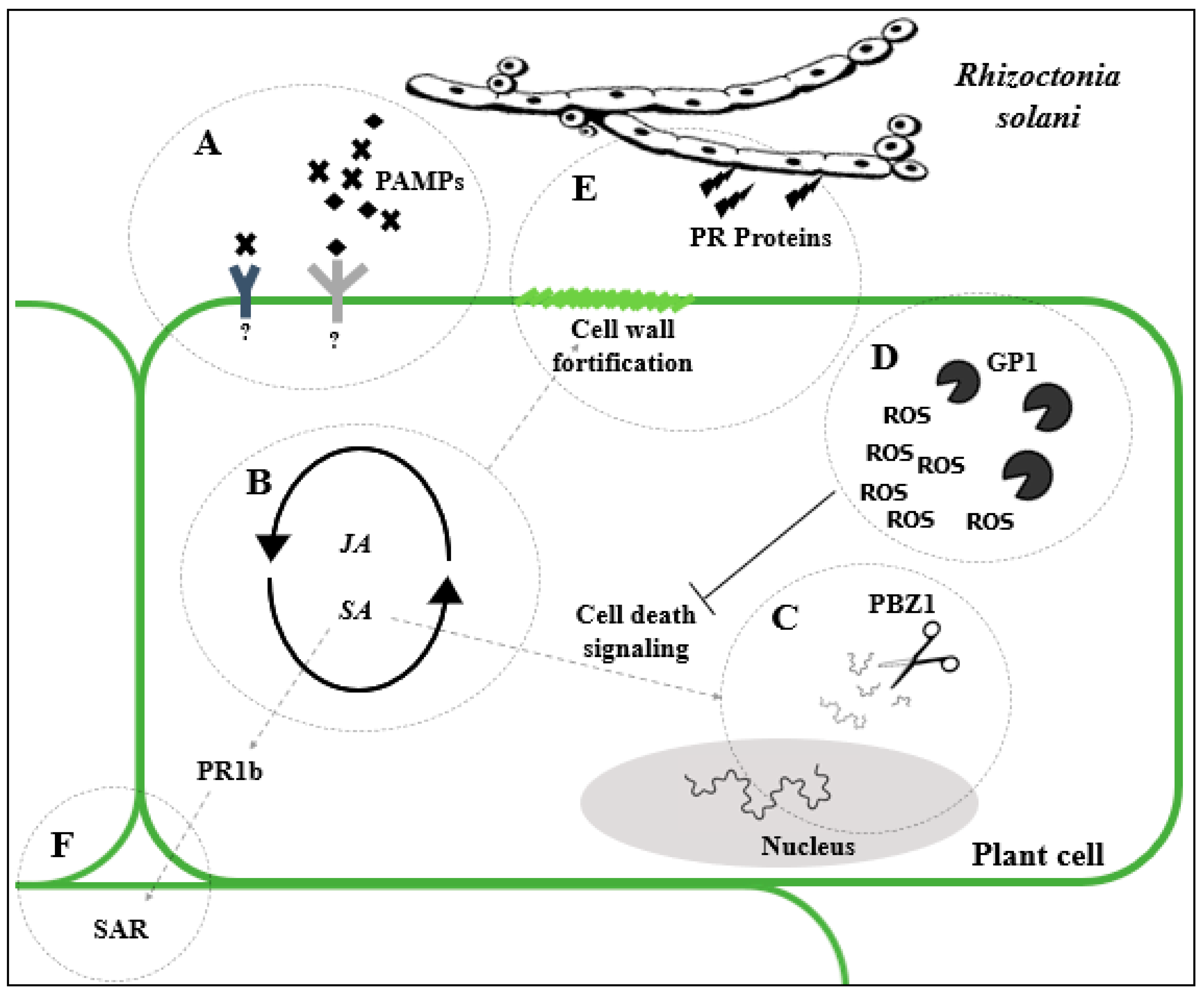
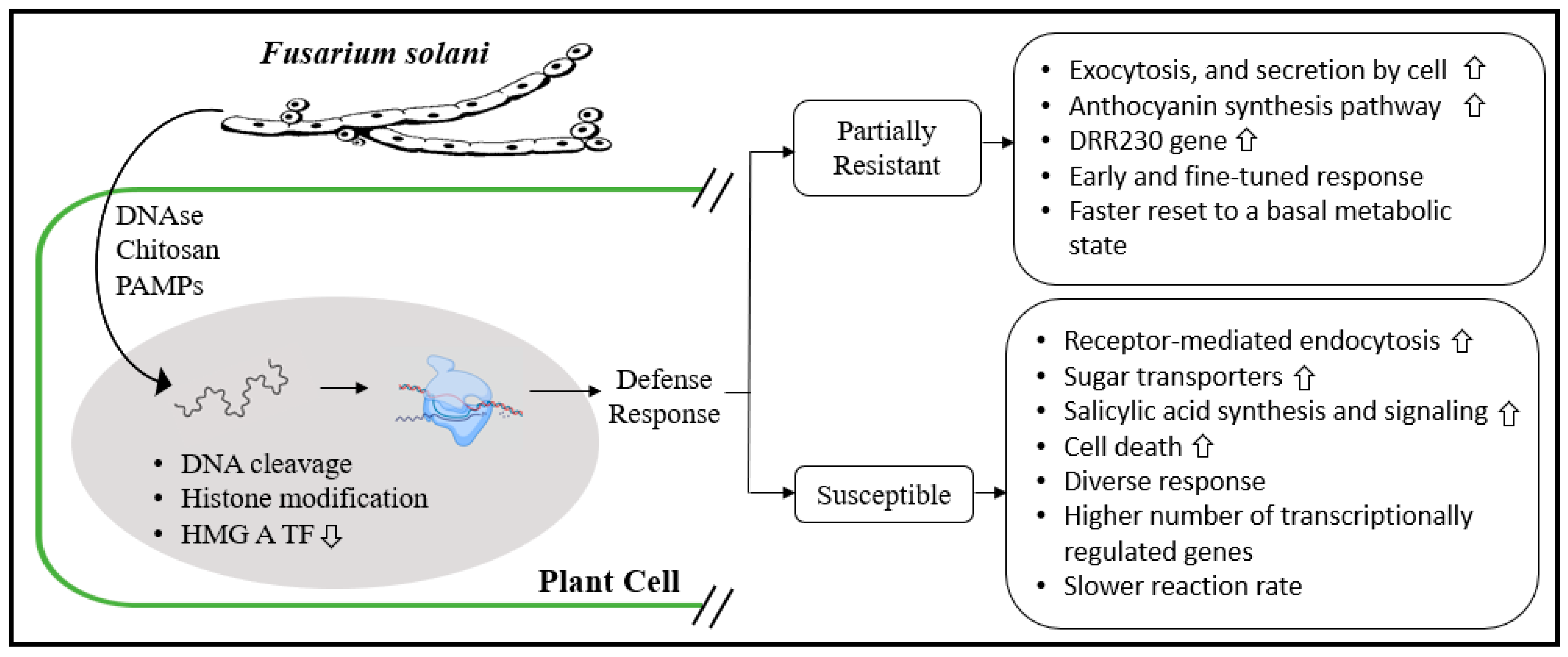

| Crops spp. | Disease Name | AG | Resistance | References |
|---|---|---|---|---|
| Barley (Hordeum vulgare) | Root rot | 5 | No reports | [36,37,38,39] |
| 8 | Moderate and high levels of resistance. High resistant transgenic lines | |||
| Bean (Phaseolus vulgaris) | Root rot | 2 | Moderate and high levels of resistance | [40,41,42,43] |
| 4 | Moderate and high levels of resistance | |||
| 5 | Moderate levels of resistance | |||
| Cabbage (Brassica oleracea var. capitata L.) | Wirestem | 2 | Moderate levels of resistance | [44,45,46] |
| 4 | Moderate levels of resistance | |||
| Carrot (Daucus carota) | Crown and brace root rot | 1 | Moderate levels of resistance | [47,48,49] |
| 2 | Moderate levels of resistance | |||
| 4 | Moderate levels of resistance. Moderately resistant transgenic line | |||
| Faba bean (Vicia faba) | Root rot and stem canker | 2 | Moderate levels of resistance | [50,51,52] |
| 4 | Moderate and high levels of resistance | |||
| 5 | Moderate levels of resistance | |||
| Lettuce (Lactuca sativa) | Bottom rot | 1 | No reports | [53] |
| Maize (Zea mays) | Banded leaf and sheath blight disease | 1 | Moderate and high levels of resistance | [52,54,55,56,57] |
| 2 | Moderate levels of resistance | |||
| Oat (Avena sativa) | Root rot, Bare patch | 8 | Moderate levels of resistance | [38] |
| Oilseed rape (Brassica napus) | Root rot and damping-off | 2 | Moderate levels of resistance | [58,59,60] |
| 4 | No reports | |||
| Onion (Allium cepa) | Stunting | 4 | No reports | [61,62] |
| 8 | Moderate levels of resistance | |||
| Pea (Pisum sativum) | Root rot | 4 | Moderate levels of resistance | [63,64] |
| Potato (Solanum tuberosum) | Black scurf and Stem canker | 2 | Moderate levels of resistance | [65,66,67,68] |
| 3 | Moderate and high levels of resistance | |||
| Rice (Oryza sativa) | Sheath blight | 1 | Moderate levels of resistance. Moderate levels of resistant transgenic lines. | [69,70,71,72] |
| Rye (Secale cereale) | Root rot, Bare patch | 8 | Moderate levels of resistance | [38,73] |
| Soybean (Glycine max) | Root rot | 1 | Moderate levels of resistance | [74,75,76,77] |
| 2 | Moderate and high levels of resistance | |||
| 4 | Moderate and high levels of resistance | |||
| Sugar beet (Beta vulgaris) | Root rot | 2 | Moderate levels of resistance | [78,79,80] |
| Tomato (Solanum lycopersicum) | Foot and Root rot | 3 | Moderate levels of resistance | [81,82,83] |
| Foot and Root rot | 4 | Moderate levels of resistance | ||
| Triticale (Triticale hexaploide) | Root rot, Bare patch | 8 | Moderate levels of resistance | [38] |
| Wheat (Triticum aestivum) | Root rot, Bare patch | 8 | Moderate levels of resistance | [38,39,84,85] |
