A Type I and a Type II Metacaspase Are Differentially Regulated during Corolla Development and in Response to Abiotic and Biotic Stresses in Petunia × hybrida
Abstract
:1. Introduction
2. Materials and Methods
2.1. Identifying a Type II Metacaspase (PhMC2) from Petunia
2.2. Expression, Purification, and Activity of Recombinant PhMC2 Proteins
2.3. Plant Materials
2.4. Treatments and Tissue Collection
2.4.1. Plant and Flower Tissue
2.4.2. Botrytis Inoculations
2.4.3. Salinity Stress
2.4.4. Nutrient Deprivation
2.4.5. Drought Stress
2.5. RNA Extraction and Gene Expression Analysis by Quantitative PCR
2.6. Statistical Analysis
3. Results
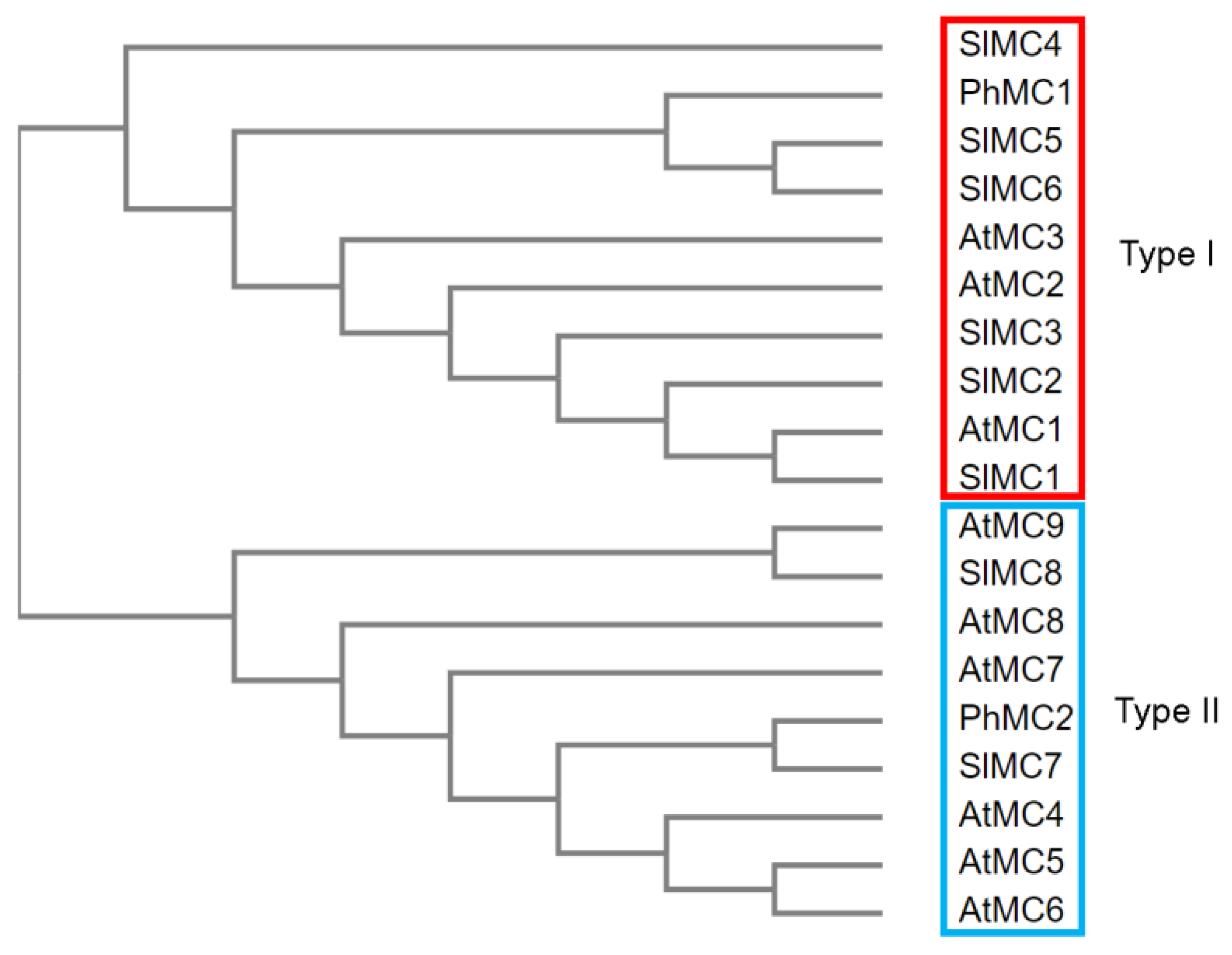
3.1. Identification of a Type II Metacaspase from Petunia
3.2. Activity of Recombinant Metacaspases
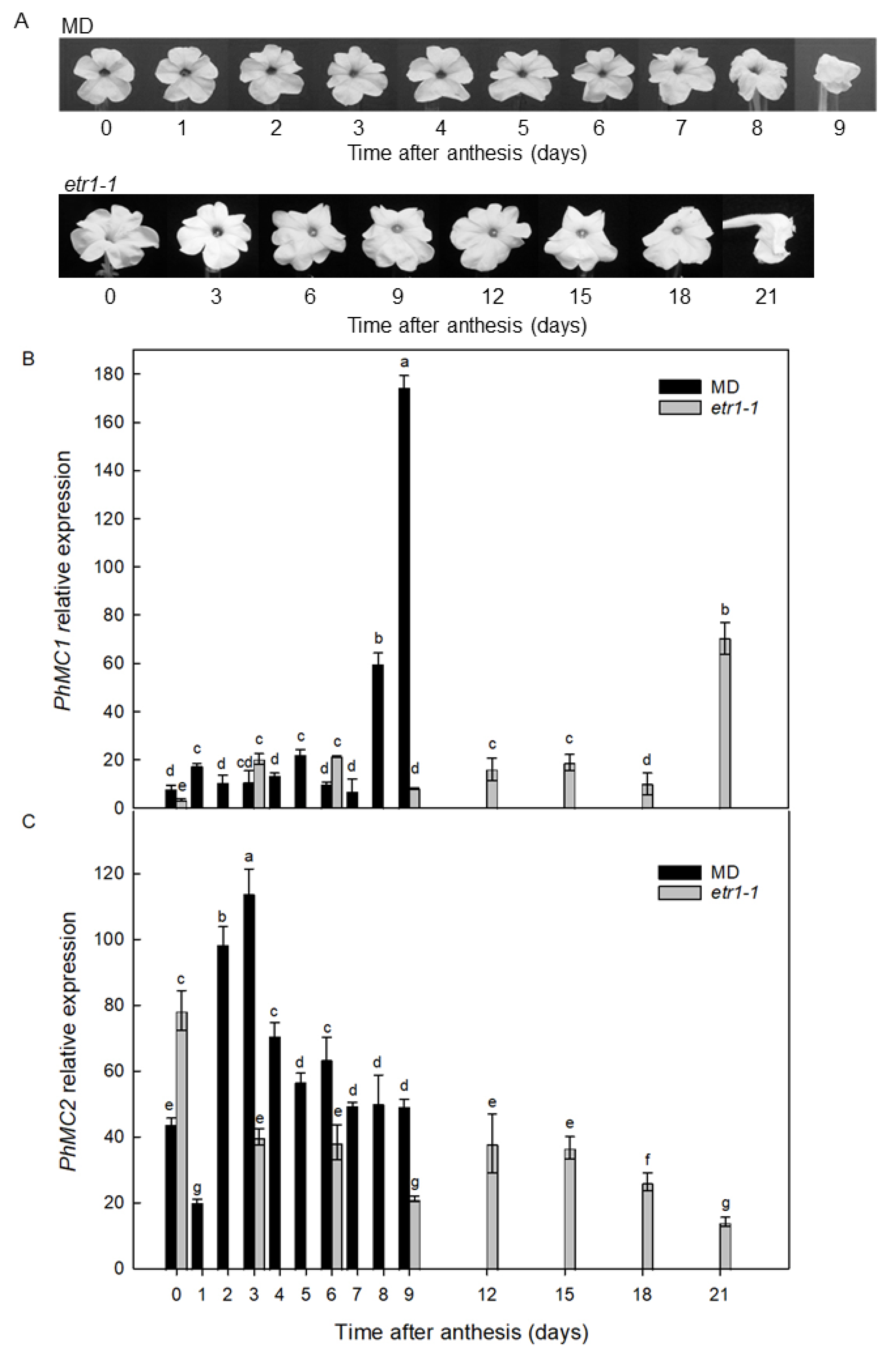
3.3. PhMC1 and PhMC2 Expression in Corollas from Wild Type and Ethylene Insensitive Petunias
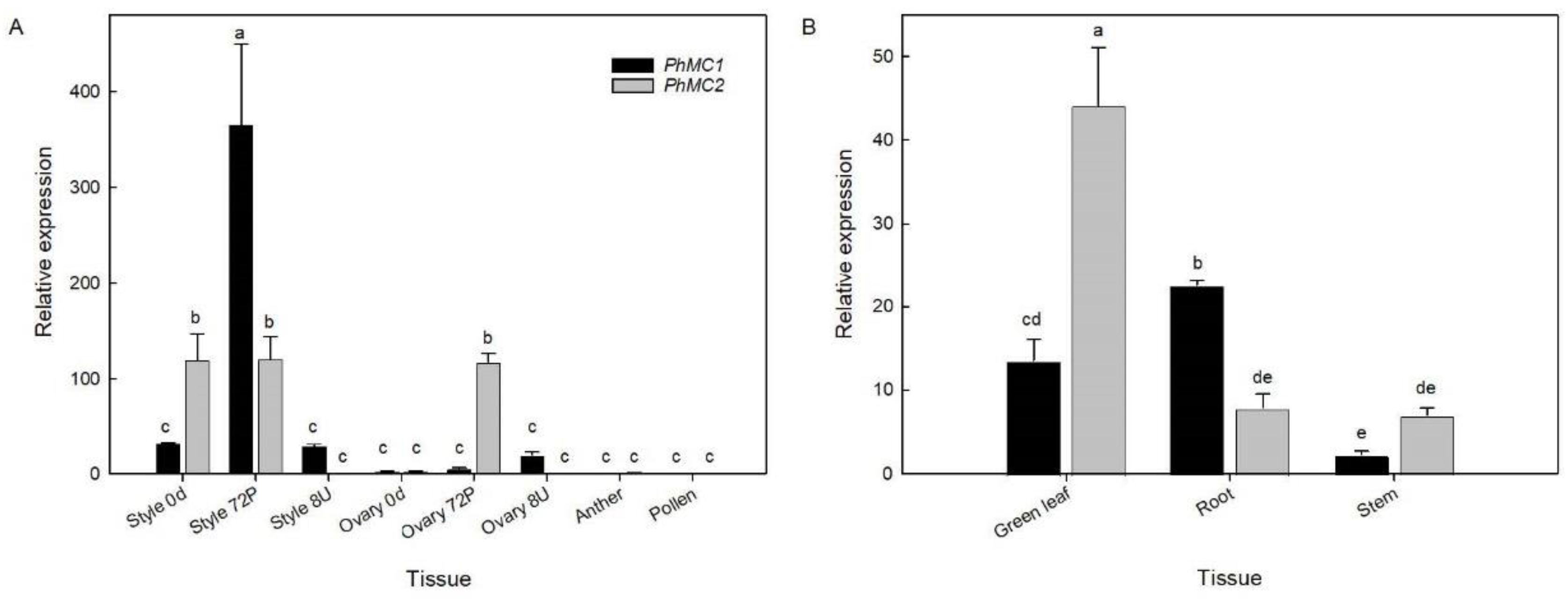
3.4. PhMC1 and PhMC2 Expression in Different Tissues
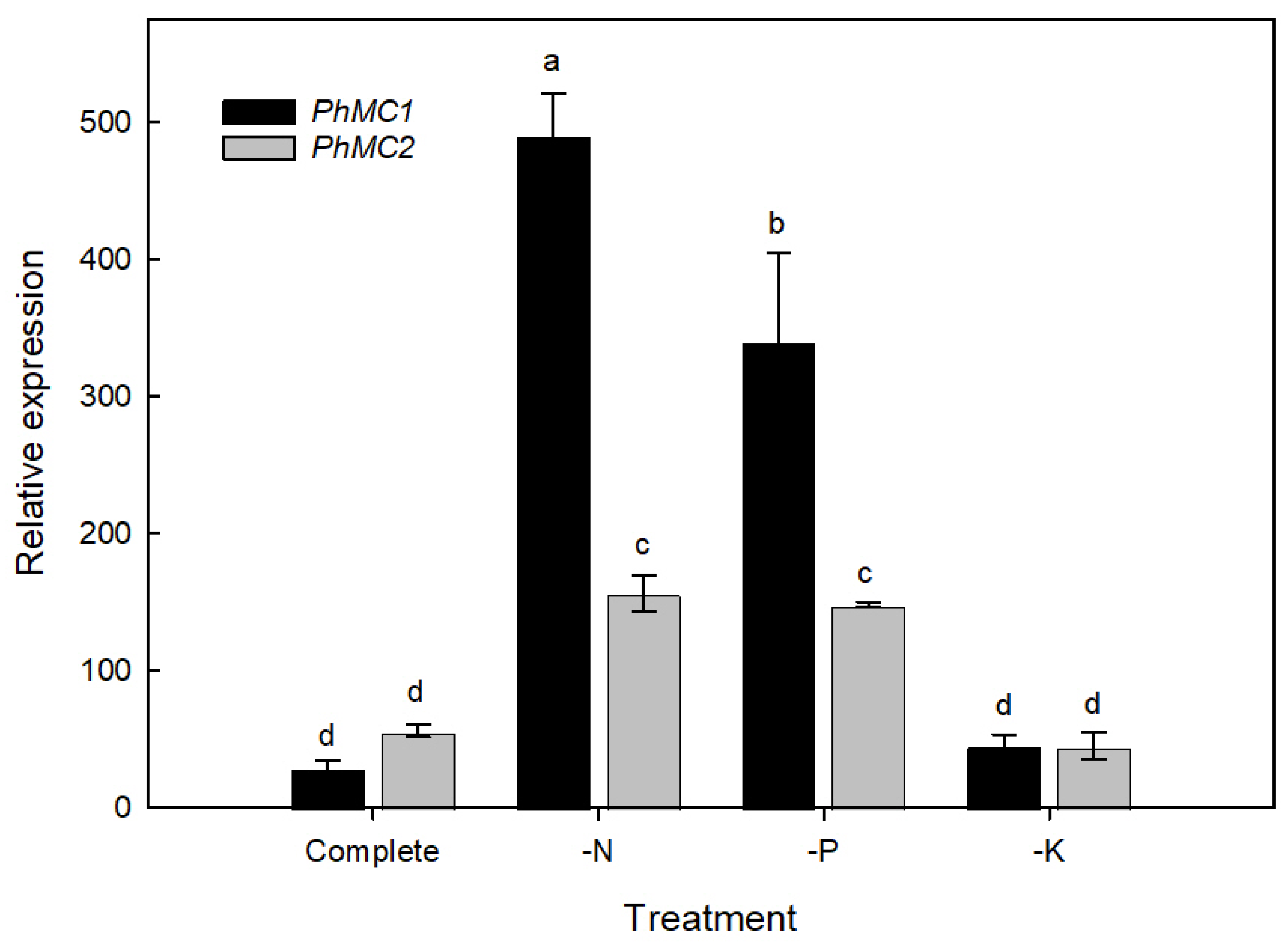
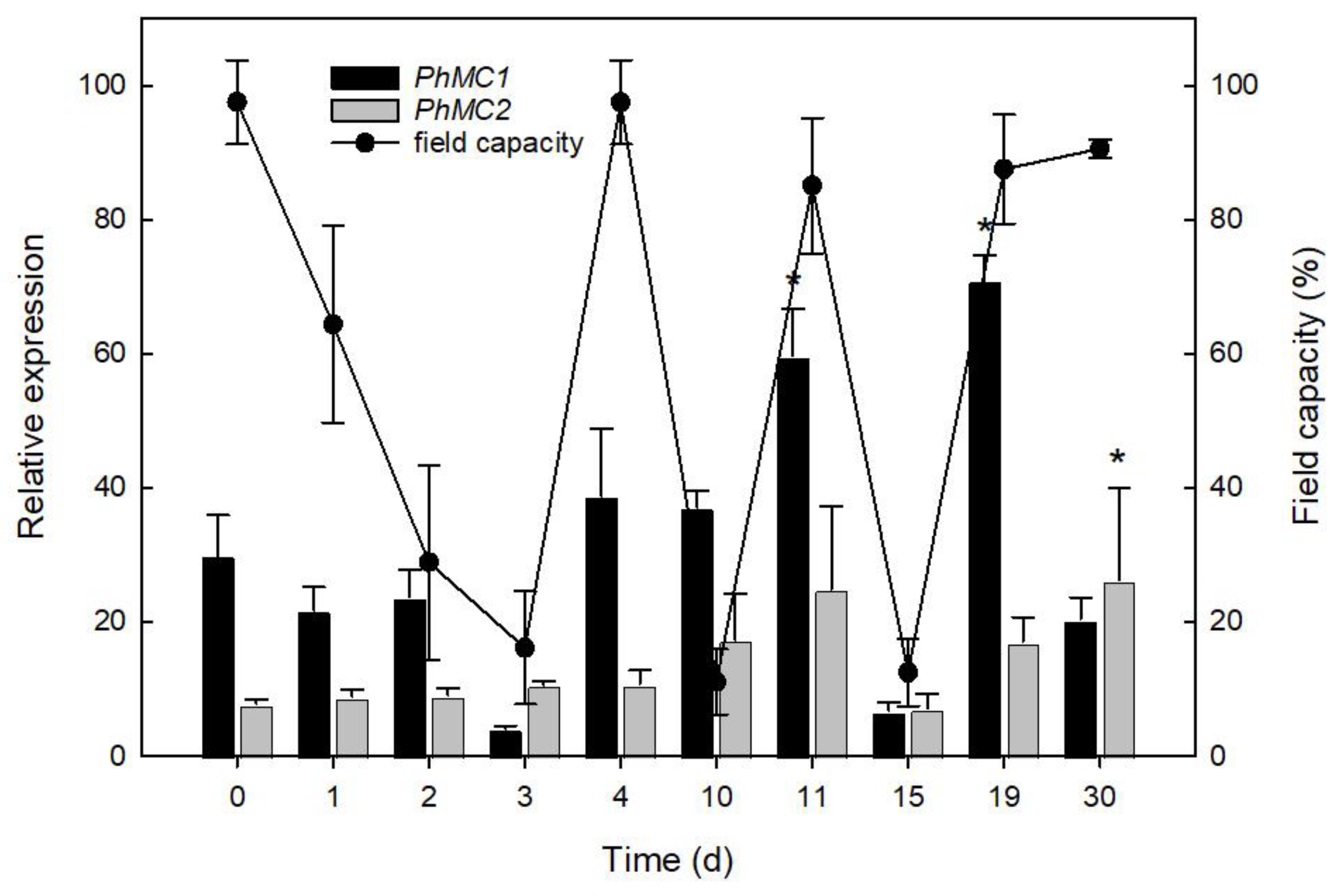
3.5. PhMC1 and PhMC2 Expression during Biotic and Abiotic Stress Responses in Petunia
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fischer, U.; Jänicke, R.U.; Schulze-Osthoff, K. Many Cuts to Ruin: A Comprehensive Update of Caspase Substrates. Cell Death Differ. 2003, 10, 76–100. [Google Scholar] [CrossRef] [Green Version]
