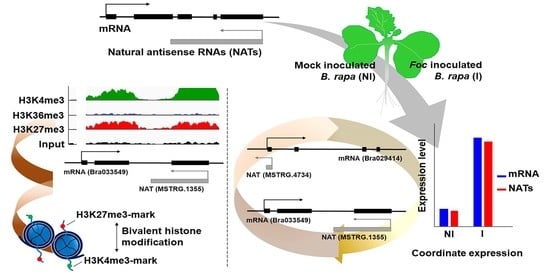
Transcriptional Association between mRNAs and Their Paired Natural Antisense Transcripts Following Fusarium oxysporum Inoculation in Brassica rapa L.
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Growth Conditions
2.2. RNA Extraction and Gene Expression Analysis
2.3. DNA Extraction and PCR
2.4. Detection of Epigenetic States in lncRNA Coding Regions
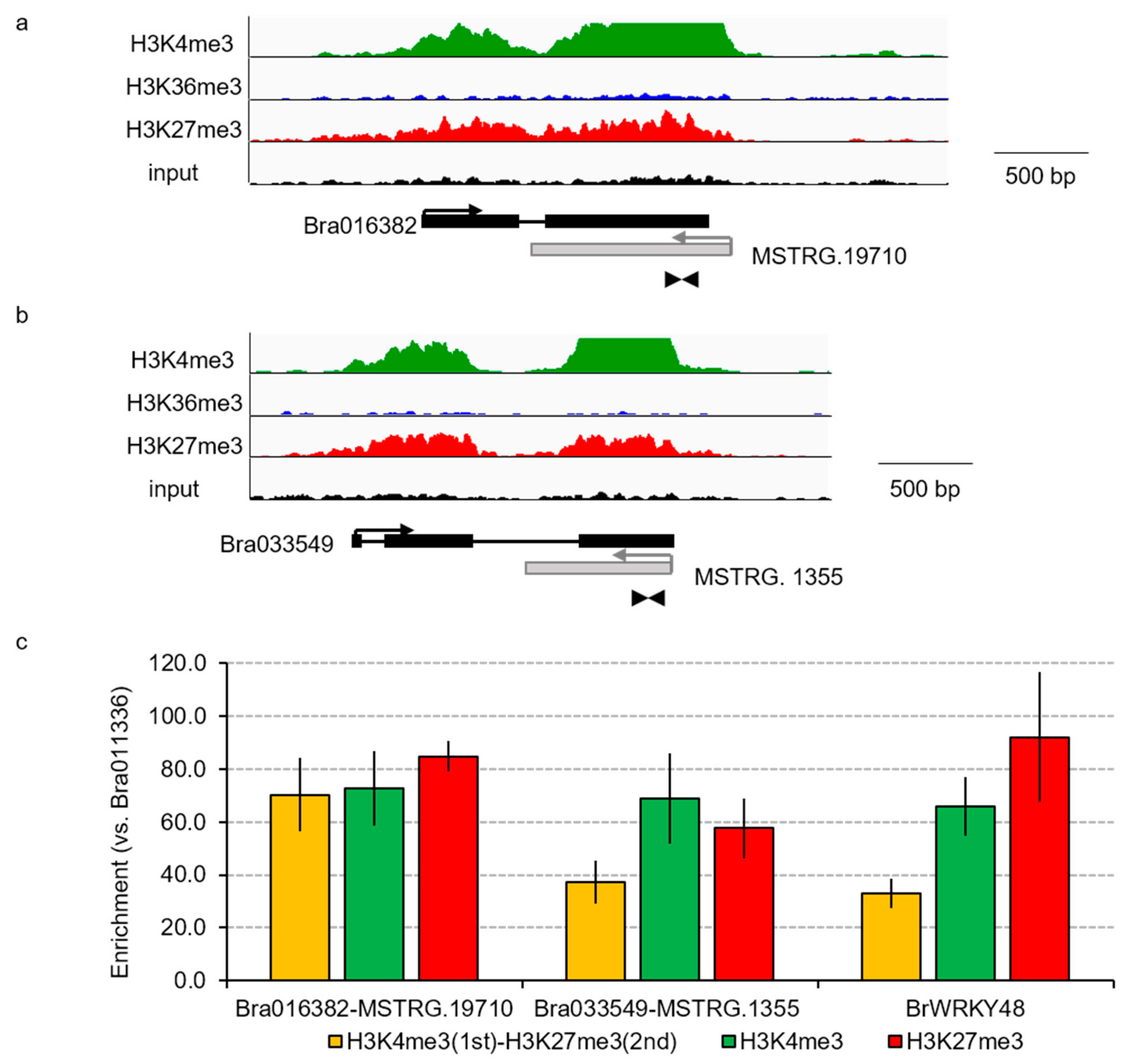
2.5. Sequential ChIP-qPCR
3. Results
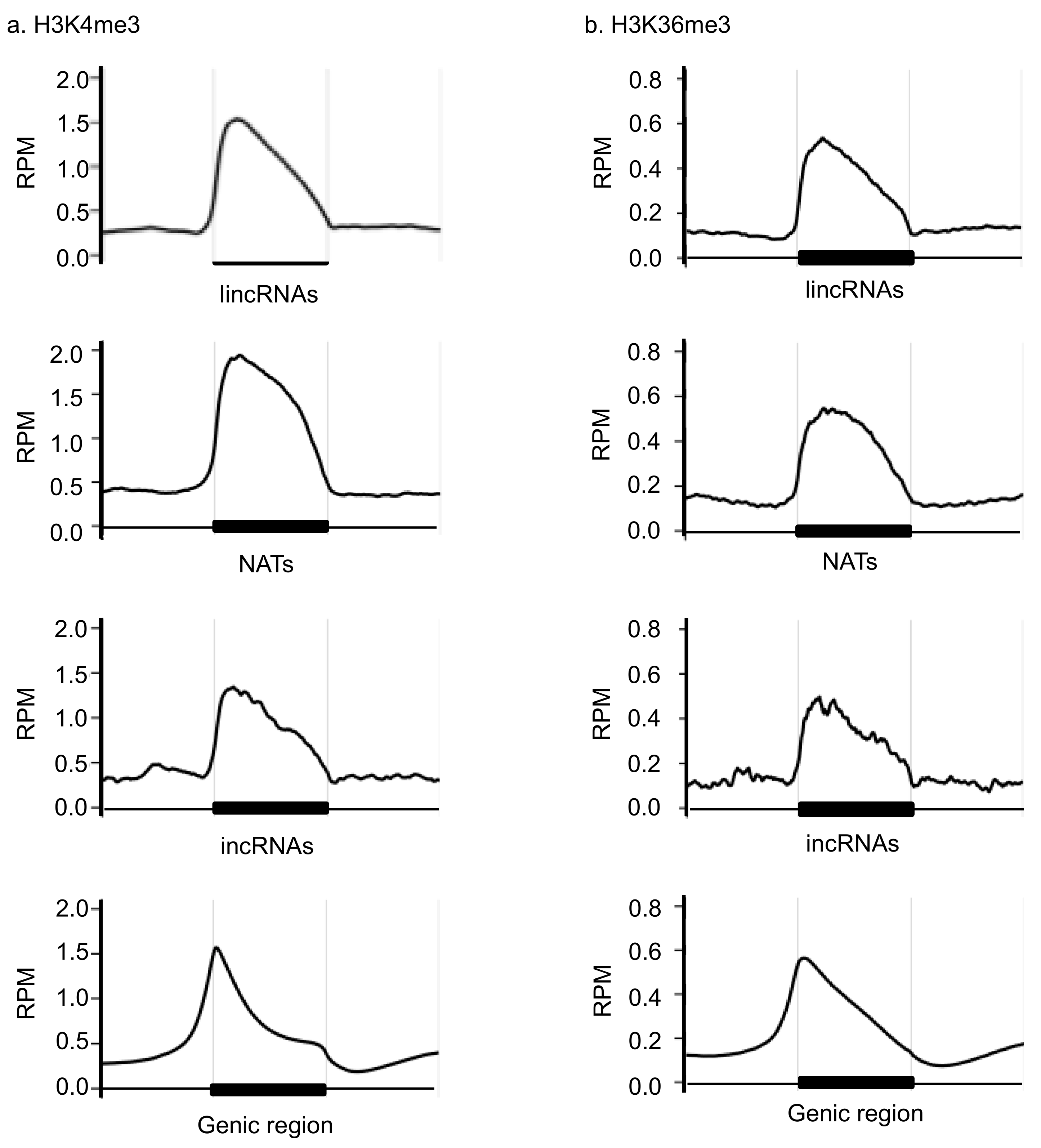
3.1. Comparison of the H3K4me3 and H3K36me3 States in the lncRNA Coding Region
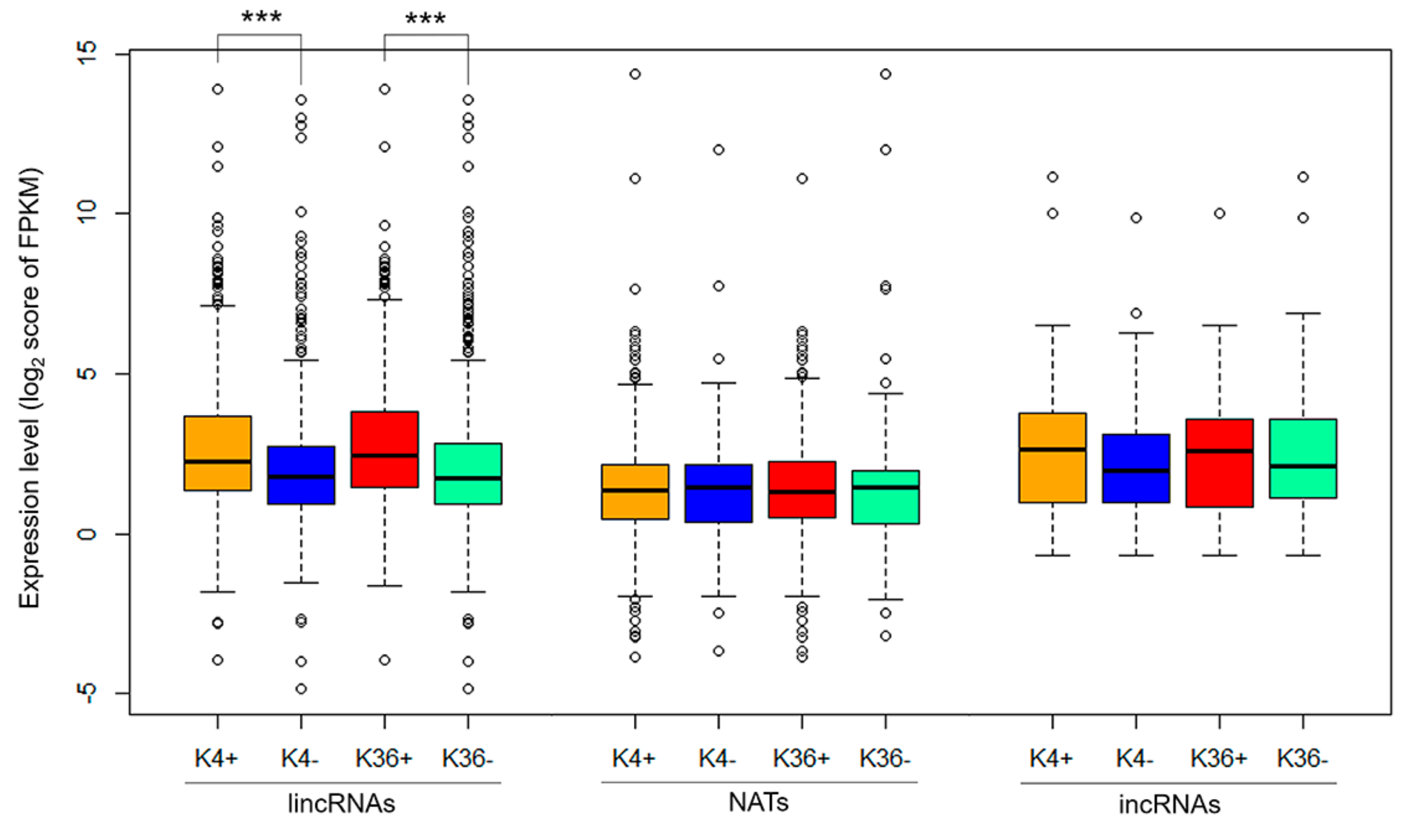
3.2. Characterization of Histone Modification States in Differentially Expressed Genes Following F. oxysporum f. sp. conglutinans (Foc) Inoculation and in Their Overlapping lncRNAs
3.3. Relationship between mRNA and Their Paired NAT Transcription Following F. oxysporum f. sp. conglutinans (Foc) Inoculation
3.4. Transcriptional Conservation of NATs among B. rapa Varieties
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lv, H.; Miyaji, N.; Osabe, K.; Akter, A.; Mehraj, H.; Shea, D.J.; Fujimoto, R. The importance of genetic and epigenetic research in the Brassica vegetables in the face of climate change. In Genomic Designing of Climate-Smart Vegetable Crops; Kole, C., Ed.; Springer: Cham, Switzerland, 2020; pp. 161–255. [Google Scholar]
- Fujimoto, R.; Uezono, K.; Ishikura, S.; Osabe, K.; Peacock, W.J.; Dennis, E.S. Recent research on the mechanism of heterosis is important for crop and vegetable breeding systems. Breed. Sci. 2018, 68, 145–158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mehraj, H.; Akter, A.; Miyaji, N.; Miyazaki, J.; Shea, D.J.; Fujimoto, R.; Doullah, M.A.U. Genetics of clubroot and Fusarium wilt disease resistance in Brassica vegetables: The application of marker assisted breeding for disease resistance. Plants 2020, 9, 726. [Google Scholar] [CrossRef] [PubMed]
