Abstract
Strawberry species (Fragaria spp.) are known as the “queen of fruits” and are cultivated around the world. Over the past few years, eight strawberry genome sequences have been released. The reuse of these large amount of genomic data, and the more large-scale comparative analyses are very challenging to both plant biologists and strawberry breeders. To promote the reuse and exploration of strawberry genomic data and enable extensive analyses using various bioinformatics tools, we have developed the Genome Database for Strawberry (GDS). This platform integrates the genome collection, storage, integration, analysis, and dissemination of large amounts of data for researchers engaged in the study of strawberry. We collected and formatted the eight published strawberry genomes. We constructed the GDS based on Linux, Apache, PHP and MySQL. Different bioinformatic software were integrated. The GDS contains data from eight strawberry species, as well as multiple tools such as BLAST, JBrowse, synteny analysis, and gene search. It has a designed interface and user-friendly tools that perform a variety of query tasks with a few simple operations. In the future, we hope that the GDS will serve as a community resource for the study of strawberries.
1. Introduction
Strawberries (Fragaria spp.), comprising of approximately 25 species [1], are plants from the Rosaceae. Their ploidy types range from diploid to decaploid [2,3], while wild members of the genus distributed throughout the northern hemisphere and parts of western South America [4]. The main cultivated and commercial strawberry species is the octoploid Fragaria ×ananassa (2n = 8x = 56) [5,6,7,8]. The first strawberry genome sequence from woodland strawberry (Fragaria vesca) was released in 2010 [9]. Since then, more and more strawberry species have been sequenced and annotated. In 2013, the cultivated strawberry (Fragaria ×ananassa) genome was sequenced using the Illumina and Roche 454 sequencing platforms [10], and was re-sequenced using a combination of short- and long-read approaches, producing a higher-quality assembly [11]. Strawberry genomics research not only promotes our understanding of the origin and evolution of strawberries but also has benefits for strawberry breeding [12].
Given these recent advances in strawberry genomics, it is necessary to establish a free online resource center for the integration of strawberry genome data. Therefore, we integrated the genomes and other related data of eight strawberry species (Fragaria ×ananassa, Fragaria iinumae, Fragaria nilgerrensis, Fragaria nipponica, Fragaria nubicola, Fragaria orientalis, Fragaria vesca, and Fragaria viridis) referring to the databases of other species, such as Arabidopsis, kiwifruit, and walnut [13,14,15,16,17,18,19]. We excavated, analyzed, and appropriately clustered these data into the online platform Genome Database for Strawberry (GDS). The GDS provides a user-friendly web interface; it also integrates a series of practical bioinformatics tools that enable researchers to search, browse, or retrieve specific information.
Genomics, transcriptomics and proteomic technology has developed rapidly. The GDS developed here will greatly benefit future application of high-throughput and -omics technologies. In addition, our achievement provides a directly resource for strawberry breeders and research communities, which will further facilitate the development of new strawberry cultivars with improved flavor. Nowadays, the phylogenomic relationships among the strawberry genomes is unclear. The current debate on the evolutionary of strawberries is one of the most important issues in the world. Our database could promote research on strawberry evolution.
2. Materials and Methods
2.1. Web Server and Code
The GDS is based on the web server software Apache (v2.4.41) on Linux operating system. PHP (v7.4) and MySQL (v8.0) were used for back-end code and HTML5, CSS3, and JavaScript for front-end codes. All codes have been submitted to Github (https://github.com/, accessed on 25 December 2021) and can be accessed for free by entering “Han-Oscar/GDS-code”. Data were deposited into the mysql database in batches and displayed on the website upon searching using Navicat (version 15) software.
2.2. Formatting the Genomic Data of Strawberries
The GDS cover the genomic sequences from eight strawberry species from GDR (Genome Database for Rosaceae) and Kazusa (Strawberry GARDEN). Fragaria nipponica, Fragaria viridis, and Fragaria orientalis each has one version of the genome data, Fragaria ×ananassa, Fragaria nilgerrensis, Fragaria nubicola, and Fragaria iinumae has two, and Fragaria vesca has three. Only the latest version of genome data was used for downstream analysis. One can then download the genome sequence and protein sequence and gene annotations of the eight strawberry species which were analyzed and classified well. Specifically, the gene and protein IDs of Fragaria xananassa and Fragaria vesca remain the same, and those of Fragaria iinumae, Fragaria nilgerrensis, Fragaria nipponica, Fragaria nubicola, and Fragaria orientalis should start with “FII_”, “FNil_”, “FNI_”, “FNub_”, and “FOR_” before the data can be analyzed by different bioinformatics software.
3. Results
3.1. Overview of the GDS
We created a user-friendly website for the GDS to make it easier for the scientific community to use. The domain name of the GDS is http://eplant.njau.edu.cn/strawberry (accessed on 25 December 2021), and it currently has two terabytes of server space. We implemented the GDS in Apache httpd, HTML5, PHP, and MySQL. The GDS web pages were created using HTML and Bootstrap, and were connected to the database through Apache, PHP, and MySQL to allow for the query of gene-related information by users.
3.2. The Homepage of GDS
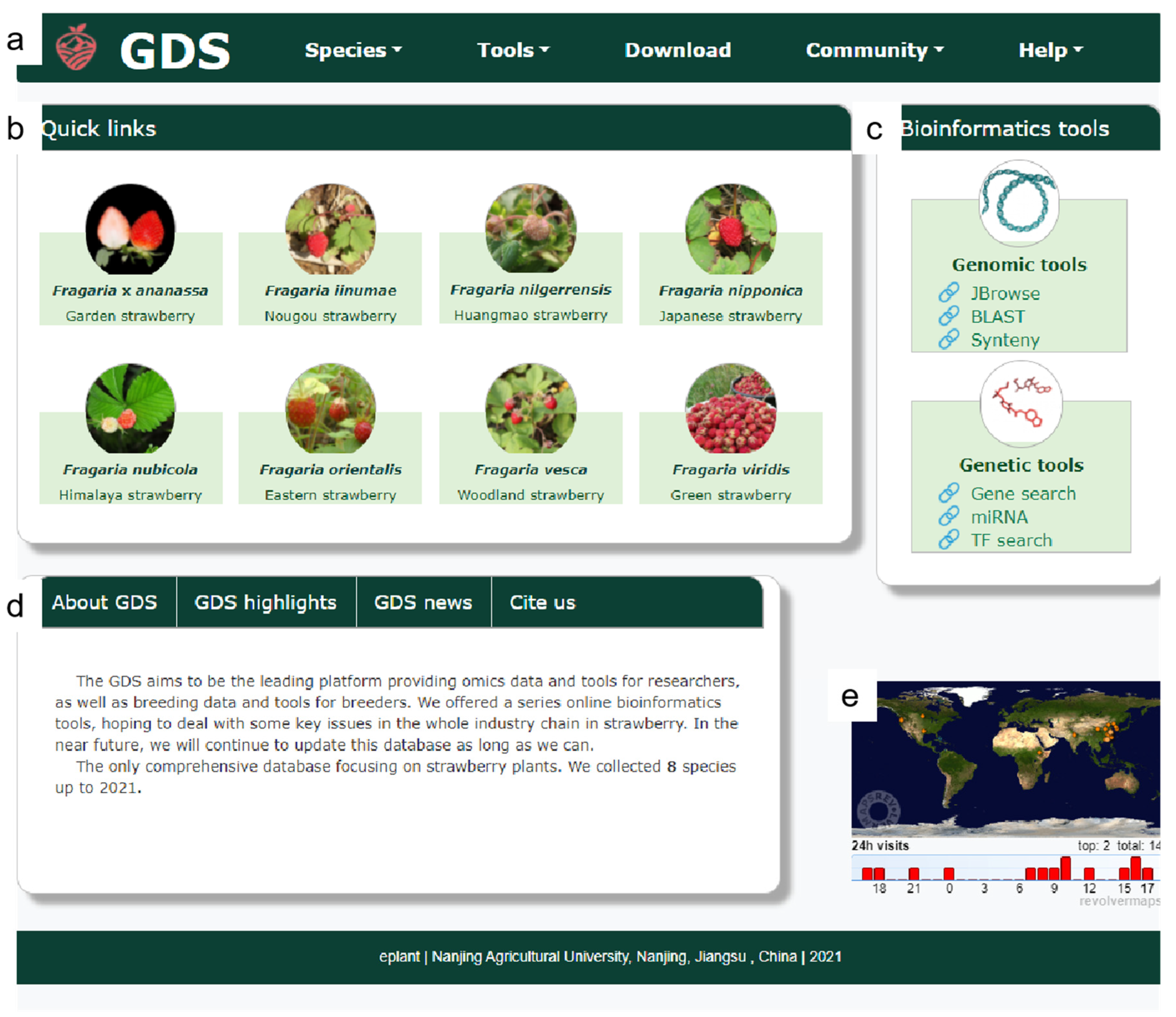
The interface of the GDS included five parts (Figure 1): the “navigation bar”, the “species gallery”, the “tool sets”, the “brief introduction”, and the “live visitor statistics”. At the top of the homepage, the navigation bar (Figure 1a) consists of six labels: Logo, Species, Tools, Download, Community, and Help. Below the navigation bar are quick links to the eight strawberry species. A suite of bioinformatics tools is on the right (Figure 1b,c). Below the species gallery and tool set is a brief introduction to the GDS (Figure 1d). Finally, live statistics (Figure 1e) tools were implemented to collect the number and location of visits.
