Selection on BrFLC1 Is Related to Intraspecific Diversity of Brassica rapa Vegetables
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sequence Variation in Genome Region of BrFLC1
2.2. Plant Materials and Growth Conditions
2.3. Evaluation of Bolting Time and Flowering Time
2.4. KASP Assay for BrFLC1 Genotyping
2.5. Statistical Analysis
3. Results
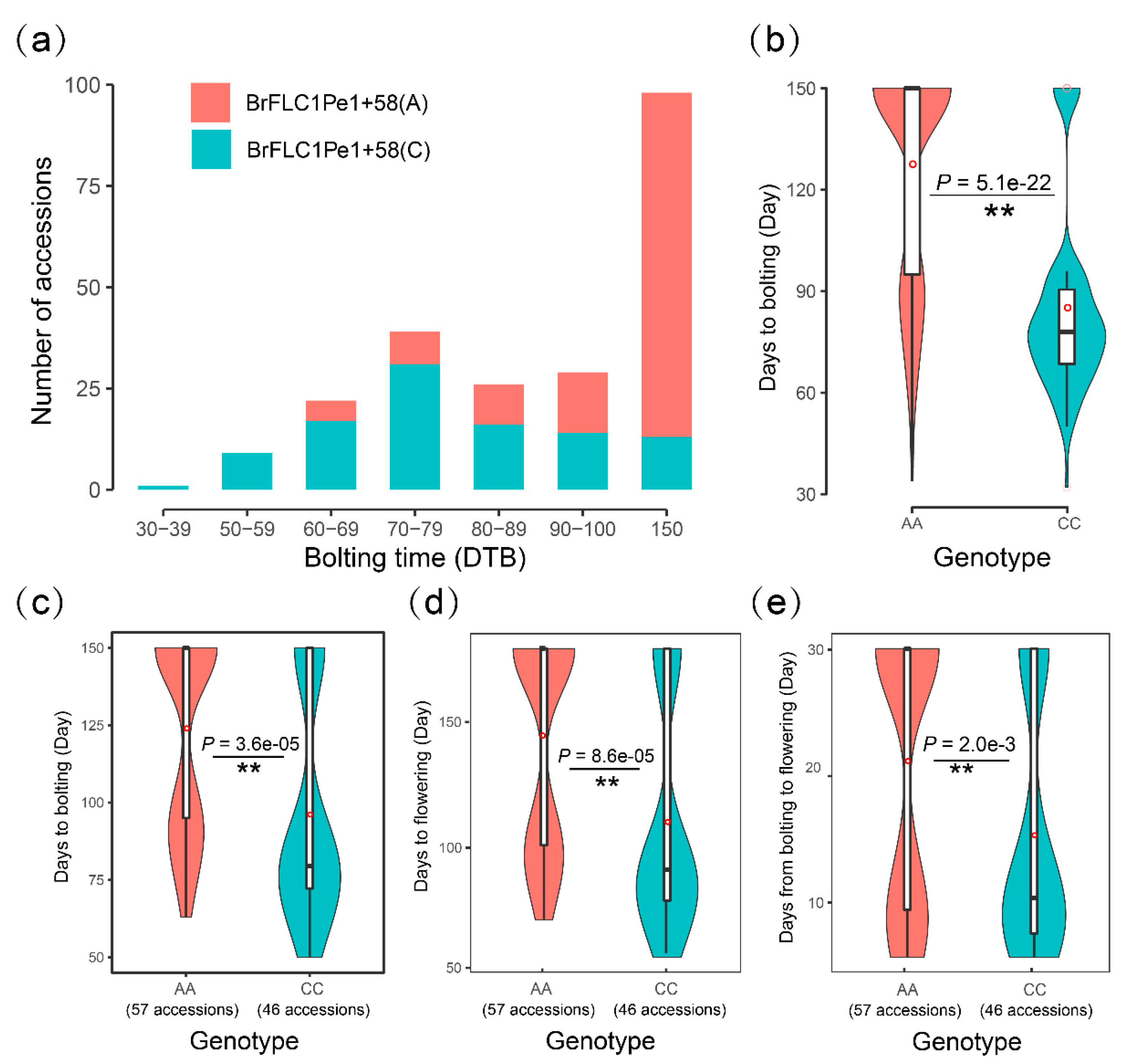
3.1. A Nonsynonymous Mutation Pe1+58(A/C) of BrFLC1 Differently Appeared among B. rapa Subspecies
3.2. Pe1+58(A/C) of BrFLC1 Is Related to Flowering Time Variations among a Germplasm Collection of 236 B. Rapa
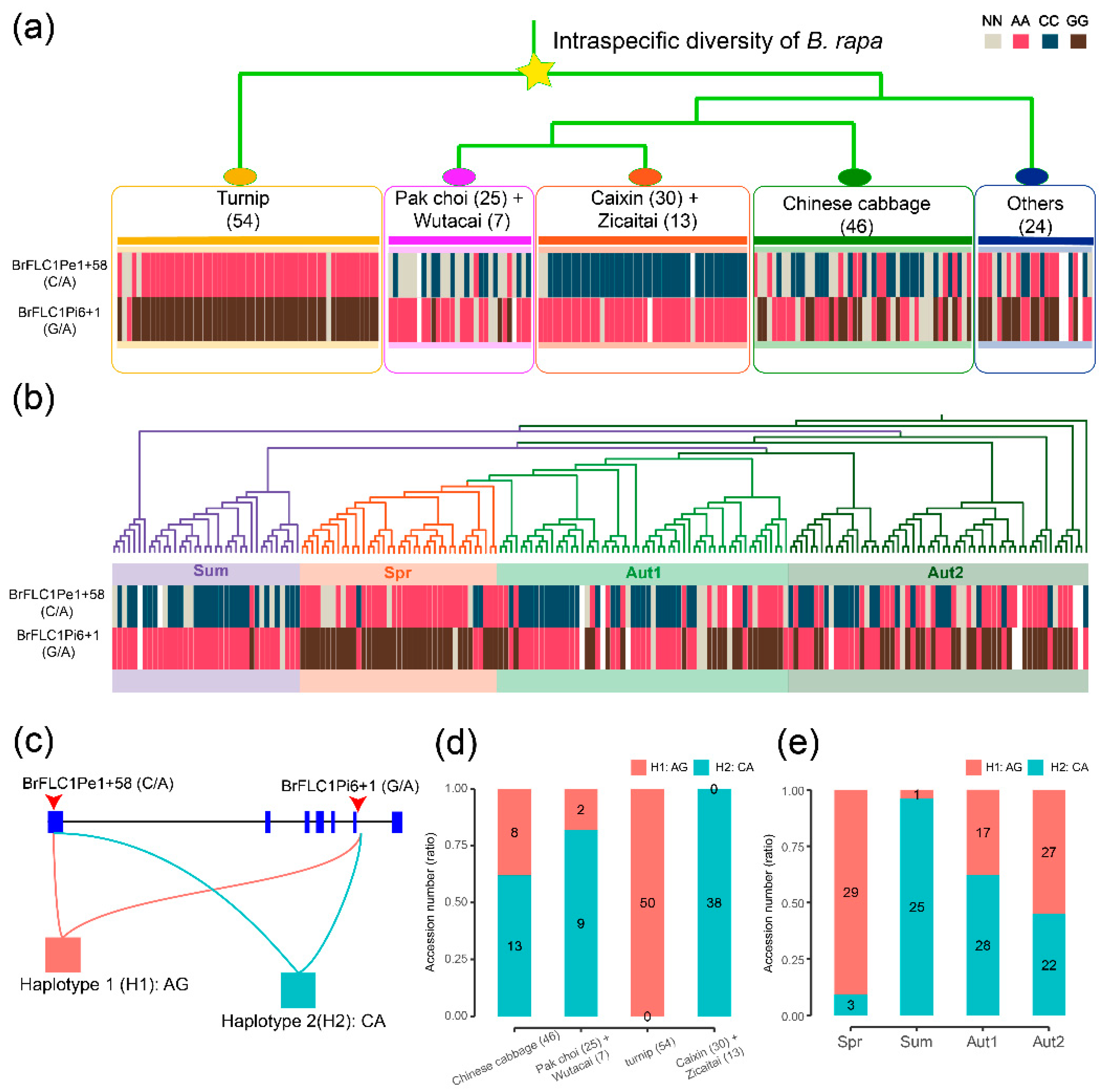
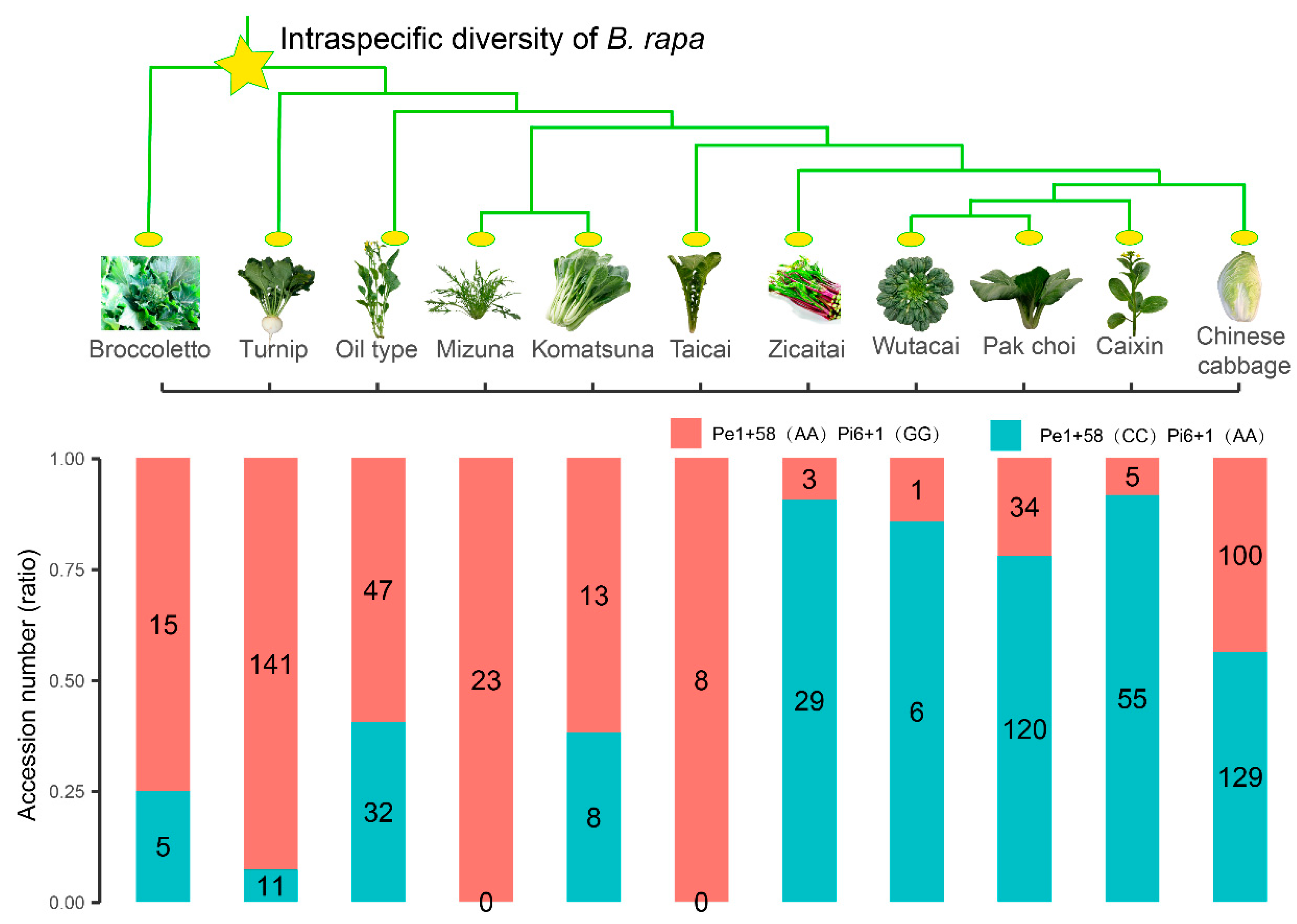
3.3. BrFLC1 Haplotype Consisting of BrFLC1Pe1+58 (A/C) and BrFLC1Pi6+1(G/A) Was Associated with the Intraspecific Diversification and Ecotype Diversification in B. rapa
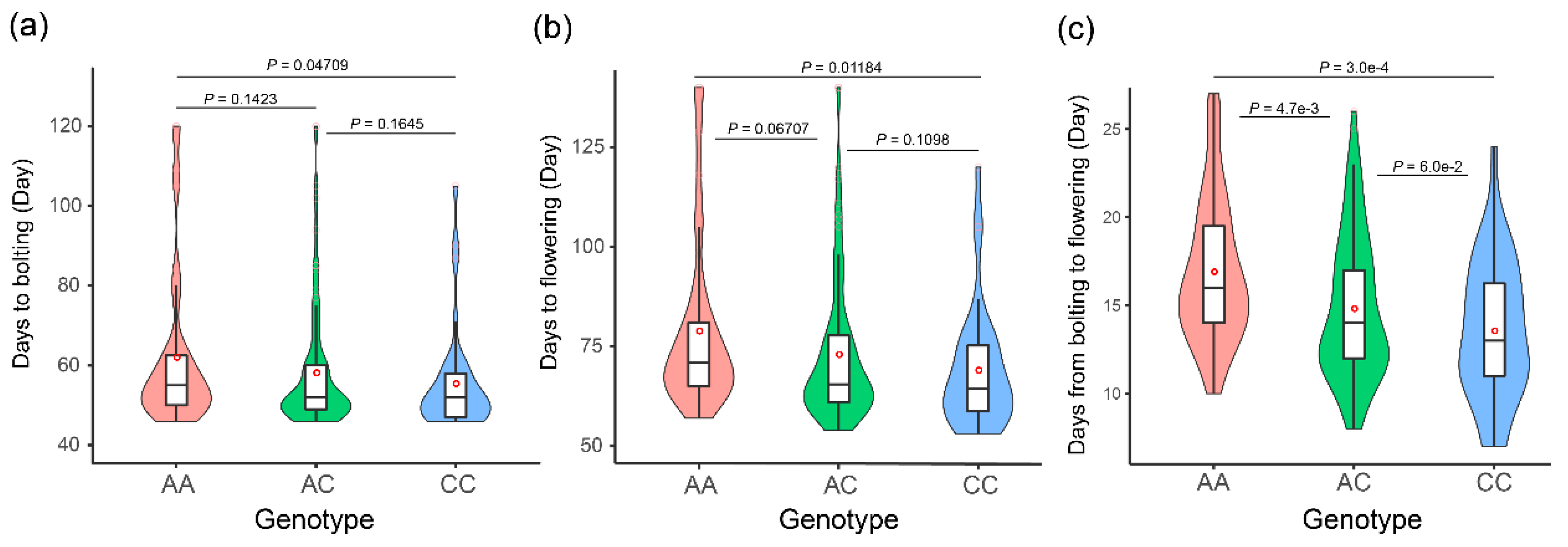
3.4. The BrFLC1Pe1+58 (A/C) Locus Poses Independent Genetic Effects on Flowering Time in B. rapa
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Fornara, F.; de Montaigu, A.; Coupland, G. SnapShot: Control of flowering in Arabidopsis. Cell 2010, 141, 550. [Google Scholar] [CrossRef] [Green Version]
- Deng, W.W.; Ying, H.; Helliwell, C.A.; Taylor, J.M.; Peacock, W.J.; Dennis, E.S. FLOWERING LOCUS C (FLC) regulates development pathways throughout the life cycle of Arabidopsis. Proc. Natl. Am. Sci. USA 2011, 108, 6680–6685. [Google Scholar] [CrossRef] [Green Version]