- Enya, J.; Togawa, M.; Takeuchi, T.; Yoshida, S.; Tsushima, S.; Arie, T.; Sakai, T. Biological and phylogenetic characterization of Fusarium oxysporum complex, which causes yellows on Brassica spp., and proposal of F. oxysporum f. sp rapae, a novel forma specialis pathogenic on B. rapa in Japan. Phytopathology 2008, 98, 475–483. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akter, M.A.; Mehraj, H.; Itabashi, T.; Shindo, T.; Osaka, M.; Akter, A.; Miyaji, N.; Chiba, N.; Miyazaki, J.; Fujimoto, R. Breeding for disease resistance in Brassica vegetables using DNA marker selection. In Brassica Breeding and Biotechnology; Islam, A.K.M.A., Hossain, M.A., Islam, A.K.M.M., Eds.; IntechOpen: London, UK, 2021. [Google Scholar]
- Miyaji, N.; Akter, M.A.; Suzukamo, C.; Mehraj, H.; Shindo, T.; Itabashi, T.; Okazaki, K.; Shimizu, M.; Kaji, M.; Katsumata, M.; et al. Development of a new DNA marker for Fusarium yellows resistance in Brassica rapa vegetables. Plants 2021, 10, 1082. [Google Scholar] [CrossRef]
- Shimizu, M.; Fujimoto, R.; Ying, H.; Pu, Z.J.; Ebe, Y.; Kawanabe, T.; Saeki, N.; Taylor, J.M.; Kaji, M.; Dennis, E.S.; et al. Identification of candidate genes for Fusarium yellows resistance in Chinese cabbage by differential expression analysis. Plant Mol. Biol. 2014, 85, 247–257. [Google Scholar] [CrossRef]
- Kawamura, K.; Kawanabe, T.; Shimizu, M.; Okazaki, K.; Kaji, M.; Dennis, E.S.; Osabe, K.; Fujimoto, R. Genetic characterization of inbred lines of Chinese cabbage by DNA markers; towards the application of DNA markers to breeding of F1 hybrid cultivars. Data Brief 2016, 6, 229–237. [Google Scholar] [CrossRef] [Green Version]
- Miyaji, N.; Shimizu, M.; Miyazaki, J.; Osabe, K.; Sato, M.; Ebe, Y.; Takada, S.; Kaji, M.; Dennis, E.S.; Fujimoto, R.; et al. Comparison of transcriptome profiles by Fusarium oxysporum inoculation between Fusarium yellows resistant and susceptible lines in Brassica rapa L. Plant Cell Rep. 2017, 36, 1841–1854. [Google Scholar] [CrossRef] [Green Version]
- Miyaji, N.; Shimizu, M.; Takasaki-Yasuda, T.; Dennis, E.S.; Fujimoto, R. The transcriptional response to salicylic acid plays a role in Fusarium yellows resistance in Brassica rapa L. Plant Cell Rep. 2021, 40, 605–619. [Google Scholar] [CrossRef]
- Itabashi, E.; Osabe, K.; Fujimoto, R.; Kakizaki, T. Epigenetic regulation of agronomical traits in Brassicaceae. Plant Cell Rep. 2018, 37, 87–101. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.H. Multifaceted chromatin structure and transcription changes in plant stress response. Int. J. Mol. Sci. 2021, 22, 2013. [Google Scholar] [CrossRef]