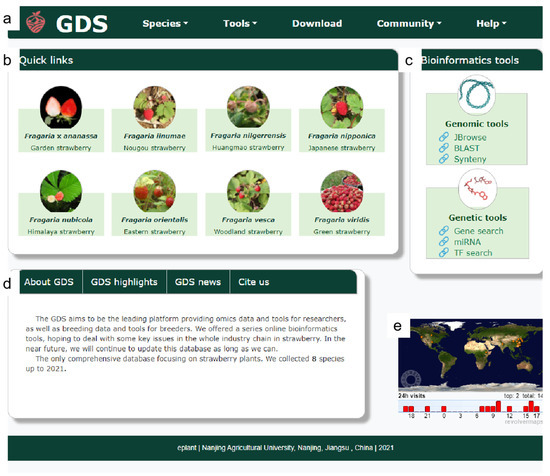
Figure 1.
The GDS homepage. (a) Navigation bar; (b) species gallery; (c) tool set; (d) brief introduction, and (e) live visitor statistics.
3.3. Introduction to the Strawberry Species and Genomes
In the individual species module, we provide visitors with the eight species of strawberry. The first part of the module includes the Latin, English, and Chinese names of the species. The second part provides taxonomic information. In the third part, we summarize detailed and accurate information for the species. The fourth part lists genome assembly details such as genome size, contig N50, and sequencing technology, and the final part provides references to the relevant genome report.
3.4. Data Sets
The reference genome sequence and general feature format (GFF), coding sequence (CDS), protein sequence (PEP) files, and expression data are included in the GDS [20]. A summary of the genomic data currently available in the GDS is presented in Table 1. The versions of the genomes from top to bottom in the table are v1.0a2, r1.1, v1.0, r1.1, r1.1, r1.1, v4.0a2, and v1.0 [21,22,23,24,25].

Table 1.
Statistics of the genome features for eight strawberry species.
3.5. Completeness of the Genomes
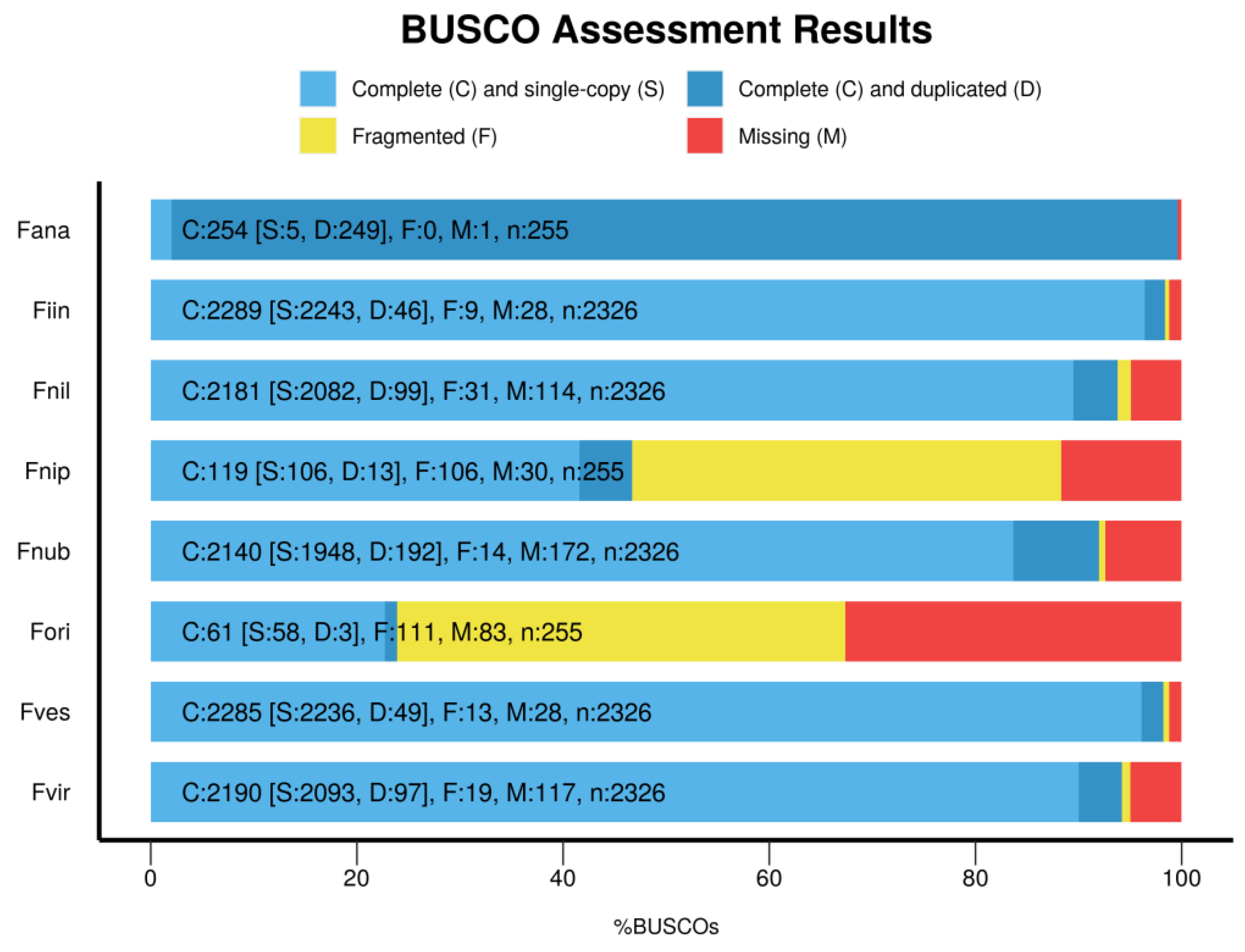
BUSCO provides measures for the quantitative assessment of genome assembly, gene set, and transcriptome completeness based on evolutionarily informed expectations of gene content from near-universal single-copy orthologs selected from OrthoDB [26]. On the species introduction page, we have integrated BUSCO5 results of the eight species, including complete and single-copy, complete and duplicated, fragmented, and missing orthologs. The results are presented as a bar graph (Figure 2), showing that F. ananassa currently has the most complete gene assembly of the eight strawberry species.
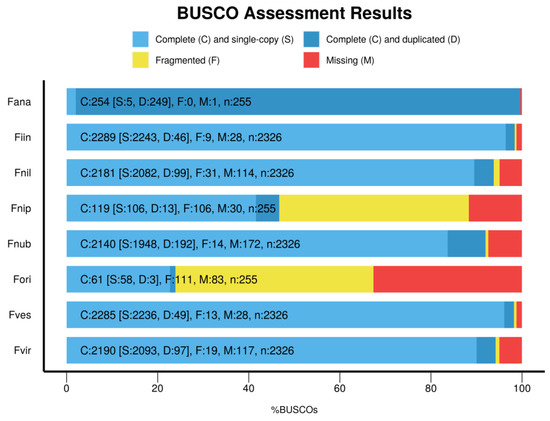
Figure 2.
Genomic completeness of eight strawberry species evaluated using BUSCO V5 software. Fana, Fragaria ×ananassa; Fiin, Fragaria iinumae; Fnil, Fragaria nilgerrensis; Fnip, Fragaria nipponica; Fnub, Fragaria nubicola; Fori, Fragaria orientalis; Fves, Fragaria vesca; and Fvir, Fragaria viridis.