- Uren, A.; O’Rourke, K.; Aravind, L.; Pisabarro, M.; Seshagiri, S.; Koonin, E.; Dixit, V. Identification of Paracaspases and Metacaspases: Two Ancient Families of Caspase-like Proteins, One of Which Plays a Key Role in MALT Lymphoma. Mol. Cell 2000, 6, 961–967. [Google Scholar] [CrossRef] [PubMed]
- Vercammen, D.; van de Cotte, B.; de Jaeger, G.; Eeckhout, D.; Casteels, P.; Vandepoele, K.; Vandenberghe, I.; van Beeumen, J.; Inzé, D.; van Breusegem, F. Type II Metacaspases Atmc4 and Atmc9 of Arabidopsis thaliana Cleave Substrates after Arginine and Lysine. J. Biol. Chem. 2004, 279, 45329–45336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bozhkov, P.V.; Suarez, M.F.; Filonova, L.H.; Daniel, G.; Zamyatnin, A.A.; Rodriguez-Nieto, S.; Zhivotovsky, B.; Smertenko, A. Cysteine Protease McII-Pa Executes Programmed Cell Death during Plant Embryogenesis. Proc. Natl. Acad. Sci. USA 2005, 4, 14463–14468. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Watanabe, N.; Lam, E. Two Arabidopsis Metacaspases AtMCP1b and AtMCP2b Are Arginine/Lysine-Specific Cysteine Proteases and Activate Apoptosis-like Cell Death in Yeast. J. Biol. Chem. 2005, 280, 14691–14699. [Google Scholar] [CrossRef] [Green Version]
- Vercammen, D.; Belenghi, B.; van de Cotte, B.; Beunens, T.; Gavigan, J.A.; de Rycke, R.; Brackenier, A.; Inzé, D.; Harris, J.L.; van Breusegem, F. Serpin1 of Arabidopsis thaliana is a Suicide Inhibitor for Metacaspase 9. J. Mol. Biol. 2006, 364, 625–636. [Google Scholar] [CrossRef]
- Choi, C.J.; Berges, J.A. New Types of Metacaspases in Phytoplankton Reveal Diverse Origins of Cell Death Proteases. Cell Death Dis. 2013, 4, e490. [Google Scholar] [CrossRef] [Green Version]