- Talbert, P.B.; Henikoff, S. Histone variants at a glance. J. Cell Sci. 2021, 134, jcs244749. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Carey, M.; Workman, J.L. The role of chromatin during transcription. Cell 2007, 128, 707–719. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bannister, A.; Kouzarides, T. Regulation of chromatin by histone modifications. Cell Res. 2011, 21, 381–395. [Google Scholar] [CrossRef] [PubMed]
- Zhao, T.; Zhan, Z.; Jiang, D. Histone modifications and their regulatory roles in plant development and environmental memory. J. Genet. Genom. 2019, 46, 467–476. [Google Scholar] [CrossRef] [PubMed]
- Demetriadou, C.; Koufaris, C.; Kirmizis, A. Histone N-alpha terminal modifications: Genome regulation at the tip of the tail. Epigenetics Chromatin 2020, 13, 29. [Google Scholar] [CrossRef]
- Kim, J.M.; Sasaki, T.; Ueda, M.; Sako, K.; Seki, M. Chromatin changes in response to drought, salinity, heat, and cold stresses in plants. Front. Plant Sci. 2015, 6, 114. [Google Scholar] [CrossRef] [Green Version]
- Meyer, P. Epigenetic variation and environmental change. J. Exp. Bot. 2015, 66, 3541–3548. [Google Scholar] [CrossRef] [Green Version]
- Fujimoto, R.; Sasaki, T.; Ishikawa, R.; Osabe, K.; Kawanabe, T.; Dennis, E.S. Molecular mechanisms of epigenetic variation in plants. Int. J. Mol. Sci. 2012, 13, 9900–9922. [Google Scholar] [CrossRef] [Green Version]
- Quadrana, L.; Colot, V. Plant transgenerational epigenetics. Annu. Rev. Genet. 2016, 50, 467–491. [Google Scholar] [CrossRef]
- Sequeira-Mendes, J.; Aragüez, I.; Peiró, R.; Mendez-Giraldez, R.; Zhang, X.; Jacobsen, S.E.; Bastolla, U.; Gutierrez, C. The functional topography of the Arabidopsis genome is organized in a reduced number of linear motifs of chromatin states. Plant Cell 2014, 26, 2351–2366. [Google Scholar] [CrossRef] [Green Version]
- Qian, S.; Lv, X.; Scheid, R.N.; Lu, L.; Yang, Z.; Chen, W.; Liu, R.; Boersma, M.D.; Denu, J.M.; Zhong, X.; et al. Dual recognition of H3K4me3 and H3K27me3 by a plant histone reader SHL. Nat. Commun. 2018, 9, 2425. [Google Scholar] [CrossRef]