- Michaels, S.D.; Amasino, R.M. FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 1999, 11, 949–956. [Google Scholar] [CrossRef] [Green Version]
- Johanson, U.; West, J.; Lister, C.; Michaels, S.; Amasino, R.; Dean, C. Molecular analysis of FRIGIDA, a major determinant of natural variation in Arabidopsis flowering time. Science 2000, 290, 344–347. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wang, H.; Wang, J.; Sun, R.; Wu, J.; Liu, S.; Bai, Y.; Mun, J.H.; Bancroft, I.; Cheng, F.; et al. The genome of the mesopolyploid crop species Brassica rapa. Nat. Genet. 2011, 43, 1035–1039. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, T.J.; Kim, J.S.; Kwon, S.J.; Lim, K.B.; Choi, B.S.; Kim, J.A.; Jin, M.; Park, J.Y.; Lim, M.H.; Kim, H.I.; et al. Sequence-level analysis of the diploidization process in the triplicated FLOWERING LOCUS C region of Brassica rapa. Plant Cell 2006, 18, 1339–1347. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.Y.; Park, B.S.; Kwon, S.J.; Kim, J.; Lim, M.H.; Park, Y.D.; Kim, D.Y.; Suh, S.C.; Jin, Y.M.; Ahn, J.H.; et al. Delayed flowering time in Arabidopsis and Brassica rapa by the overexpression of FLOWERING LOCUS C (FLC) homologs isolated from Chinese cabbage (Brassica rapa ssp. pekinensis). Plant Cell Rep. 2007, 26, 327–336. [Google Scholar] [CrossRef] [PubMed]