- Klemenčič, M.; Funk, C. Type III Metacaspases: Calcium-Dependent Activity Proposes New Function for the P10 Domain. New Phytol. 2018, 218, 1179–1191. [Google Scholar] [CrossRef] [Green Version]
- Huh, S.U. Evolutionary Diversity and Function of Metacaspases in Plants: Similar to but not Caspases. Int. J. Mol. Sci. 2022, 23, 4588. [Google Scholar] [CrossRef]
- Tsiatsiani, L.; van Breusegem, F.; Gallois, P.; Zavialov, A.; Lam, E.; Bozhkov, P. Metacaspases. Cell Death Differ. 2011, 18, 1279–1288. [Google Scholar] [CrossRef] [Green Version]
- Castillo-Olamendi, L.; Bravo-Garcìa, A.; Morán, J.; Rocha-Sosa, M.; Porta, H. AtMCP1b, a Chloroplast-Localised Metacaspase, Is Induced in Vascular Tissue after Wounding or Pathogen Infection. Funct. Plant Biol. 2007, 34, 1061–1071. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, R.; Drury, G.E.; Rotari, V.I.; Gordon, A.; Willer, M.; Farzaneh, T.; Woltering, E.J.; Gallois, P. Metacaspase-8 Modulates Programmed Cell Death Induced by Ultraviolet Light and H2O2 in Arabidopsis. J. Biol. Chem. 2008, 283, 774–783. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Basak, S.; Kundu, P. Plant Metacaspases: Decoding Their Dynamics in Development and Disease. Plant Physiol. Biochem. 2022, 180, 50–63. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Liu, J.; Wei, Y. Identification and Analysis of the Metacaspase Gene Family in Tomato. Biochem. Biophys. Res. Commun. 2016, 479, 523–529. [Google Scholar] [CrossRef] [PubMed]