- Blanco, E.; González-Ramírez, M.; Alcaine-Colet, A.; Aranda, S.; Croce, L.D. The bivalent genome: Characterization, structure, and regulation. Trends Genet. 2020, 36, 118–131. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Z.; Zhang, W.; Marand, A.P.; Zhu, B.; Buell, C.R.; Jiang, J. Cold stress induces enhanced chromatin accessibility and bivalent histone modifications H3K4me3 and H3K27me3 of active genes in potato. Genome Biol. 2019, 20, 123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mehraj, H.; Takahashi, S.; Miyaji, N.; Akter, A.; Suzuki, Y.; Seki, M.; Dennis, E.S.; Fujimoto, R. Characterization of histone H3 lysine 4 and 36 tri-methylation in Brassica rapa L. Front. Plant Sci. 2021, 12, 659634. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Liu, K.; Yu, R.; Zhou, B.; Huang, P.; Cao, Z.; Zhou, Y.; Wang, J. From “Dark Matter” to “Star”: Insight into the regulation mechanisms of plant functional long non-coding RNAs. Front. Plant Sci. 2021, 12, 650926. [Google Scholar] [CrossRef] [PubMed]
- Collins, L.J.; Penny, D. The RNA infrastructure: Dark matter of the eukaryotic cell? Trends Genet. 2009, 25, 120–128. [Google Scholar] [CrossRef]
- Cech, T.R.; Steitz, J.A. The noncoding RNA revolution-trashing old rules to forge new ones. Cell 2014, 157, 77–94. [Google Scholar] [CrossRef] [Green Version]
- Ariel, F.; Romero-Barrios, N.; Jégu, T.; Benhamed, M.; Crespi, M. Battles and hijacks: Noncoding transcription in plants. Trends Plant Sci. 2015, 20, 362–371. [Google Scholar] [CrossRef]
- Ponting, C.P.; Oliver, P.L.; Reik, W. Evolution and functions of long noncoding RNAs. Cell 2009, 136, 629–641. [Google Scholar] [CrossRef] [Green Version]
- Chekanova, J.A. Long non-coding RNAs and their functions in plants. Curr. Opin. Plant Biol. 2015, 27, 207–216. [Google Scholar] [CrossRef] [Green Version]