- Lou, P.; Zhao, J.; Kim, J.S.; Shen, S.; Carpio, D.P.D.; Song, X.; Jin, M.; Vreugdenhil, D.; Wang, X.; Koornneef, M.; et al. Quantitative trait loci for flowering time and morphological traits in multiple populations of Brassica rapa. J. Exp. Bot. 2007, 58, 4005–4016. [Google Scholar] [CrossRef] [Green Version]
- Wu, J.; Wei, K.; Cheng, F.; Li, S.; Wang, Q.; Zhao, J.; Bonnema, G.; Wang, X. A naturally occurring InDel variation in BraA.FLC.b (BrFLC2) associated with flowering time variation in Brassica rapa. BMC Plant Biol. 2012, 12, 151. [Google Scholar] [CrossRef] [Green Version]
- Yuan, Y.X.; Wu, J.; Sun, R.F.; Zhang, X.W.; Xu, D.H.; Bonnema, G.; Wang, X.W. A naturally occurring splicing site mutation in the Brassica rapa FLC1 gene is associated with variation in flowering time. J. Exp. Bot. 2009, 60, 1299–1308. [Google Scholar] [CrossRef] [Green Version]
- Zhao, J.J.; Kulkarni, V.; Liu, N.; Carpio, D.P.D.; Bucher, J.; Bonnema, G. BrFLC2 (FLOWERING LOCUS C) as a candidate gene for a vernalization response QTL in Brassica rapa. J. Exp. Bot. 2010, 61, 1817–1825. [Google Scholar] [CrossRef] [Green Version]
- Kitamoto, N.; Yui, S.; Nishikawa, K.; Takahata, Y.; Yokoi, S. A naturally occurring long insertion in the first intron in the Brassica rapa FLC2 gene causes delayed bolting. Euphytica 2014, 196, 213–223. [Google Scholar] [CrossRef]