- Dubey, N.; Trivedi, M.; Varsani, S.; Vyas, V.; Farsodia, M.; Singh, S.K. Genome-Wide Characterization, Molecular Evolution and Expression Profiling of the Metacaspases in Potato (Solanum tuberosum L.). Heliyon 2019, 5, 1162. [Google Scholar] [CrossRef] [Green Version]
- Lee, R.E.C.; Puente, L.G.; Kærn, M.; Megeney, L.A. A Non-Death Role of the Yeast Metacaspase: Yca1p Alters Cell Cycle Dynamics. PLoS ONE 2008, 3, e2956. [Google Scholar] [CrossRef] [Green Version]
- Lee, R.E.C.; Brunette, S.; Puente, L.G.; Megeney, L.A. Metacaspase Yca1 Is Required for Clearance of Insoluble Protein Aggregates. Proc. Natl. Acad. Sci. USA 2010, 107, 13348–13353. [Google Scholar] [CrossRef] [Green Version]
- Tsiatsiani, L.; Timmerman, E.; de Bock, P.J.; Vercammen, D.; Stael, S.; van de Cotte, B.; Staes, A.; Goethals, M.; Beunens, T.; van Damme, P.; et al. The Arabidopsis METACASPASE9 Degradome. Plant Cell 2013, 25, 2831–2847. [Google Scholar] [CrossRef] [Green Version]
- Wrzaczek, M.; Vainonen, J.P.; Stael, S.; Tsiatsiani, L.; Help-Rinta-Rahko, H.; Gauthier, A.; Kaufholdt, D.; Bollhöner, B.; Lamminmäki, A.; Staes, A.; et al. GRIM REAPER Peptide Binds to Receptor Kinase PRK 5 to Trigger Cell Death in Arabidopsis. EMBO J. 2015, 34, 55–66. [Google Scholar] [CrossRef] [Green Version]
- Coll, N.S.; Smidler, A.; Puigvert, M.; Popa, C.; Valls, M.; Dangl, J.L. The Plant Metacaspase AtMC1 in Pathogen-Triggered Programmed Cell Death and Aging: Functional Linkage with Autophagy. Cell Death Differ. 2014, 21, 1399–1408. [Google Scholar] [CrossRef]
- Chapin, L.; Jones, M.L. Nutrient Remobilization during Pollination-Induced Corolla Senescence in Petunia. Acta Hortic. 2007, 755. [Google Scholar] [CrossRef]