3.6. Phylogenomic Relationships among the Strawberry Genomes
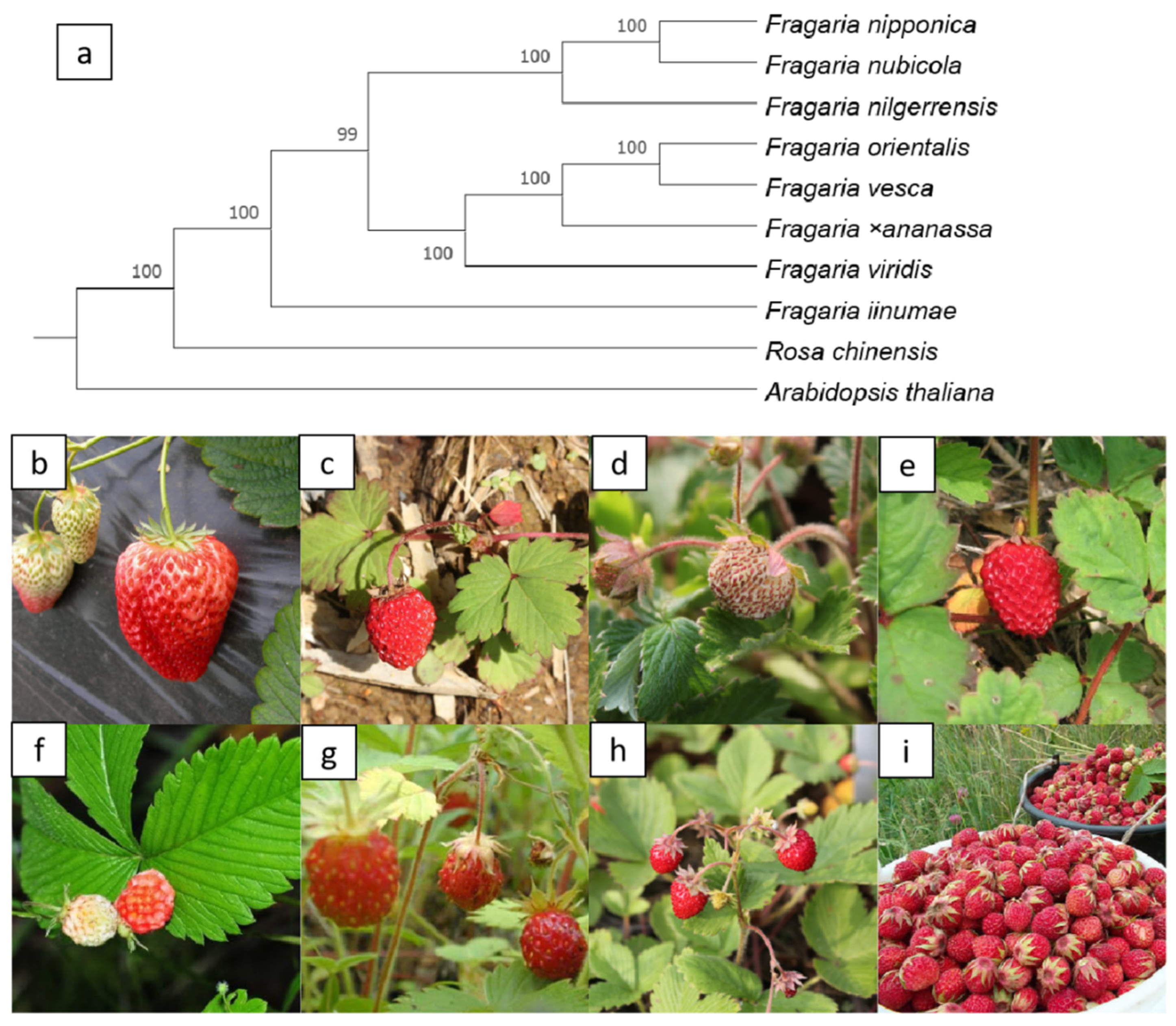
OrthoFinder [27,28] is a fast, accurate, and comprehensive platform for comparative genomics. It identifies orthogroups and orthologs, infers rooted gene trees for all orthogroups, and identifies all of the gene duplication events in the gene trees. To analyze the relationships among eight strawberry genomes, we constructed a phylogenetic tree using OrthoFinder (V2.5.2) software and included two additional species, Rosa chinensis, and Arabidopsis thaliana (Figure 3a).
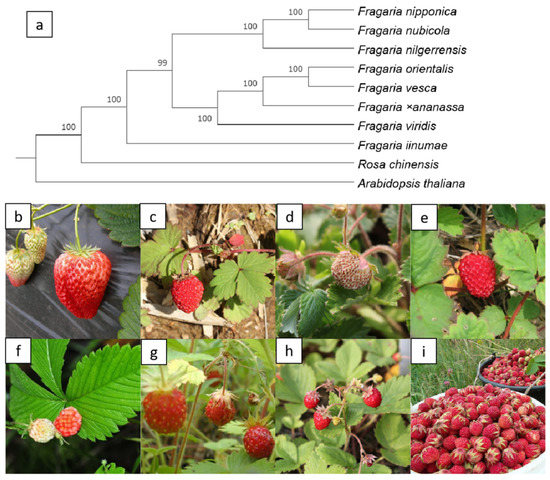
Figure 3.
Phylogenomic analysis of eight strawberry species. (a) A phylogenomic species tree of eight strawberry species; (b) F. ×ananassa; (c) F. iinumae; (d) F. nilgerrensis; (e) F. nipponica; (f) F. nubicola; (g) F. orientalis; (h) F. vesca, and (i) F. viridis. The picture (c) is from https://en.wikipedia.org/wiki/File:Fragaria_iinumae_(fruits).jpg (accessed on 10 November 2021); picture (i) is from https://en.wikipedia.org/wiki/File:Клубника_(Fragaria_viridis).jpeg (accessed on 10 November 2021); picture (f) is from http://www.fpcn.net/uploads/allimg/131107/2-13110G30231V8.JPG (accessed on 10 November 2021).
Fragaria virginiana and Fragaria chiloensis are the genomes of the progenitor species of Fragaria ×ananassa. However, the dispute over its diploid ancestor has lasted for more than half a century and is still unresolved. In 2019, Edger et al. speculated that it has four different diploid ancestors, F. vesca, F. iinumae, F. viridis and F. nipponica [11]. Unexpectedly, just a few months later, Liston and others completed a reanalysis of the same set of data, but they came to a completely different conclusion. They believed that the octoploid strawberry has only two existing ancestors, F. vesca and F. iinumae [29]. Edger et al. insisted on the previous conclusion [30]. The structure of our phylogenetic tree [31] clearly indicates that Fragaria vesca is closest to Fragaria ×ananassa. However, because of the low sequencing and assembled technology or gene introgression, there is no direct evidence indicating the origin of the cultivated strawberry.
The pictures below the tree (Figure 3b–i) shows F. ×ananassa, F. iinumae, F. nilgerrensis, F. nipponica, F. nubicola, F. orientalis, F. vesca, and F. viridis. Photographs b, d, e, g and h were provided by our colleague, Dr. Qiao and the others were obtained from Wikipedia or Baidu.
3.7. Genomic Comparison of Gene Orthogroups
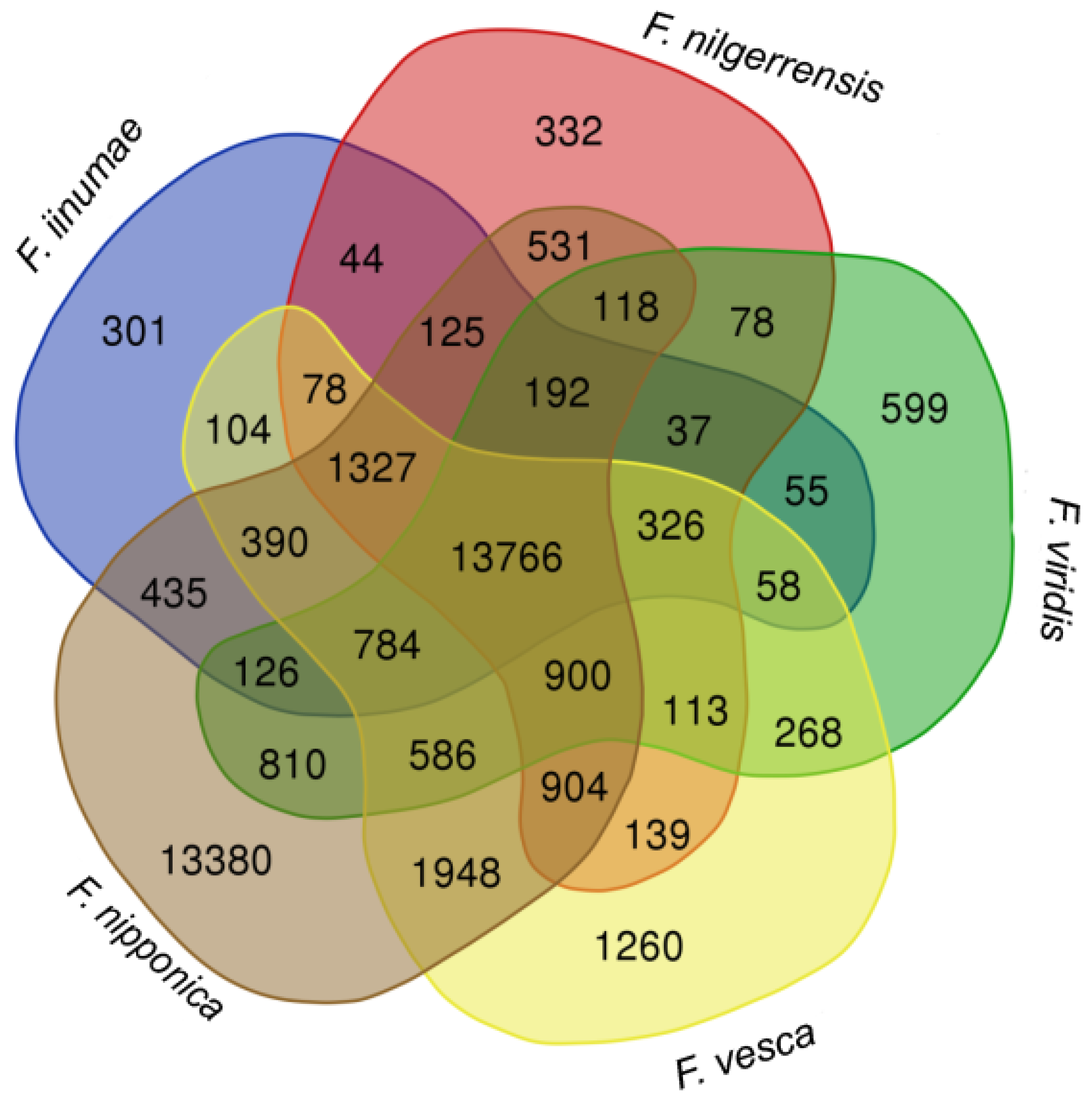
To provide an overview of the comparison among these strawberry genomes, we compared the number of gene orthogroups identified by Orthofinder in the strawberry genomes. We uploaded the orthogroup data to an online Venn diagram tool (https://www.vandepeerlab.org/?q=tools/venn-diagrams, accessed on 10 November 2021) to generate a Venn diagram showing the shared and unique gene orthogroups in F. iinumae, F. nilgerrensis, F. nipponica, F. vesca, and F. viridis. The gene orthogroup numbers are shown in each segment of the diagram (Figure 4); 13,766 gene orthogroups were shared among the five species, and 13,380 gene families appeared to be unique to F. nipponica.