| Fungi spp. | Crop spp. | Disease Name | Resistance Reported in the Literature | Host Range | Distribution | References |
|---|---|---|---|---|---|---|
| Fusarium avenaceum | Alfalfa (Medicago sativa) | Root and crown rot | Moderate levels of resistance | Wide range with over 200 species, including pulses, cereals, canola (Brassica napus), flax (Linum spp.), and alfalfa (Medicago truncatula) | Worldwide | [94] |
| Barley (Hordeum vulgare) | Head blight | Moderate levels of resistance | [95,96] | |||
| Clover (Trifolium subterraneum) | Root rot | Moderate levels of resistance | [97] | |||
| Pea (Pisum sativum) | Root rot | Moderate levels of resistance | [98,99,100] | |||
| Wheat (Triticum aestivum) | Head blight | Moderate levels of resistance | [101,102,103] | |||
| F. culmorum | Barley (Hordeum vulgare) | Head blight | Moderate levels of resistance | Wide range of host plants, including rye (Secale cereale), corn (Zea mays), sorghum (Sorghum spp.), various grasses, sugar beet (Beta vulgaris), bean (Phaseolus spp.), pea (Pisum sativum), asparagus (Asparagus spp.), hop (Humulus lupulus), strawberry (Fragaria × ananassa), and potato (Solanum tuberosum) | Worldwide | [104,105] |
| Oat (Avena sativa) | Head blight | Moderate levels of resistance | [106] | |||
| Wheat (Triticum aestivum) | Root rot and head blight | Moderate levels of resistance | [101,102,107,108,109] | |||
| F. graminearum | Barley (Hordeum vulgare) | Head blight | Moderate levels of resistance | Wide range, especially many species of cereals and grasses such as oat (Avena sativa), rice (Oryza), cucumber (Cucumis sativus), soy (Glycine max), tomato (Lycopersicon spp.), alfalfa (Medicago truncatula), sorghum (Sorghum spp.) | Worldwide | [104,110,111,112] |
| Maize (Zea mays) | Ear mold and root rot | Moderate levels of resistance | [113,114,115,116,117] | |||
| Soybean (Glycine max) | Pod blight and root rot | High levels of resistance | [118,119,120] | |||
| Wheat (Triticum aestivum) | Head blight | Moderate levels of resistance | [104,121,122,123,124,125] | |||
| F. pseudograminearum | Barley (Hordeum vulgare) | Crown rot | Moderate levels of resistance | All major winter cereals barley (Hordeum vulgare), oats (Avena sativa) and grass genera, such as Phalaris, Agropyron and Bromus | All areas cultivated with wheat and barley | [126,127] |
| Wheat (Triticum aestivum) | Crown rot | Moderate levels of resistance | [128,129] | |||
| F. solani f sp. batatas | Sweetpotato (Ipomoea batatas) | Storage root | Moderate levels of resistance | Not well known | China | [130] |