- Jones, M.L.; Stead, A.D.; Clark, D.G. Petunia Flower Senescence. In Petunia: A Model System for Comparative Research; Gerats, T., Strommer, J., Eds.; Springer: New York, NY, USA, 2009; pp. 301–324. [Google Scholar]
- Chapin, L.J.; Moon, Y.; Jones, M.L. Downregulating a Type I Metacaspase in Petunia Accelerates Flower Senescence. J. Am. Soc. Hortic. Sci. 2017, 142, 405–414. [Google Scholar] [CrossRef] [Green Version]
- Goujon, M.; McWilliam, H.; Li, W.; Valentin, F.; Squizzato, S.; Paern, J.; Lopez, R. A New Bioinformatics Analysis Tools Framework at EMBL-EBI. Nucleic. Acids. Res. 2010, 38, W695–W699. [Google Scholar] [CrossRef] [PubMed]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, Scalable Generation of High-Quality Protein Multiple Sequence Alignments Using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef] [PubMed]
- Heckman, K.L.; Pease, L.R. Gene Splicing and Mutagenesis by PCR-Driven Overlap Extension. Nat. Protoc. 2007, 2, 924–932. [Google Scholar] [CrossRef]
- Hoeberichts, F.A.; Have, A.T.; Woltering, E.J. A Tomato Metacaspase Gene is Upregulated during Programmed Cell Death in Botrytis Cinerea-Infected Leaves. Planta 2003, 217, 517–522. [Google Scholar] [CrossRef]
- Quijia Pillajo, J.O.; Chapin, L.J.; Jones, M.L. Senescence and Abiotic Stress Induce Expression of Autophagy-Related Genes in Petunia. J. Am. Soc. Hortic. Sci. 2018, 143, 154–163. [Google Scholar] [CrossRef] [Green Version]