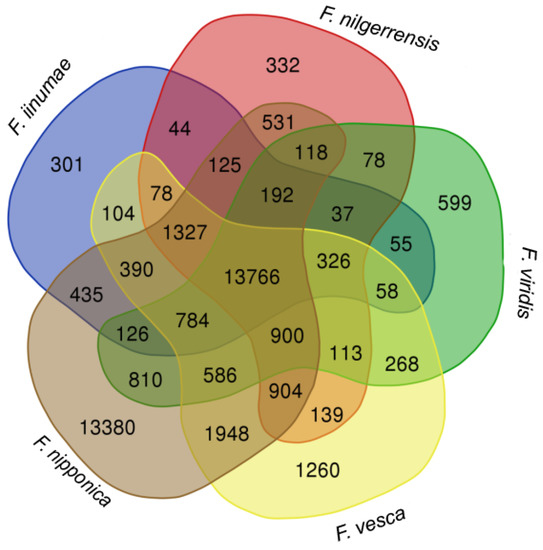
Figure 4.
Venn diagram of gene orthogroups in five diploid and wild Fragaria species. Comparison of the number of shared gene families among five diploid strawberries, F. iinumae, F. nilgerrensis, F. nipponica, F. vesca, and F. viridis.
The number of predicted genes was quite higher in F. nipponica, compared with the other four species, and the number of specific genes for F. nipponica was also extremely high (13,380). The reason why so many genes were predicted in F. nipponica is because there were more than 80,000 proteins annotated, only using Illumina sequencing technology, in 2014. In the future, with the improvement of the technology of sequencing, this problem will disappear.
3.8. Gene Annotations
There are tens of thousands of genes and proteins in the eight strawberry species, and these sequences contain large amounts of valuable species information for which researchers are searching. Consequently, we have integrated millions of data into the GDS Gene Search tool for obtaining detailed information on target genes. The following are the types of detailed gene information that our tool integrates.
1. Gene family annotation. The ancestral genes of strawberries have undergone genomic duplication and mutation during their long evolutionary history [32], resulting in a series of related genes with similar conserved sequence motifs. The Pfam [33] (http://pfam.xfam.org/, accessed on 10 November 2021) database is a large collection of protein families. Each family is represented by multiple sequence alignments and a hidden Markov model (HMMs) [34]. We have analyzed the proteins of the eight strawberry species using the Pfam 34.0 database and hmmscan (version 3.3) software.
2. KEGG (Kyoto Encyclopedia of Genes and Genomes) annotation. KEGG is a resource for understanding the functions and utilities of biological systems, such as the cell, organism, and ecosystem. It contains molecular-level information, especially large-scale datasets generated by genome sequencing and other high-throughput technologies [35]. KofamKOALA is a web server that assigns KEGG Orthologs (KOs) to protein sequences by homology search against a database of profile hidden Markov models (KOfam) with precomputed adaptive thresholds. KofamKOALA was installed using Ruby (v2.4 and above, v2.7 was used in this study), HMMER (v3.1 and above, v3.3 was used here), and Parallel (the latest version). The GDS uses KofamKOALA [36] software to make KEGG predictions that contain the KO IDs and more exhaustive information from the official website (https://www.kegg.jp/, accessed on 10 November 2021). We use KofamKOALA (v1.2), which relies on a file named exec_annotation, to analyze the protein files of eight kinds of strawberries.
3. GO annotation. GO [37] is a database established by the Gene Ontology Consortium. It aims to establish a database that is applicable to various species and that limits and describes the functions of genes and proteins. The updated semantic vocabulary standard is applicable to all species. By establishing a set of controlled vocabulary terms with a dynamic form, GO annotations can describe the roles of genes and proteins in cells and organisms. InterPro [38,39] was developed based on Java and aggregates data resources from multiple functional annotation databases such as Pfam, Panther, SMART, SUPERFAMILY, and tmhmm. It predicts the biological functions of proteins by classifying their sequences into protein families and predicting protein domains. InterProscan (v5.5) was used to annotate proteins from the eight strawberry species. A comparison library is available upon downloading the latest version of InterProscan. Instructions on InterProscan can be obtained by entering “./interproscan.sh” in the terminal. The final data can be obtained from the MySQL database.
4. Signal peptide prediction [40]. Signal peptides are short (5–30 amino acid) peptide chains that guide the transfer of newly synthesized proteins to the secretory pathway. The SignalP [41] software tool predicts whether there is a potential signal peptide cleavage site and identifies its location in a given amino acid sequence. Users may enter the “singalP” folder of the download interface to download data. SignalP (5.0 version) is used here with command “signalp -batch 30,000 -org euk -fasta proteins” for the analysis of proteins from the eight strawberry species.
To date, eight nuclear genomes, 436,160 protein sequences, 3,107,804 GO annotations, 27,481 signal peptides, and 1918 transcription factors [42] (Table 2) have been downloaded, analyzed, and organized in the GDS MySQL database.

Table 2.
Statistics of whole datasets in GDS.
3.9. Sequence Searches Using Basic Local Alignment Search Tool (BLAST)
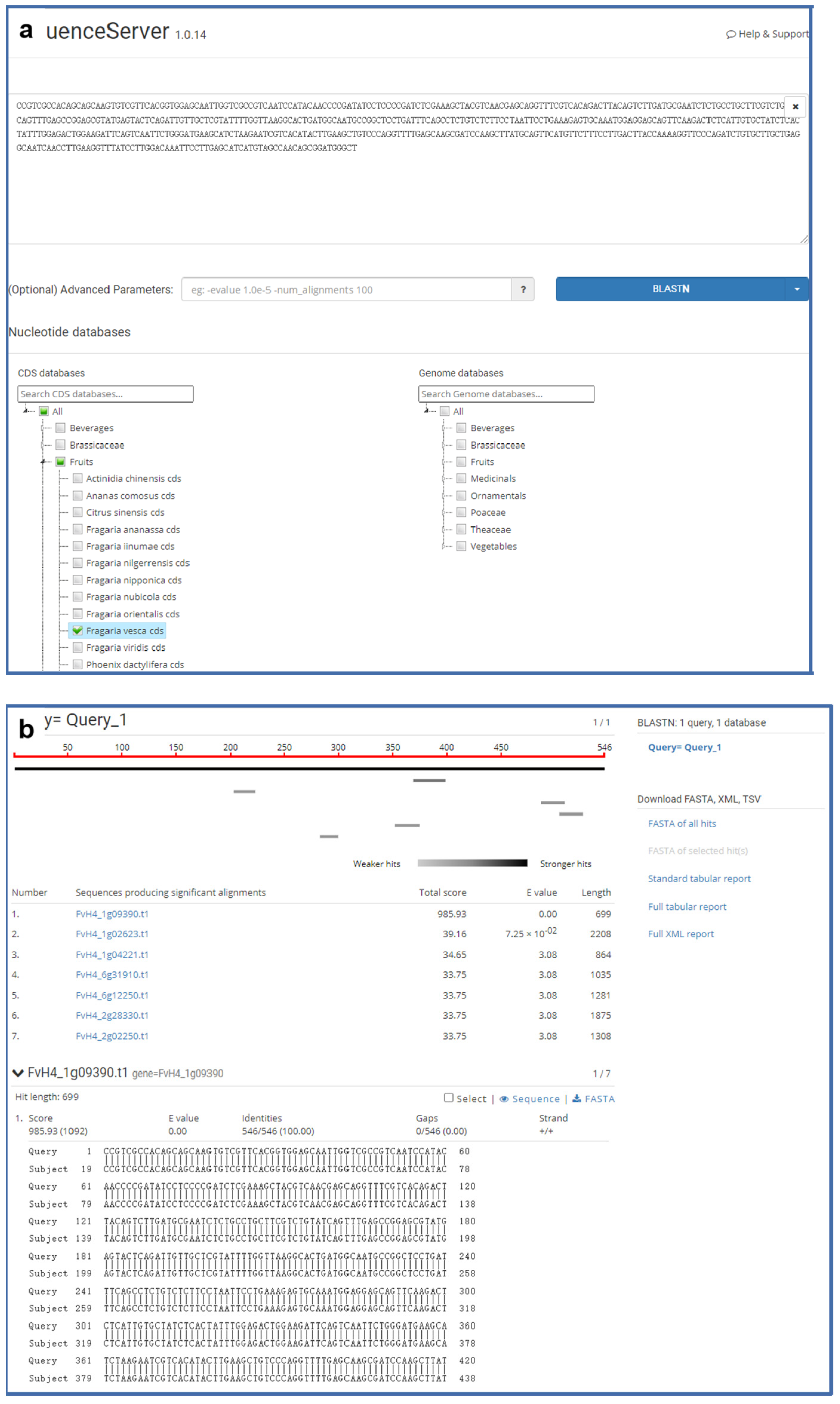
Sequence similarity comparison is a widely used basic bioinformatics tool for the identification of possible homology between sequences and potential similarities in gene function [43]. Hence, it is necessary for most users to find regions of similarity between biological sequences in gene information databases. GDS employs the free, open-source, and powerful Sequenceserver software for BLAST searches [44]. This interface of Sequenceserver is simple, user-friendly, and powerful (Figure 5). SequenceServer has a simple interface, it performs BLAST and visually inspect BLAST results for biological interpretation. It uses simple algorithms to prevent potential errors during analysis and provides flexible text-based, visual outputs to support researchers’ work efficiency. SequenceServer is a BLAST+ server for personal use with a clear and thoughtful design. It contains genomic sequences, CDS, and protein sequences of strawberries, and uses jstree to optimize BLAST to offer clear visualization of complicated results.