| F. solani f. sp. glycines | Soybean (Glycine max) | Sudden death syndrome | Moderate levels of resistance | Broad range including bean (Phaseolus spp.), Soybean (Glycine max), alfalfa (Medicago truncatula), clover (Trifolium spp.), pea (Pisum sativum), corn (Zea mays), wheat (Triticum spp.), ryegrass (Lolium spp.), pigweed (Amaranthus spp.), and lambsquarters (Chenopodium album) | All areas cultivated with soybean in America, Asia, and Africa | [131,132,133] |
| F. solani f. sp. phaseoli | Bean (Phaseolus vulgaris) | Root rot | Moderate and high levels of resistance | Pea (Pisum sativum) | Areas cultivated with bean in all continents except Australia | [134,135,136] |
| F. solani f. sp. pisi | Pea (Pisum sativum) | Root rot | Moderate levels of resistance | Chickpea (Cicer arietinum), clover, soybean, as well as several other non-legume hosts, such as ryegrass (Lolium spp.), potato (Solanum tuberosum) ginseng (Panax ginseng) and mulberry tree (Morus alba) | Worldwide | [137,138] |
| F. verticillioides | Maize (Zea mays) | Root and ear rot | Moderate levels of resistance | Wide range of hosts such as rice (Oryza sativa), sorghum (Sorghum spp.), soybean (Glycine max), alfalfa (Medicago truncatula), bean (Phaseolus spp.), wheat (Triticum spp.), ryegrass (Lolium spp.) | Worldwide | [139] |
| Fungi spp. | Crop spp. | Disease Name | Resistance Reported in the Literature | Host Range | Distribution | References |
|---|---|---|---|---|---|---|
| Phoma betae | Sugar beet (Beta vulgaris) | Crown and root rot | Moderate levels of resistance | Different varieties of Beta vulgaris such as table beet, sugar beet, Swiss chard | World-wide distribution, found in all beet-growing areas. | [151] |
| P. terrestris (Setophoma terrestris) | Corn (Zea mays) | Red root rot | Moderate levels of resistance | 45 genera including cereals, vegetables and grasses such as soybean (Glycine max), pea (Pisum sativum), sugarcane (Saccharum spp.), oats (Avena sativa), barley (Hordeum vulgare), wheat (Triticum spp.), cucumber (Cucumis sativus), tomato (Lycopersicon spp.), pepper (Capsicum annuum) | World-wide distribution | [152] |
| Onion (Allium cepa) | Pink root rot | Moderate levels of resistance | [153,154] | |||
| P. sclerotioides | Alfalfa (Medicago sativa) | Brown root rot | Moderate levels of resistance | Wheat (Triticum spp.), barley (Hordeum vulgare), and oat (Avena sativa). | All areas cultivated with alfalfa in North America and Australia | [155,156] |
| Thielaviopsis basicola | Cotton (Gossypium herbaceum) | Black root rot | Moderate levels of resistance. Moderate levels of resistance in transgenic lines | Wide range of hosts, plants from at least 15 families including horseradish (Armoracia rusticana), carrot (Daucus carota), strawberry (Fragaria × ananassa), tomato (Lycopersicon spp.), bean (Phaseolus spp), pea (Pisum sativum) | World-wide distribution | [157,158,159,160] |