- Xi, X.; Wei, K.; Gao, B.; Liu, J.; Liang, J.; Feng, C.; Wang, X.; Wu, J. BrFLC5: A weak regulator of flowering time in Brassica rapa. Theor. Appl. Genet. 2018, 131, 2107–2116. [Google Scholar] [CrossRef] [PubMed]
- Okazaki, K.; Sakamoto, K.; Kikuchi, R.; Saito, A.; Togashi, E.; Kuginuki, Y.; Matsumoto, S.; Hirai, M. Mapping and characterization of FLC homologs and QTL analysis of flowering time in Brassica oleracea. Theor. Appl. Genet. 2007, 114, 595–608. [Google Scholar] [CrossRef]
- Chen, L.; Dong, F.; Cai, J.; Xin, Q.; Fang, C.; Liu, L.; Wan, L.; Yang, G.; Hong, D. A 2.833-kb insertion in BnFLC.A2 and its homeologous exchange with BnFLC.C2 during breeding selection generated early-flowering rapeseed. Mol. Plant 2017, 11, 222–225. [Google Scholar] [CrossRef] [PubMed]
- Cheng, F.; Sun, R.; Hou, X.; Zheng, H.; Zhang, F.; Zhang, Y.; Liu, B.; Liang, J.; Zhuang, M.; Liu, Y.; et al. Subgenome parallel selection is associated with morphotype diversification and convergent crop domestication in Brassica rapa and Brassica oleracea. Nat. Genet. 2016, 48, 1218–1224. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Bassetti, N.; Petrasch, S.; Zhang, N.; Bucher, J.; Shen, S.; Zhao, J.; Bonnema, G. What makes turnip: Anatomy, physiology and transcriptome during early stages of its hypocotyl-tuber development. Horti Res. 2019, 6, 38. [Google Scholar] [CrossRef] [Green Version]