- Rai, M.I.; Alam, M.; Lightfoot, D.A.; Gurha, P.; Afzal, A.J. Classification and experimental identification of plant long non-coding RNAs. Genomics 2019, 111, 997–1005. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Zhu, Q.H.; Kaufmann, K. Long non-coding RNAs in plants: Emerging modulators of gene activity in development and stress responses. Planta 2020, 252, 92. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Guo, H.; Hu, W.; Ji, W. The emerging role of long non-coding RNAs in plant defense against fungal stress. Int. J. Mol. Sci. 2020, 21, 2659. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Hu, J.; Wu, T.; Yang, Q.; Feng, X.; Lin, H.; Feng, S.; Cui, C.; Yu, Y.; Zhou, R.; et al. Comparative analysis of long noncoding RNAs in angiosperms and characterization of long noncoding RNAs in response to heat stress in Chinese cabbage. Hortic. Res. 2021, 8, 48. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Liu, S.; Qi, H.; Cai, H.; Xu, M. Research progress on plant long non-coding RNA. Plants 2020, 9, 408. [Google Scholar] [CrossRef] [Green Version]
- Asensi-Fabado, M.A.; Amtmann, A.; Perrella, G. Plant responses to abiotic stress: The chromatin context of transcriptional regulation. Biochim. Biophys. Acta Gene Regul. Mech. 2017, 1860, 106–122. [Google Scholar] [CrossRef] [Green Version]
- Akter, A.; Takahashi, S.; Deng, W.; Shea, D.J.; Itabashi, E.; Shimizu, M.; Miyaji, N.; Osabe, K.; Nishida, N.; Suzuki, Y.; et al. The histone modification H3 lysine 27 tri-methylation has conserved gene regulatory roles in the triplicated genome of Brassica rapa L. DNA Res. 2019, 26, 433–443. [Google Scholar] [CrossRef]
- Hung, F.Y.; Chen, C.; Yen, M.R.; Hsieh, J.W.A.; Chenlong, L.; Shih, Y.H.; Chen, F.F.; Chen, P.Y.; Cui, Y.; Wu, K. The expression of long non-coding RNAs is associated with H3Ac and H3K4me2 changes regulated by the HDA6-LDL1/2 histone modification complex in Arabidopsis. NAR Genom. Bioinfor. 2020, 2, lqaa066. [Google Scholar] [CrossRef]
- Mehraj, H.; Shea, D.J.; Takahashi, S.; Miyaji, N.; Akter, A.; Seki, M.; Dennis, E.S.; Fujimoto, R.; Osabe, K. Genome-wide analysis of long noncoding RNAs, 24-nt siRNAs, DNA methylation and H3K27me3 marks in Brassica rapa. PLoS ONE 2021, 16, e0242530. [Google Scholar]
- Zhu, Q.H.; Stephen, S.; Taylor, J.; Helliwell, C.A.; Wang, M.B. Long noncoding RNAs responsive to Fusarium oxysporum infection in Arabidopsis thaliana. New Phytol. 2014, 201, 574–584. [Google Scholar] [CrossRef]