| Tobacco (Nicotiana spp.) | Black root rot | Moderate levels of resistance | [161,162] |
| Oomycetes spp. | Crop spp. | Reported Symptoms | Resistance Reported in the Literature | Host Range | Distribution | References |
|---|---|---|---|---|---|---|
| Aphanomyces cochlioides | Sugar beet (Beta vulgaris) | Root rot and damping off. Infected hypocotyl and root rapidly turn black. Undersized plants with yellowed lower leaves. Severely infected plants die. Postharvest reduction in sugar yield | High levels of resistance | Spinach (Spinacia oleracea), and several wild species of Beta, including B. maritima and B. patellaris | Across all sugar beet plantations | [170,171,172,173,174] |
| Aphanomyces euteiches | Alfalfa (Medicago sativa) | Damping off and root rot. Root tissue becomes honey-brown or blackish-brown. Chlorosis, necrosis, and wilting of the foliage. Severely infected plants die. | High levels of resistance | Faba bean (Vicia faba), red clover (Trifolium pratense), white clover (Trifolium repens), Medicago truncatula, lentil (Lens culinaris) | All areas cultivated with alfalfa, bean, and pea in Asia, Europe, Oceania, North America | [6,135,175,176,177,178] |
| Bean (Phaseolus vulgaris) | Levels of partial and complete resistance | |||||
| Pea (Pisum sativum) | High levels of partial resistance | |||||
| Phytophthora citrophthora | Citrus spp. | Serious gummosis of citrus trees, root rot, stem necrosis, canker, fruit rot, twig blight, and seedling blight. | Tolerant transgenic C. sinensis. Partial levels of resistance in citrus rootstocks. High levels of resistant citrus rootstocks | 88 genera including: kiwifruit (Actinidia deliciosa), watermelon (Citrullus lanatus) strawberry (Fragaria ananassa), walnut (Juglans regia), apricot (Prunus armeniaca), sweet cherry (P. avium), almond (P. dulcis), potato (Solanum tuberosum), cocoa (Theobroma cacao), blueberries (Vaccinium) | Worldwide | [179,180,181,182,183,184] |
| Phytophthora nicotianae | Citrus spp. | Symptoms vary per host. Damping-off, crown rot, leaf blight, fruit rot. Occasionally, it attacks aerial parts of the plant and can cause brown rot of fruit. | Tolerant transgenic C. limonia. Partial levels of resistance in citrus rootstocks | 255 genera in 90 families. including tobacco, citrus, cotton, and orchids | Worldwide | [180,181,185,186,187,188,189] |
| Tomato (Solanum lycopersicum) | Partial levels of resistance | |||||
| Phytophthora cactorum | Apple (Malus domestica) | Damping off of seedlings, fruit rot, leaf, stem and root rot, collar and crown rot, stem canker. | Partial levels of resistance | 154 genera of vascular plants in 54 families | Worldwide | [181,190,191,192,193,194,195] |