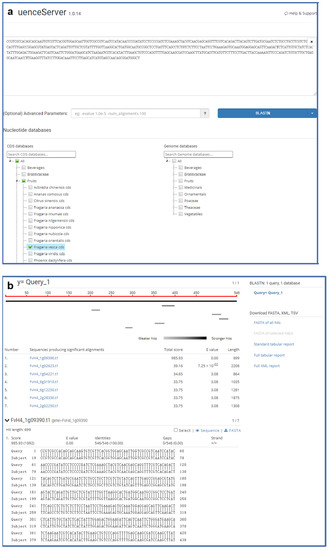
Figure 5.
The BLAST tool integrated into GDS. (a) A user can enter genomic, PEP, or CDS sequences into the text box, then select the species name below; (b) the resulting alignment scores are ranked from high to low, and the details of each sequence alignment are given below the list of scores.
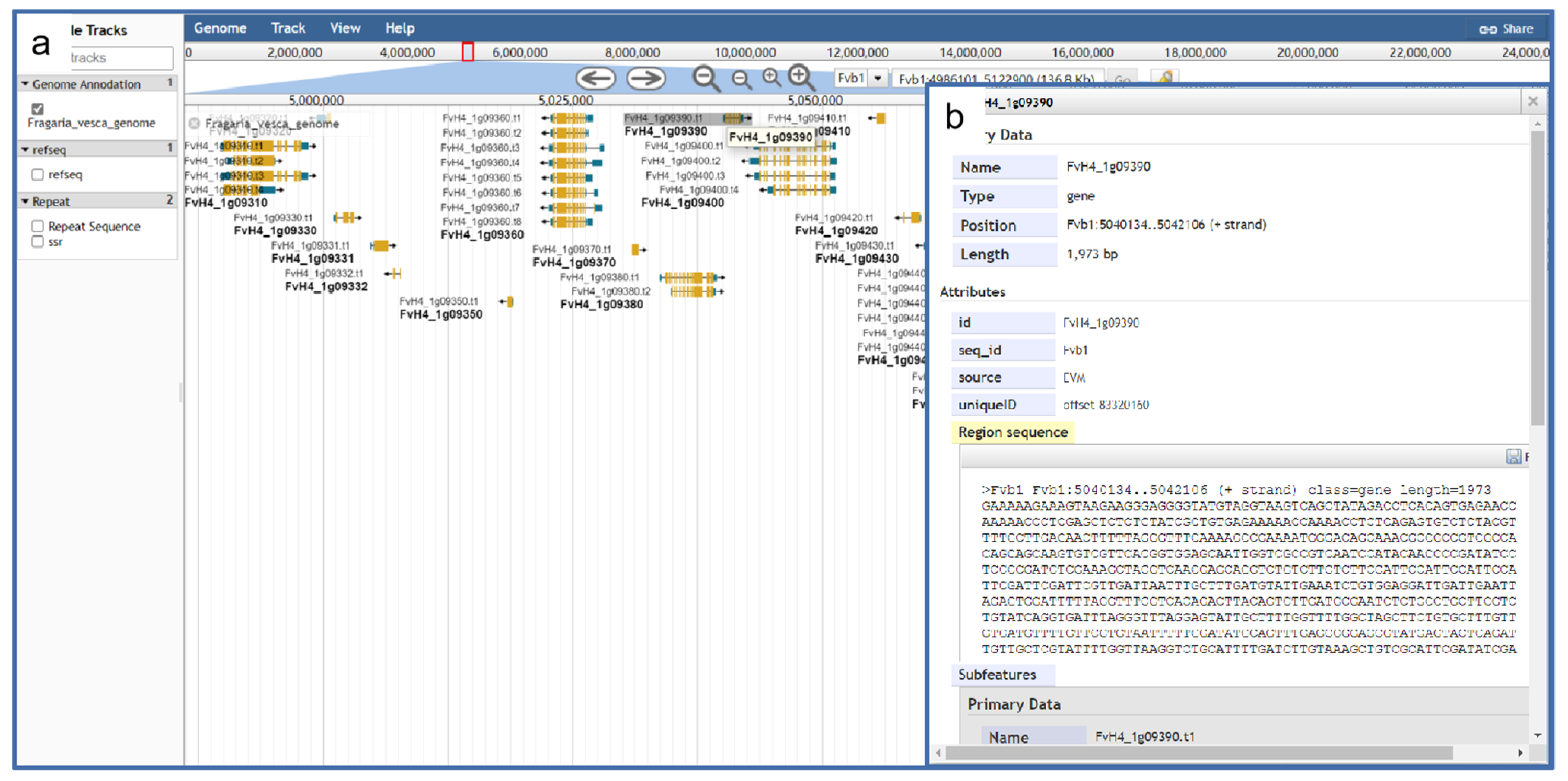
3.10. Genomic Visualization Using JBrowse
A genome browser is a software tool that can be deployed on the server side so that users can access online platforms. JBrowse is a fully featured genome browser that can visualize various types of genome-located data, located in a variety of different data stores, and interfacing to other client and server applications. We used JBrowse [45] built using HTML5 and JavaScript. It integrates and visualizes various existing genome data, including eight nuclear genomes and seven chloroplast genomes [46] so that users can visually browse and analyze the genome and various types of annotation data with strong scalability (Figure 6). In addition, the genome browser can support other types of data, such as repetitive sequences.
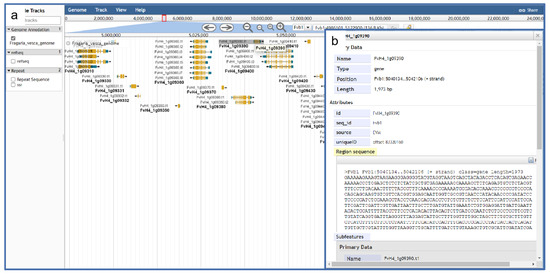
Figure 6.
The JBrowse tool integrated into GDS for visualization of strawberry genomic details. (a) Gene visualization interface; (b) detailed data on individual genes.
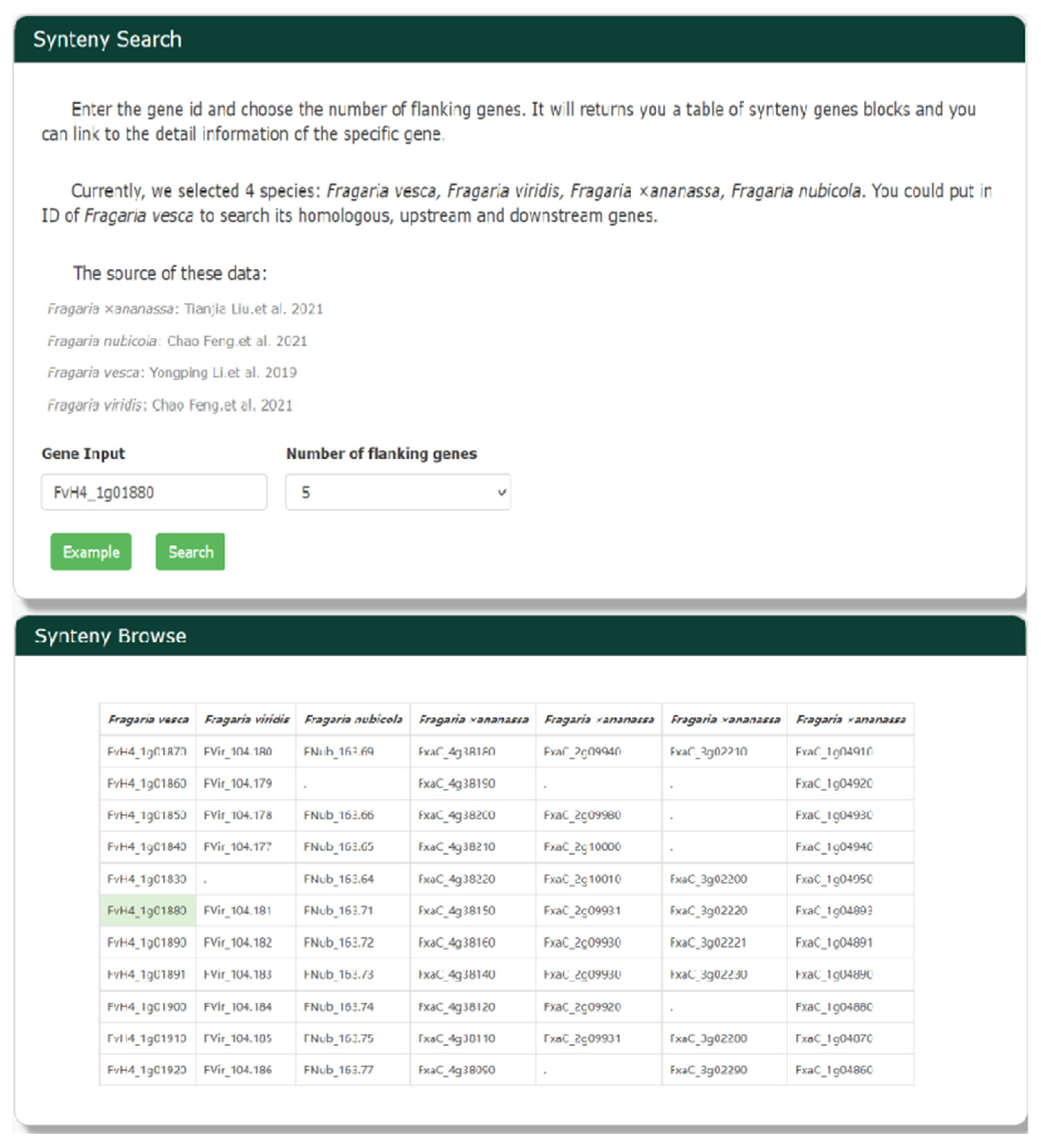
3.11. Tracing Whole-Genome Duplication Using Synteny Browse Search
Given the close phylogenetic relationships among strawberry species, there are likely to be many homologous gene blocks in their genomes. The Python version of MCScan [47] was used to identify homologous gene blocks in the genomes of the strawberry species. We selected four species (Figure 7), including cultivated strawberry, to use in searches of homologous genes, as well as upstream and downstream genes. Scientists can look for syntenic genes of F. vesca by entering a gene identifier, finding the homologous gene(s), and using them as input for a subsequent gene search.