- Su, T.; Wang, W.; Li, P.; Zhang, B.; Li, P.; Xin, X.; Sun, H.; Yu, Y.; Zhang, D.; Zhao, X.; et al. A genomic variation map provides insights into the genetic basis of spring Chinese Cabbage (Brassica rapa ssp. pekinensis) selection. Mol. Plant 2018, 11, 1360–1376. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, S.F.; Zhou, Y.Q.; Chen, Y.R.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, 884–890. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Lou, P.; Bonnema, G.; Yang, B.; He, H.; Zhang, Y.; Fang, Z. Linkage mapping of a dominant male sterility gene Ms-cd1 in Brassica oleracea. Genome 2005, 48, 848–854. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.; Chang, L.; Zhang, T.; Chen, H.; Zhang, L.; Lin, R.; Liang, J.; Wu, J.; Freeling, M.; Wang, X. Impacts of allopolyploidization and structural variation on intraspecific diversification in Brassica rapa. Genome Biol. 2021, 22, 166. [Google Scholar] [CrossRef] [PubMed]
- Fuller, D.Q. Contrasting patterns in crop domestication and domestication rates: Recent archaeobotanical insights from the Old World. Ann. Bot. 2007, 100, 903–924. [Google Scholar] [CrossRef]
- Izawa, T. Adaptation of flowering time by natural and artificial selection in Arabidopsis and rice. J. Exp. Bot. 2007, 58, 3091–3097. [Google Scholar] [CrossRef] [Green Version]
- Lu, K.; Wei, L.; Li, X.; Wang, Y.; Wu, J.; Liu, M.; Zhang, C.; Chen, Z.; Xiao, Z.; Jian, H.; et al. Whole-genome resequencing reveals Brassica napus origin and genetic loci involved in its improvement. Nat. Commun. 2019, 10, 1154. [Google Scholar] [CrossRef] [Green Version]