- Shea, D.J.; Nishida, N.; Takada, S.; Itabashi, E.; Takahashi, S.; Akter, A.; Miyaji, N.; Osabe, K.; Mehraj, H.; Shimizu, M.; et al. Long noncoding RNAs in Brassica rapa L. following vernalization. Sci. Rep. 2019, 9, 9302. [Google Scholar] [CrossRef] [PubMed]
- Fujimoto, R.; Sasaki, T.; Nishio, T. Characterization of DNA methyltransferase genes in Brassica rapa. Genes Genet. Syst. 2006, 81, 235–242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murray, M.G.; Thompson, W.F. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980, 8, 4321–4326. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Finnegan, E.J.; Bond, D.M.; Buzas, D.M.; Goodrich, J.; Helliwell, C.A.; Tamada, Y.; Yun, J.Y.; Amasino, R.M.; Dennis, E.S. Polycomb proteins regulate the quantitative induction of VERNALIZATION INSENSITIVE 3 in response to low temperatures. Plant J. 2011, 65, 382–391. [Google Scholar] [CrossRef]
- Shea, D.J.; Shimizu, M.; Itabashi, E.; Miyaji, N.; Miyazaki, J.; Osabe, K.; Kaji, M.; Okazaki, K.; Fujimoto, R. Genome re-sequencing, SNP analysis, and genetic mapping of the parental lines of a commercial F1 hybrid cultivar of Chinese cabbage. Breed. Sci. 2018, 68, 375–380. [Google Scholar] [CrossRef]
- Liu, X.; Hao, L.; Li, D.; Zhu, L.; Hu, S. Long non-coding RNAs and their biological roles in plants. Genom. Proteom. Bioinform. 2015, 13, 137–147. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.L.; Chekanova, J.A. Long noncoding RNAs in plants. Adv. Exp. Med. Biol. 2017, 1008, 133–154. [Google Scholar]
- Wang, H.; Chung, P.J.; Liu, J.; Jang, I.C.; Kean, M.J.; Xu, J.; Chua, N.H. Genome-wide identification of long noncoding natural antisense transcripts and their responses to light in Arabidopsis. Genome Res. 2014, 24, 444–453. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Niu, Q.W.; Wu, H.W.; Liu, J.; Ye, J.; Yu, N.; Chua, N.H. Analysis of non-coding transcriptome in rice and maize uncovers roles of conserved lncRNAs associated with agriculture traits. Plant J. 2015, 84, 404–416. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Dong, H.; Zhou, D.; Li, M.; Liu, Y.; Zhang, F.; Feng, Y.; Yu, D.; Lin, S.; Cao, J. Systematic identification of long non-coding RNAs during pollen development and fertilization in Brassica rapa. Plant J. 2018, 96, 203–222. [Google Scholar] [CrossRef] [Green Version]