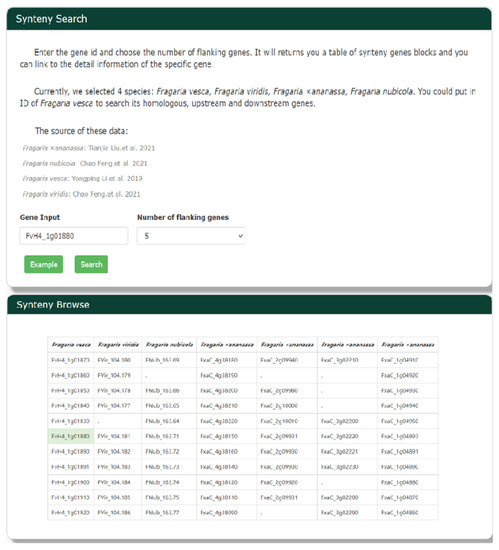
Figure 7.
The synteny search tool in GDS is designed for whole-genome duplication analyses. Researchers can use Synteny Browse Search to look for syntenic genes by entering a gene identifier and selecting a number of flanking genes to be presented.
3.12. microRNA Search
microRNAs (miRNAs) are a class of non-coding single-stranded RNA molecules with a length of approximately 22 nucleotides that are encoded by endogenous genes. The Rfam database [48,49] is a collection of RNA families, each represented by multiple sequence alignments, consensus secondary structures, and covariance models. The GDS use cmscan [50] from the Infernal (V1.1.4) software package to predict the miRNAs in the six species of strawberry with high-quality assemblies.
3.13. Transcription Factor Search
Transcription factors play an important role in all biological processes, from seed germination to senescence. Therefore, it is critical for researchers to gain a good understanding of the relationship between the structures and functions of various transcription-factor families. iTAK [51] is a program that can identify plant transcription factors (TFs), transcriptional regulators (TRs), and protein kinases (PKs) based on protein or nucleotide sequences. It then classifies individual TFs, TRs, and PKs into different gene families. iTAK (v1.7) is used here to identify and analyze transcription factors from the six highly assembled strawberry species.
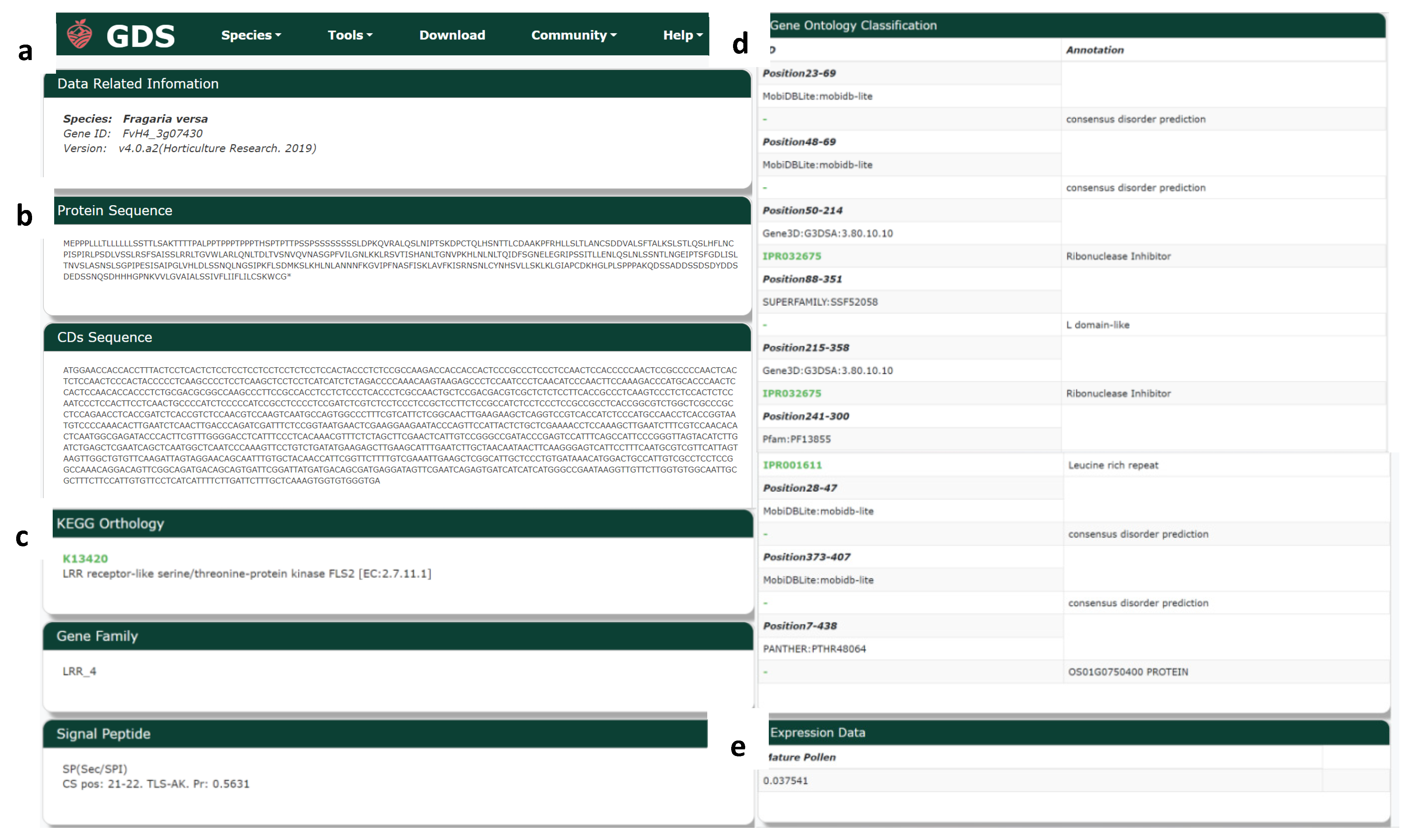
3.14. Gene Search
From the search results of BLAST and JBrowse, scientists can enter a gene identifier to search for information about the gene version (Figure 8a), protein and CDS sequence (Figure 8b), KEGG annotation, gene family, signal peptides (Figure 8c), and GO annotation (Figure 8d). The results include links to the corresponding annotation databases for more information, as well as gene expression data (Figure 8e) from mature pollen.
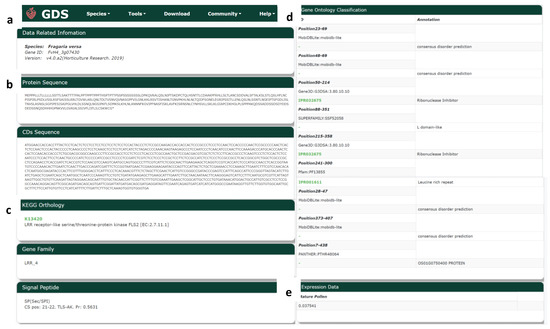
Figure 8.
Gene search tool in GDS. (a) Related information; (b) protein and CDS sequence; (c) KEGG annotation, gene family, and signal peptides; (d) GO annotation, and (e) expression.
3.15. Download
The Download module provides access to the genome assembly, CDS and PEP sequences, annotation data, and miRNA downloads in FASTA and GFF3 formats. Chloroplast genomes of seven strawberry species and related publications are also available. We implemented an FTP site to store and share the data, which users can download at a rapid speed.
3.16. Community of Strawberry Researchers
In the Community module, we provided links to 39 horticultural conferences and 11 relevant publications on strawberry genome research. We also included an FAQ section to explain how to use the database and a contact list for the researchers who established it.