- Warwick, S.I. Brassicaceae in Agriculture. In Genetics and Genomics of the Brassicaceae; Schmidt, R., Bancroft, I., Eds.; Plant Genetics and Genomics: Crops and Models; Springer: New York, NY, USA, 2011; Volume 9, pp. 34–38. [Google Scholar]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, J.; Cai, X.; Li, Y.; Chen, Y.; Gao, B.; Lin, R.; Liang, J.; Wang, X.; Wu, J. Selection on BrFLC1 Is Related to Intraspecific Diversity of Brassica rapa Vegetables. Horticulturae 2021, 7, 247. https://doi.org/10.3390/horticulturae7080247
Liu J, Cai X, Li Y, Chen Y, Gao B, Lin R, Liang J, Wang X, Wu J. Selection on BrFLC1 Is Related to Intraspecific Diversity of Brassica rapa Vegetables. Horticulturae. 2021; 7(8):247. https://doi.org/10.3390/horticulturae7080247
Chicago/Turabian StyleLiu, Jiahe, Xu Cai, Yufang Li, Yue Chen, Baozhen Gao, Runmao Lin, Jianli Liang, Xiaowu Wang, and Jian Wu. 2021. "Selection on BrFLC1 Is Related to Intraspecific Diversity of Brassica rapa Vegetables" Horticulturae 7, no. 8: 247. https://doi.org/10.3390/horticulturae7080247
APA StyleLiu, J., Cai, X., Li, Y., Chen, Y., Gao, B., Lin, R., Liang, J., Wang, X., & Wu, J. (2021). Selection on BrFLC1 Is Related to Intraspecific Diversity of Brassica rapa Vegetables. Horticulturae, 7(8), 247. https://doi.org/10.3390/horticulturae7080247