- Vercammen, D.; Declercq, W.; Vandenabeele, P.; van Breusegem, F. Are Metacaspases Caspases? J. Cell Biol. 2007, 179, 375–380. [Google Scholar] [CrossRef]
- Kwon, S.I.; Hwang, D.J. Expression Analysis of the Metacaspase Gene Family in Arabidopsis. J. Plant Biol. 2013, 56, 391–398. [Google Scholar] [CrossRef]
- Zhang, C.; Gong, P.; Wei, R.; Li, S.; Zhang, X.; Yu, Y.; Wang, Y. The Metacaspase Gene Family of Vitis vinifera L.: Characterization and Differential Expression during Ovule Abortion in Stenospermocarpic Seedless Grapes. Gene 2013, 528, 267–276. [Google Scholar] [CrossRef]
- Wang, L.; Zhang, H. Genomewide Survey and Characterization of Metacaspase Gene Family in Rice (Oryza sativa). J. Genet. 2014, 93, 93–102. [Google Scholar] [CrossRef] [PubMed]
- Broderick, S.R.; Wijeratne, S.; Wijeratn, A.J.; Chapin, L.J.; Meulia, T.; Jones, M.L. RNA-Sequencing Reveals Early, Dynamic Transcriptome Changes in the Corollas of Pollinated Petunias. BMC Plant Biol. 2014, 14, 1–21. [Google Scholar] [CrossRef] [Green Version]
- Bollhöner, B.; Zhang, B.; Stael, S.; Denancé, N.; Overmyer, K.; Goffner, D.; van Breusegem, F.; Tuominen, H. Post Mortem Function of AtMC9 in Xylem Vessel Elements. New Phytol. 2013, 200, 498–510. [Google Scholar] [CrossRef] [PubMed]
- Coll, N.S.; Vercammen, D.; Smidler, A.; Clover, C.; van Breusegem, F.; Dangl, J.L.; Epple, P. Arabidopsis Type I Metacaspases Control Cell Death. Science 2010, 330, 1393–1397. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, N.; Lam, E. Arabidopsis Metacaspase 2d Is a Positive Mediator of Cell Death Induced during Biotic and Abiotic Stresses. Plant J. 2011, 66, 969–982. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.M.; Bae, C.; Oh, S.K.; Choi, D. A Pepper (Capsicum annuum L.) Metacaspase 9 (Camc9) Plays a Role in Pathogen-Induced Cell Death in Plants. Mol. Plant Pathol. 2013, 14, 557–566. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Zhang, H.; Hong, Y.; Liu, S.; Li, D.; Song, F. Stress-Responsive Expression, Subcellular Localization and Protein–Protein Interactions of the Rice Metacaspase Family. Int. J. Mol. Sci. 2015, 16, 16216–16241. [Google Scholar] [CrossRef] [Green Version]
- Hao, Y.; Wang, X.; Wang, K.; Li, H.; Duan, X.; Tang, C.; Kang, Z. TaMCA1, a Regulator of Cell Death, Is Important for the Interaction between Wheat and Puccinia striiformis. Sci. Rep. 2016, 6, 26946. [Google Scholar] [CrossRef] [Green Version]
- Ma, S.; Shi, H.; Wang, G.F. The Potential Roles of Different Metacaspases in Maize Defense Response. Plant Signal. Behav. 2021, 16, 1906574. [Google Scholar] [CrossRef]
- Watanabe, N.; Lam, E. Calcium-Dependent Activation and Autolysis of Arabidopsis Metacaspase 2d. J. Biol. Chem. 2011, 286, 10027–10040. [Google Scholar] [CrossRef]
- Yue, J.Y.; Wang, Y.J.; Jiao, J.L.; Wang, W.W.; Wang, H.Z. The Metacaspase TaMCA-Id Negatively Regulates Salt-Induced Programmed Cell Death and Functionally Links with Autophagy in Wheat. Front. Plant Sci. 2022, 13. [Google Scholar] [CrossRef] [PubMed]