4. Discussion
The rapid development of genome sequencing technology has enabled the sequencing, assembly, and annotation of many plant genomes, providing genetic information on plant growth, development, and evolution. Genome sequencing and analysis technologies have not only deepened our understanding of plant species but also accelerated gene functional studies and molecular breeding. There are many strawberry species and varieties with complicated genomes, and their genome data are refined and updated very often. For example, the Fragaria vesca genome (214.4 Mb) was the first sequenced strawberry genome [10]. However, its quality was not ideal. Later on, Edger and associates from the University of California sequenced the genome of woodland strawberry Hawaii-4 using single-molecule real-time (SMRT) sequencing [22] and constructed a more complete genome map (V4.0). The SMRT sequencing can produce much longer contigs, greatly facilitating genome assembly and annotation. Specifically, the length of contig N50 of V4.0 reached 7.9 Mb, 300 times longer than those of V1.0, and >99.8% of the contigs were successfully mapped to the seven chromosomes. This new strawberry genome map offers more accurate sequences and detailed location information. The polyploidization of strawberries, which contain diploid, tetraploid (Fragaria orientalis) [5], and octoploid (Fragaria ×ananassa) varieties, has made the sharing, analysis, and integration of their genomic data a difficult task. A more convenient online database with multiple integrated and classified strawberry genomes is urgently needed. It will facilitate the gene-functional studies, thereby promoting the improvement of the yield and quality of strawberries [52]. To our knowledge, GDS is currently the only up-to-date database for strawberries that integrates multiple bioinformatics tools.
The storage and analysis of strawberry genome data are also hot research topics, and databases such as GDR (Genome Database for Rosaceae) and Kazusa (Strawberry garden) were created for these purposes. Although GDR and Kazusa have developed databases for strawberries, these databases have a number of problems that require urgent solutions. First, most of the genomic data are unprocessed and scattered. The data lack functional annotations, are not clustered in gene families, and are not preformatted for searching. After downloading data from these public databases, one must process the data oneself, which is a challenge for those with less expertise in bioinformatics. More importantly, some websites are difficult to access in China, and data downloads are also greatly restricted sometimes. To date, there is no specific, widely available database for strawberry research. Here, we use the latest version of the software for the analysis of strawberry proteins that are not all available on other websites. In addition, our laboratory specializes in strawberry research and has extensive collaboration with other strawberry research groups. In the future, newly released strawberry sequencing data will be updated in the database and made available to all researchers in a timely manner.
GDS stores the genome sequences of eight strawberry species and related gene annotations. The advanced and popular BLAST and JBrowse tools have been implemented, as well as a syntenic block search tool and an miRNA finder. This database serves as a central portal for the strawberry research community, enabling researchers to download genomes, protein sets, transcription data, and recently published articles on the strawberry genome. The GDS will be constantly updated when new genomes, transcriptomes, and other types of genetic datasets are published. In the future, we will develop and establish more gene online analysis tools to facilitate strawberry researchers in conducting online analysis. We will do our best to develop and deploy new omics tools in the GDS to provide a better user experience. Furthermore, GDS will contain studies and statistics on strawberries’ breeding. In summary, this new database incorporates published strawberry plant genomes, multiple analysis tools, new features for strawberry plant genomic data analysis, gene function characterization, synteny and miRNA search, and publication, which is easily accessed and can potentially benefit the strawberry plant research community.
Author Contributions
Z.C. and F.C. designed and led this project. Y.Z. constructed the GDS. Y.Q., J.D., Z.N. and J.X. analyzed the data. Y.Z. wrote the draft manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
Fei Chen acknowledges funding from the Fundamental Research Fund for the Central University (KYXJ202004) and a starting fund from Nanjing Agricultural University (804012). Zongming Cheng and Jinsong Xiong acknowledge funding from Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD). This work was supported by the National Natural Science Foundation of China (Grant no. 32072540, 31872056).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All code about the database is available on Github (https://github.com/Han-Oscar/GDS-code, accessed on 10 November 2021).
Acknowledgments
We thank Yushan Qiao, Zhiyou Ni, Jianke Du for valuable comments and suggestions on our database. We thank Xiaogang Lei and Xiaojiang Li for providing technical assistance to our database development and Fei Chen, Zongming Cheng for assistance with the correction of the English language in the manuscript.
Conflicts of Interest
The authors declare that they have no conflict of interest.
References
- Kim, E.H. A New Species of Fragaria (Rosaceae) from Oregon. J. Bot. Res. Inst. Tex. 2012, 6, 9–15. [Google Scholar]
- Kim, E.H.; Preeda, N.; Tomohiro, Y. Decaploidy in Fragaria iturupensis (Rosaceae). Am. J. Bot. 2009, 96, 713–719. [Google Scholar] [CrossRef]
- Van de Peer, Y.; Mizrachi, E.; Marchal, K. The evolutionary significance of polyploidy. Nat. Rev. Genet. 2017, 18, 411–424. [Google Scholar] [CrossRef] [PubMed]
- Lei, J.J.; Xue, L.; Guo, R.X.; Dai, H.P. The Fragaria species native to China and their geographical distribution. Acta Hortic. 2017, 1156, 37–46. [Google Scholar] [CrossRef]
- Detlef, U.; Klaus, O. Diversity of volatile patterns in sixteen Fragaria vesca L. accessions in comparison to cultivars of Fragaria ×ananassa. J. Appl. Bot. Food Qual. 2013, 86, 37–46. [Google Scholar] [CrossRef]
- Hendrix, B.; Stewart, J.M. Estimation of the nuclear DNA content of gossypium species. Ann. Bot. 2005, 95, 789–797. [Google Scholar] [CrossRef]
- Tennessen, J.A.; Govindarajulu, R.; Ashman, T.L.; Liston, A. Evolutionary origins and dynamics of octoploid strawberry subgenomes revealed by dense targeted capture linkage maps. Genome Biol. Evol. 2014, 6, 3295–3313. [Google Scholar] [CrossRef] [Green Version]
- Cappelletti, R.; Sabbadini, S.; Mezzetti, B. Strawberry (Fragaria ×ananassa). Methods Mol. Biol. 2015, 1224, 217–227. [Google Scholar] [CrossRef]
- Shulaev, V.; Sargent, D.J.; Crowhurst, R.N.; Mockler, T.C.; Folkerts, O.; Delcher, A.L.; Jaiswal, P.; Mockaitis, K.; Liston, A.; Mane, S.P.; et al. The genome of woodland strawberry (Fragaria vesca). Nat. Genet. 2011, 43, 109–116. [Google Scholar] [CrossRef]
- Hirakawa, H.; Shirasawa, K.; Kosugi, S.; Tashiro, K.; Nakayama, S.; Yamada, M.; Kohara, M.; Watanabe, A.; Kishida, Y.; Fujishiro, T.; et al. Dissection of the Octoploid Strawberry Genome by Deep Sequencing of the Genomes of Fragaria Species. DNA Res. 2014, 21, 169–181. [Google Scholar] [CrossRef]
- Edger, P.P.; Poorten, T.J.; VanBuren, R.; Hardigan, M.A.; Colle, M.; McKain, M.R.; Smith, R.D.; Teresi, S.J.; Nelson, A.D.L.; Wai, C.M.; et al. Origin and evolution of the octoploid strawberry genome. Nat. Genet. 2019, 51, 541–547. [Google Scholar] [CrossRef] [Green Version]
- Chen, F.; Song, Y.; Li, X.; Chen, J.; Mo, L.; Zhang, X.; Lin, Z.; Zhang, L. Genome sequences of horticultural plants: Past, present, and future. Hortic. Res. 2019, 6, 112. [Google Scholar] [CrossRef] [Green Version]
- Xiaoming, S.; Fulei, N.; Wei, C.; Xiao, M.; Ke, G.; Qihang, Y.; Jinpeng, W.; Nan, L.; Pengchuan, S.; Qiaoying, P.; et al. Coriander Genomics Database: A genomic, transcriptomic, and metabolic database for coriander. Hortic. Res. 2020, 7, 55. [Google Scholar] [CrossRef] [Green Version]
- Tam, P.S.; Peter, L.; Scott, C.E. GigaDB: Announcing the GigaScience database. Gigascience 2012, 1, 11. [Google Scholar] [CrossRef] [Green Version]