- Deforges, J.; Reis, R.S.; Jacquet, P.; Sheppard, S.; Gadekar, V.P.; Hart-Smith, G.; Tanzer, A.; Hofacker, I.L.; Iseli, C.; Xenarios, I.; et al. Control of cognate sense mRNA translation by cis-Natural Antisense RNAs. Plant Physiol. 2019, 180, 305–322. [Google Scholar] [CrossRef] [Green Version]
- Yu, Y.; Zhang, Y.; Chen, X.; Chen, Y. Plant noncoding RNAs: Hidden players in development and stress responses. Annu. Rev. Cell Dev. Biol. 2019, 35, 407–431. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Li, J.; Lian, B.; Gu, H.; Li, Y.; Qi, Y. Global identification of Arabidopsis lncRNAs reveals the regulation of MAF4 by a natural antisense RNA. Nat. Commun. 2018, 9, 5056. [Google Scholar] [CrossRef] [Green Version]
- Reis, R.S.; Poirier, Y. Making sense of the natural antisense transcript puzzle. Trends Plant Sci. 2021, 26, 1104–1115. [Google Scholar] [CrossRef] [PubMed]
- Wunderlich, M.; Gross-Hardt, R.; Schöffl, F. Heat shock factor HSFB2a involved in gametophyte development of Arabidopsis thaliana and its expression is controlled by a heat-inducible long non-coding antisense RNA. Plant Mol. Biol. 2014, 85, 541–550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cui, J.; Luan, Y.; Jiang, N.; Bao, H.; Meng, J. Comparative transcriptome analysis between resistant and susceptible tomato allows the identification of lncRNA16397 conferring resistance to Phytophthora infestans by co-expressing glutaredoxin. Plant J. 2017, 89, 577–589. [Google Scholar] [CrossRef] [Green Version]
- Van Damme, M.; Huibers, R.P.; Elberse, J.; den Ackerveken, G.V. Arabidopsis DMR6 encodes a putative 2OG-Fe(II) oxygenase that is defense-associated but required for susceptibility to downy mildew. Plant J. 2008, 54, 785–793. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhao, L.; Zhao, J.; Li, Y.; Wang, J.; Guo, R.; Gan, S.; Liu, C.J.; Zhang, K. S5H/DMR6 encodes a salicylic acid 5-hydroxylase that fine-tunes salicylic acid homeostasis. Plant Physiol. 2017, 175, 1082–1093. [Google Scholar] [CrossRef] [Green Version]

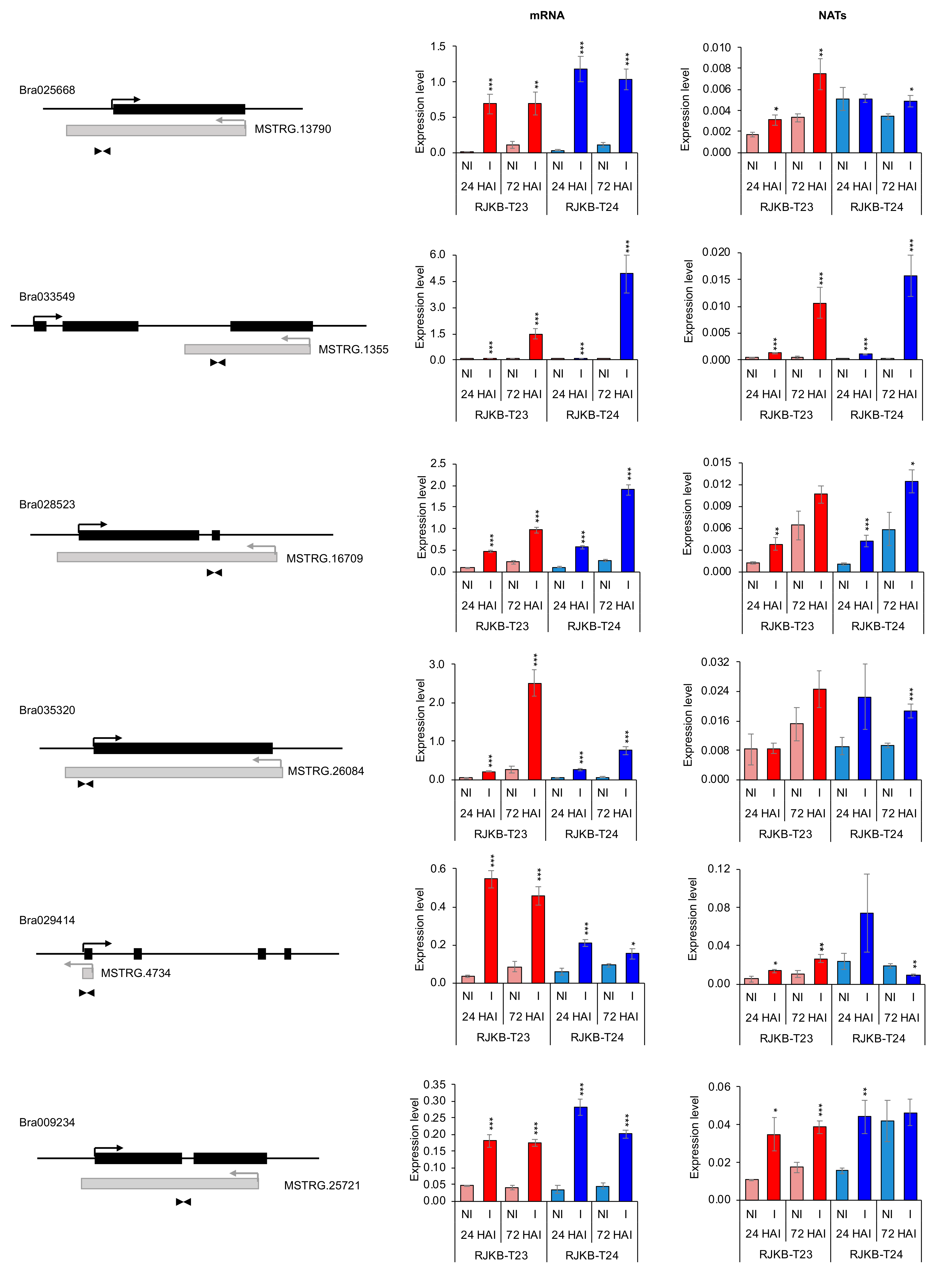
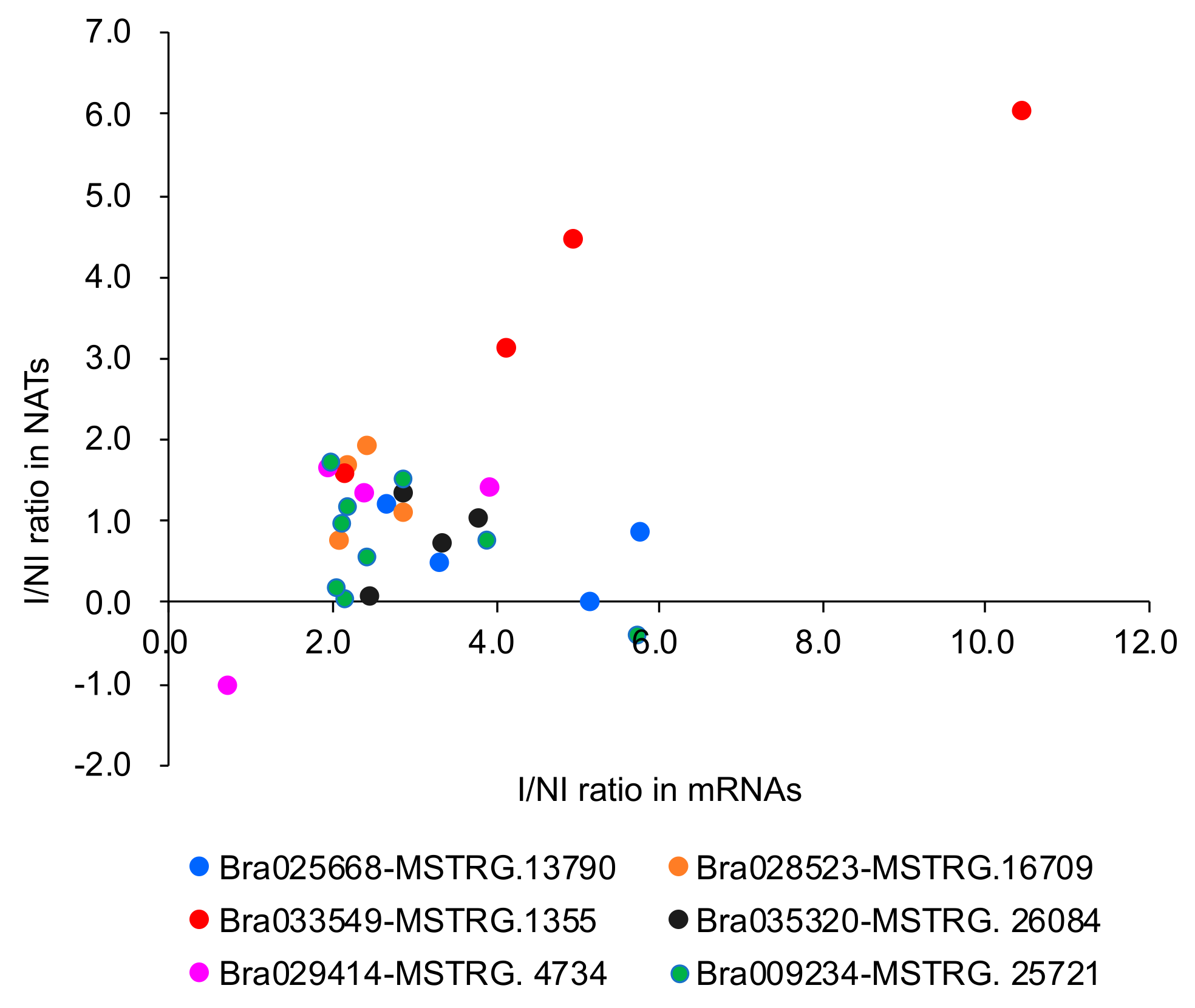