| Strawberry (Fragaria × ananassa) | Partial levels of resistance | |||||
| Phytophthora cinnamomi | Avocado (Persea americana) | Root rot, heart rot, wilt. Primary infection at the feeder roots, resulting in a brownish black and brittle appearance. | Partial levels of resistance | 266 genera in 90 families, commonly hardwood trees. | Worldwide | [181,196] |
| Phytophthora fragariae | Rasberry (Rubus spp.) | Red stele or red core root rot. Symptoms also include wilting of leaves, reduced flowering, stunting, and bitter fruit | Partial and high levels of resistance | - | All areas cultivated with rasberry and strawberry in Asia, Australia, New Zealand, Europe, North America | [181,197,198,199,200] |
| Strawberry (Fragaria × ananassa) | Partial levels of resistance | |||||
| Phytophthora sojae | Soybean (Glycine max) | Root and stem rot; pre- and post-emergence damping-off, seedling wilt, seedling blight. Plant may turn reddish-orange to orange-brown in color. | Partial levels of resistance | Lupine (Lupinus spp.); also reported in six other genera in five families | All areas cultivated with soybean in Australia, North America (Canada, USA)., South America (Chile), Asia (Korea, China) and New Zealand | [181,201,202] |
| Phytophthora capsici | Pepper (Capsicum annuum) | Fruit, stem, and root rot., seedling damping-off, and leaf wilt. Leaf tissue becomes wilted, light green or gray-green, and later tan to white. Fruit rots are olive green or light green in color. | Partial levels of resistance | 51 genera in 28 families, including tomatoes (Lycopersicon esculentum), other Solanaceae spp., Macadamia spp., cacao (Theobroma cacao) | Worldwide | [181,203,204,205] |
| Phytophthora medicaginis | Alfalfa (Medicago sativa) | Root rot, damping-off of seedlings. Reddish-brown or black root lesions. Mature plants exhibit chlorosis, desiccation of foliage, and reduced growth but may also collapse. | High levels of resistance | Sainfoin (Onobrychis viciifolia), chickpea (Cicer arietinum), cherry (Prunus mahaleb) | Cosmopolitan, throughout the range of the host | [181,206,207,208,209,210] |
| Chickpea (Cicer arietinum) | Partial levels of resistance | |||||
| Soybean (Glycine max) | Partial levels of resistance | |||||
| Pythium ultimum | Bean (Phaseolus vulgaris) | Disease can manifest as seed rot, preemergence and postemergence damping-off, root rot, dark brown or reddish roots, and sunken lesions on lower hypocotyls. Plants are stunted or chlorotic. Root tips of diseased plants appear as brown. | High levels of resistance | Cabbage (Brassica oleracea var. capitata), carrot (Daucus carota), melon (Cucumis melon), wheat (Triticum aestivum) | Worldwide | [211,212,213,214,215] |
| Cucumber (Cucumis sativus) | No records | |||||