- Junyang, Y.; Jiacheng, L.; Wei, T.; Ya, Q.W.; Xiaofeng, T.; Wei, L.; Ying, Y.; Lihuan, W.; Shengxiong, H.; Congbing, F.; et al. Kiwifruit Genome Database (KGD): A comprehensive resource for kiwifruit genomics. Hortic. Res. 2020, 7, 117. [Google Scholar] [CrossRef]
- Xiao, Q.; Li, Z.; Qu, M.; Xu, W.; Su, Z.; Yang, J. LjaFGD: Lonicera japonica functional genomics database. J. Integr. Plant Biol. 2021, 63, 1422–1436. [Google Scholar] [CrossRef]
- Wenlei, G.; Junhao, C.; Jian, L.; Jianqin, H.; Zhengjia, W.; Kean-Jin, L. Portal of Juglandaceae: A comprehensive platform for Juglandaceae study. Hortic. Res. 2020, 7, 35. [Google Scholar] [CrossRef] [Green Version]
- Lamesch, P.; Berardini, T.Z.; Li, D.; Swarbreck, D.; Wilks, C.; Sasidharan, R.; Muller, R.; Dreher, K.; Alexander, D.L.; Garcia-Hernandez, M.; et al. The Arabidopsis Information Resource (TAIR): Improved gene annotation and new tools. Nucleic Acids Res. 2012, 40, D1202–D1212. [Google Scholar] [CrossRef]
- Chen, F.; Dong, W.; Zhang, J.; Guo, X.; Chen, J.; Wang, Z.; Lin, Z.; Tang, H.; Zhang, L. The Sequenced Angiosperm Genomes and Genome Databases. Front. Plant Sci. 2018, 9, 418. [Google Scholar] [CrossRef] [Green Version]
- Bo, L.; Victor, R.; Ron, M.S.; James, A.T.; Colin, N.D. RNA-Seq gene expression estimation with read mapping uncertainty. Bioinformatics 2010, 26, 493–500. [Google Scholar] [CrossRef] [Green Version]
- Liu, T.; Li, M.; Liu, Z.; Ai, X.; Li, Y. Reannotation of the cultivated strawberry genome and establishment of a strawberry genome database. Hortic. Res. 2021, 8, 41. [Google Scholar] [CrossRef]
- Edger, P.P.; VanBuren, R.; Colle, M.; Poorten, T.J.; Wai, C.M.; Niederhuth, C.E.; Alger, E.I.; Ou, S.; Acharya, C.B.; Wang, J.; et al. Single-molecule sequencing and optical mapping yields an improved genome of woodland strawberry (Fragaria vesca) with chromosome-scale contiguity. Gigascience 2018, 7, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Lei, Y.; Wang, B.; Li, S.; Yu, S.; Wang, Y.; Li, H.; Liu, Y.; Ma, Y.; Dai, H.; et al. The high-quality genome of diploid strawberry (Fragaria nilgerrensis) provides new insights into anthocyanin accumulation. Plant Biotechnol. J. 2020, 18, 1908–1924. [Google Scholar] [CrossRef] [Green Version]
- Feng, C.; Wang, J.; Harris, A.J.; Folta, K.M.; Zhao, M.; Kang, M. Tracing the Diploid Ancestry of the Cultivated Octoploid Strawberry. Mol. Biol. Evol. 2021, 38, 478–485. [Google Scholar] [CrossRef]
- Li, Y.; Pi, M.; Gao, Q.; Liu, Z.; Kang, C. Updated annotation of the wild strawberry Fragaria vesca V4 genome. Hortic. Res. 2019, 6, 61. [Google Scholar] [CrossRef] [Green Version]
- Seppey, M.; Manni, M.; Zdobnov, E.M. BUSCO: Assessing Genome Assembly and Annotation Completeness. Methods Mol. Biol. (Clifton N.J.) 2019, 1962, 227–245. [Google Scholar] [CrossRef]
- David, M.E.; Steven, K. OrthoFinder: Phylogenetic orthology inference for comparative genomics. Genome Biol. 2019, 20, 238. [Google Scholar] [CrossRef] [Green Version]
- David, M.E.; Steven, K. OrthoFinder: Solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 2015, 16, 157. [Google Scholar] [CrossRef] [Green Version]
- Liston, A.; Wei, N.; Tennessen, J.A.; Junmin, L.; Ming, D.; Tia-Lynn, A. Revisiting the origin of octoploid strawberry. Nat. Genet. 2020, 52, 2–4. [Google Scholar] [CrossRef]
- Edger, P.P.; McKain, M.R.; Yocca, A.E.; Knapp, S.J.; Qiao, Q.; Zhang, T. Reply to: Revisiting the origin of octoploid strawberry. Nat. Genet. 2020, 52, 5–7. [Google Scholar] [CrossRef]
- Daniel, P.; James, J.L.; Richard, E.H. Phylogenetic Relationships Among Species of Fragaria (Rosaceae) Inferred from Non-coding Nuclear and Chloroplast DNA Sequences. Syst. Bot. 2000, 25, 337–348. [Google Scholar] [CrossRef]
- Chen, P.; Liu, Q.Z. Genome-wide characterization of the WRKY gene family in cultivated strawberry (Fragaria × ananassa Duch.) and the importance of several group III members in continuous cropping. Sci. Rep. 2019, 9, 8423. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sara, E.; Jaina, M.; Alex, B.; Sean, R.E.; Aurélien, L.; Simon, C.P.; Matloob, Q.; Lorna, J.R.; Gustavo, A.S.; Alfredo, S.; et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2019, 47, D427–D432. [Google Scholar] [CrossRef]
- Finn, R.D.; Clements, J.; Eddy, S.R. HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 2011, 39, 29–37. [Google Scholar] [CrossRef] [Green Version]
- Kanehisa, M.; Sato, Y. KEGG Mapper for inferring cellular functions from protein sequences. Protein Sci. 2020, 29, 28–35. [Google Scholar] [CrossRef] [Green Version]
- Aramaki, T.; Blanc-Mathieu, R.; Endo, H.; Ohkubo, K.; Kanehisa, M.; Goto, S.; Ogata, H. KofamKOALA: KEGG Ortholog assignment based on profile HMM and adaptive score threshold. Bioinformatics 2020, 36, 2251–2252. [Google Scholar] [CrossRef] [Green Version]
- The Gene Ontology Consortium. The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res. 2019, 47, D330–D338. [Google Scholar] [CrossRef] [Green Version]
- Robert, D.F.; Teresa, K.A.; Patricia, C.B.; Alex, B.; Peer, B.; Alan, J.B.; Hsin-Yu, C.; Zsuzsanna, D.; Sara, E.; Matthew, F.; et al. InterPro in 2017—beyond protein family and domain annotations. Nucleic Acids Res. 2017, 45, D190–D199. [Google Scholar] [CrossRef]
- Blum, M.; Chang, H.; Chuguransky, S.; Grego, T.; Kandasaamy, S.; Mitchell, A.; Nuka, G.; PaysanLafosse, T.; Qureshi, M.; Raj, S.; et al. The InterPro protein families and domains database: 20 years on. Nucleic Acids Res. 2020, 49, D344–D354. [Google Scholar] [CrossRef]
- Henrik, N.; Konstantinos, D.T.; Søren, B.; Gunnar, H. A Brief History of Protein Sorting Prediction. Protein J. 2019, 38, 200–216. [Google Scholar] [CrossRef] [Green Version]
- José, J.A.A.; Konstantinos, D.T.; Casper, K.S.; Thomas, N.P.; Ole, W.; Søren, B.; Gunnar, V.H.; Henrik, N. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019, 37, 420–423. [Google Scholar] [CrossRef]
- Pérez-Rodríguez, P.; Riaño-Pachón, D.M.; Corrêa, L.G.G.; Rensing, S.A.; Kersten, B.; Mueller-Roeber, B. PlnTFDB: Updated content and new features of the plant transcription factor database. Nucleic Acids Res. 2010, 38, D227–D234. [Google Scholar] [CrossRef] [Green Version]
- Christiam, C.; George, C.; Vahram, A.; Ning, M.; Jason, P.; Kevin, B.; Thomas, L.M. BLAST+: Architecture and applications. BioMed Cent. 2009, 10, 421. [Google Scholar] [CrossRef] [Green Version]
- Priyam, A.; Woodcroft, B.J.; Rai, V.; Moghul, I.; Munagala, A.; Ter, F.; Chowdhary, H.; Pieniak, I.; Maynard, L.J.; Gibbins, M.A.; et al. Sequenceserver: A Modern Graphical User Interface for Custom BLAST Databases. Mol. Biol. Evol. 2019, 36, 2922–2924. [Google Scholar] [CrossRef]
- Robert, B.; Eric, Y.; Colin, M.D.; Richard, D.H.; Monica, M.; Gregg, H.; David, M.G.; Christine, G.E.; Suzanna, E.L.; Lincoln, S.; et al. JBrowse: A dynamic web platform for genome visualization and analysis. Genome Biol. 2016, 17, 66. [Google Scholar] [CrossRef] [Green Version]
- Wambui, N.; Aaron, L.; Richard, C.; Tia-Lynn, A.; Nahla, B. Insights into phylogeny, sex function and age of Fragaria based on whole chloroplast genome sequencing. Mol. Phylogenet. Evol. 2013, 66, 17–29. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, 49. [Google Scholar] [CrossRef] [Green Version]
- Kalvari, I.; Nawrocki, E.P.; Argasinska, J.; Quinones-Olvera, N.; Finn, R.D.; Bateman, A.; Petrov, A.I. Non-Coding RNA Analysis Using the Rfam Database. Curr. Protoc. Bioinform. 2018, 62, 51. [Google Scholar] [CrossRef]
- Kalvari, I.; Nawrocki, E.P.; OntiverosPalacios, N.; Argasinska, J.; Lamkiewicz, K.; Marz, M.; GriffithsJones, S.; ToffanoNioche, C.; Gautheret, D.; Weinberg, Z.; et al. Rfam 14: Expanded coverage of metagenomic, viral and microRNA families. Nucleic Acids Res. 2020, 49, 192–200. [Google Scholar] [CrossRef]
- Nawrocki, E.P.; Kolbe, D.L.; Eddy, S.R. Infernal 1.0: Inference of RNA alignments. Bioinformatics 2009, 25, 1335–1337. [Google Scholar] [CrossRef] [Green Version]
- Zheng, Y.; Jiao, C.; Sun, H.; Rosli, H.G.; Pombo, M.A.; Zhang, P.; Banf, M.; Dai, X.; Martin, G.B.; Giovannoni, J.J.; et al. iTAK: A Program for Genome-wide Prediction and Classification of Plant Transcription Factors, Transcriptional Regulators, and Protein Kinases. Mol. Plant 2016, 9, 1667–1670. [Google Scholar] [CrossRef] [Green Version]
- Sook, J.; Taein, L.; Chun-Huai, C.; Katheryn, B.; Ping, Z.; Jing, Y.; Jodi, H.; Stephen, P.F.; Ksenija, G.; Kristin, S.; et al. 15 years of GDR: New data and functionality in the Genome Database for Rosaceae. Nucleic Acids Res. 2019, 47, 1137–1145. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).