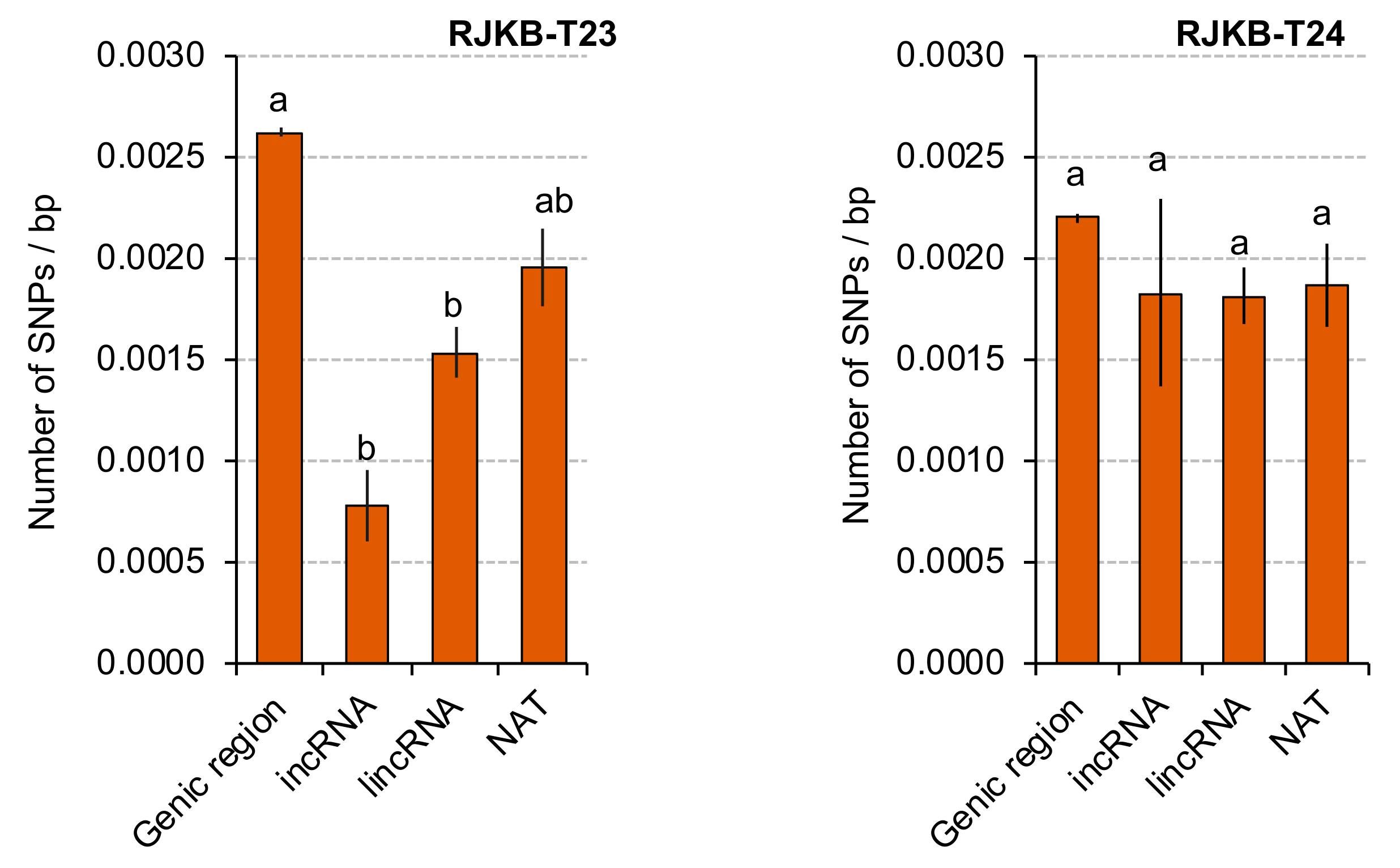

| mRNA and NAT Pair | H3K4me3 | H3K36me3 | H3K27me3 |
|---|---|---|---|
| Bra009234 | YES | YES | NO |
| MSTRG.25721 | |||
| Bra025668 | YES | YES | NO |
| MSTRG.13790 | |||
| Bra029414 | NO | NO | NO |
| MSTRG.4734 | |||
| Bra033549 | YES | NO | YES |
| MSTRG.1355 | |||
| Bra034404 | YES | YES | NO |
| MSTRG.11674 | |||
| Bra035320 | NO | NO | NO |
| MSTRG.26084 | |||
| Bra039006 | NO | NO | NO |
| MSTRG.15696 | |||
| Bra028523 | YES | YES | NO |
| MSTRG.16709 | |||
| Bra029946 | YES | YES | YES |
| MSTRG.1546 | |||
| Bra020438 | NO | NO | YES |
| MSTRG.3318 | |||
| Bra003511 | YES | NO | NO |
| MSTRG.17011 | |||
| Bra016382 | YES | NO | YES |
| MSTRG.19710 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Akter, M.A.; Mehraj, H.; Miyaji, N.; Takahashi, S.; Takasaki-Yasuda, T.; Seki, M.; Dennis, E.S.; Fujimoto, R.; Osabe, K. Transcriptional Association between mRNAs and Their Paired Natural Antisense Transcripts Following Fusarium oxysporum Inoculation in Brassica rapa L. Horticulturae 2022, 8, 17. https://doi.org/10.3390/horticulturae8010017
Akter MA, Mehraj H, Miyaji N, Takahashi S, Takasaki-Yasuda T, Seki M, Dennis ES, Fujimoto R, Osabe K. Transcriptional Association between mRNAs and Their Paired Natural Antisense Transcripts Following Fusarium oxysporum Inoculation in Brassica rapa L. Horticulturae. 2022; 8(1):17. https://doi.org/10.3390/horticulturae8010017
Chicago/Turabian StyleAkter, Mst. Arjina, Hasan Mehraj, Naomi Miyaji, Satoshi Takahashi, Takeshi Takasaki-Yasuda, Motoaki Seki, Elizabeth S. Dennis, Ryo Fujimoto, and Kenji Osabe. 2022. "Transcriptional Association between mRNAs and Their Paired Natural Antisense Transcripts Following Fusarium oxysporum Inoculation in Brassica rapa L." Horticulturae 8, no. 1: 17. https://doi.org/10.3390/horticulturae8010017
APA StyleAkter, M. A., Mehraj, H., Miyaji, N., Takahashi, S., Takasaki-Yasuda, T., Seki, M., Dennis, E. S., Fujimoto, R., & Osabe, K. (2022). Transcriptional Association between mRNAs and Their Paired Natural Antisense Transcripts Following Fusarium oxysporum Inoculation in Brassica rapa L. Horticulturae, 8(1), 17. https://doi.org/10.3390/horticulturae8010017