| Sorghum (Sorghum bicolor) | No records | |||||
| Soybean (Glycine max) | Partial levels of resistance | |||||
| Sugar beet (Beta vulgaris) | Partial levels of resistance | |||||
| Tomato (Solanum lycopersicum) | No records | |||||
| Pythium irregulare | Clover (Trifolium subterraneum) | Pre- and post-emergence damping off of seedlings (greenhouse) and root rot (field) of older plants. Contaminated seeds and seedlings will quickly turn brown and soft before decomposing. Foliage may turn chlorotic or a greenish-grey and wilt. | Moderate resistance | Over 200 host species including pineapple (Ananas comosus), cereals, grasses, celery (Apium graveolens), pepper (Capsicum annuum), pecan trees (Carya illinoinensis), Citrus spp, strawberries (Fragaria × ananassa), lentils (Lens culinaris), corn (Zea mays), soybean (Glycine max), cucumber (Cucumis sativus), onion (Allium cepa), carrot (Daucus carota) and a number of floricultural crops | Cosmopolitan in greenhouses and field systems | [97,216,217,218] |
| Soybean (Glycine max) | Moderate to high levels of resistance | |||||
| Pythium aphanidermatum | Cucumber (Cucumis sativus) | Causes root and stem rots, as well as pre- and post- emergence damping off. It causes blights of grasses and fruit. Roots are blackened, mushy and rotten. It causes wilting, loss of vigor, stunting, chlorosis and leaf drop. Beets and other fleshy plant organs are susceptible to rot in the field and during storage. | No reports | Broad host range, including cotton (Gossypium spp.), grasses, papaya (Carica papaya), cereals, Brassica species and beans (Phaseolus vulgaris) | Cosmopolitan in greenhouses and field systems | [219,220,221,222,223] |
| Lettuce (Lactuca sativa) | No reports | |||||
| Pepper (Capsicum annuum) | Moderate levels of tolerance | |||||
| Soybean (Glycine max) | High resistance | |||||
| Sugar beet (Beta vulgaris) | Partial levels of resistance | |||||
| Tomato (Solanum lycopersicum) | No reports |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Williamson-Benavides, B.A.; Dhingra, A. Understanding Root Rot Disease in Agricultural Crops. Horticulturae 2021, 7, 33. https://doi.org/10.3390/horticulturae7020033
Williamson-Benavides BA, Dhingra A. Understanding Root Rot Disease in Agricultural Crops. Horticulturae. 2021; 7(2):33. https://doi.org/10.3390/horticulturae7020033
Chicago/Turabian StyleWilliamson-Benavides, Bruce A., and Amit Dhingra. 2021. "Understanding Root Rot Disease in Agricultural Crops" Horticulturae 7, no. 2: 33. https://doi.org/10.3390/horticulturae7020033
APA StyleWilliamson-Benavides, B. A., & Dhingra, A. (2021). Understanding Root Rot Disease in Agricultural Crops. Horticulturae, 7(2), 33. https://doi.org/10.3390/horticulturae7020033