- Shamloo-Dashtpagerdi, R.; Lindlöf, A.; Aliakbari, M.; Pirasteh-Anosheh, H. Plausible Association between Drought Stress Tolerance of Barley (Hordeum vulgare L.) and Programmed Cell Death via MC1 and TSN1 Genes. Physiol. Plant. 2020, 170, 46–59. [Google Scholar] [CrossRef] [PubMed]
- Ohsumi, Y. Historical Landmarks of Autophagy Research. Cell Res. 2014, 24, 9–23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, Y.; Jones, M.L. Silencing ATG6 and PI3K Accelerates Petal Senescence and Reduces Flower Number and Shoot Biomass in Petunia. Plant Sci. 2021, 302, 110713. [Google Scholar] [CrossRef]
- Fagundes, D.; Bohn, B.; Cabreira, C.; Leipelt, F.; Dias, N.; Bodanese-Zanettini, M.H.; Cagliari, A. Caspases in Plants: Metacaspase Gene Family in Plant Stress Responses. Funct. Integr. Genom. 2015, 15, 639–649. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chapin, L.J.; Jones, M.L. A Type I and a Type II Metacaspase Are Differentially Regulated during Corolla Development and in Response to Abiotic and Biotic Stresses in Petunia × hybrida. Horticulturae 2022, 8, 1151. https://doi.org/10.3390/horticulturae8121151
Chapin LJ, Jones ML. A Type I and a Type II Metacaspase Are Differentially Regulated during Corolla Development and in Response to Abiotic and Biotic Stresses in Petunia × hybrida. Horticulturae. 2022; 8(12):1151. https://doi.org/10.3390/horticulturae8121151
Chicago/Turabian StyleChapin, Laura J., and Michelle L. Jones. 2022. "A Type I and a Type II Metacaspase Are Differentially Regulated during Corolla Development and in Response to Abiotic and Biotic Stresses in Petunia × hybrida" Horticulturae 8, no. 12: 1151. https://doi.org/10.3390/horticulturae8121151




