PpMYB39 Activates PpDFR to Modulate Anthocyanin Biosynthesis during Peach Fruit Maturation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Sample Collection
2.2. RNA Extraction and Library Construction
2.3. Mapping of Reads and Gene Expression Quantification
2.4. Analysis of Differential Expressions of Genes (DEGs)
2.5. Selected DEGs Validation by RT-qPCR
2.6. Anthocyanin Quantification in Red- and White-Fleshed Fruits
2.7. Dual Luciferase Reporter Assay
2.8. Yeast One-Hybrid Assay
3. Results
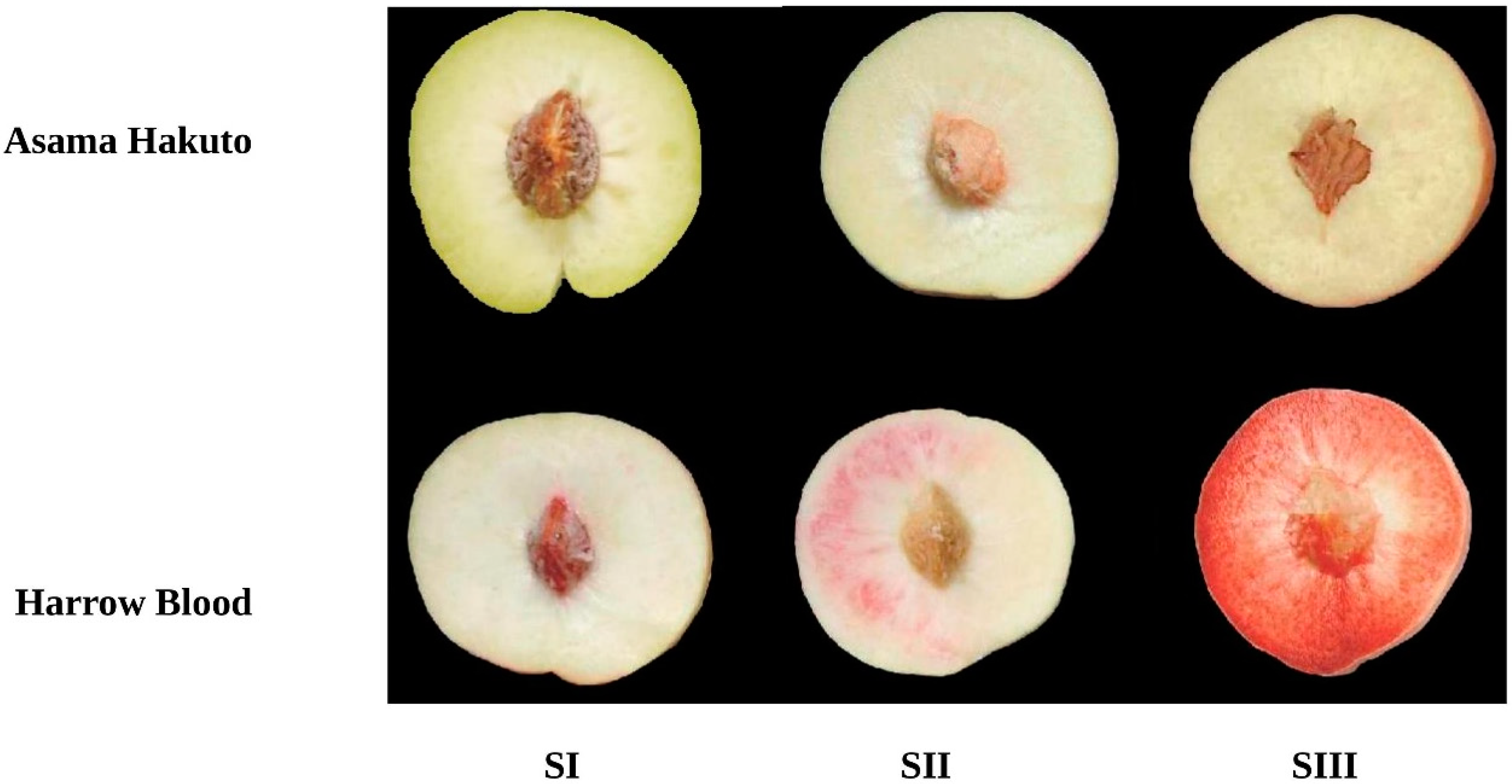
3.1. Anthocyanin Accumulation during Peach Fruit Development
3.2. Transcriptome Assembly
3.3. Differentially Expressed Genes (DEGs between Red- and White-Fleshed Fruit
3.4. Identification of Genes Associated with Anthocyanin Biosynthesis
3.5. Selection of Key Genes Controlling Flesh Color
3.6. PpMYB39 Plays a Crucial Role in Red Flesh Formation in Peaches
3.7. PpMYB39 Function as a Transcriptional Activator of PpDFR
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhao, X.; Zhang, W.; Yin, X.; Su, M.; Sun, C.; Li, X.; Chen, K. Phenolic composition and antioxidant properties of different peach [Prunus persica (L.) Batsch] cultivars in China. Int. J. Mol. Sci. 2015, 16, 5762–5778. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Folta, K.M.; Gardiner, S.E. Genetics and Genomics of Rosaceae; Springer: New York, NY, USA, 2009; Volume 6, pp. 411–506. [Google Scholar] [CrossRef]
- Arús, P.; Verde, I.; Sosinski, B.; Zhebentyayeva, T.; Abbott, A.G. The peach genome. Tree Genet. Genomes 2012, 8, 531–547. [Google Scholar] [CrossRef]
- Werner, D.J.; Creller, M.A.; Chaparro, J.X. Inheritance of the blood-flesh trait in peach. HortScience 1998, 33, 1243–1246. [Google Scholar] [CrossRef] [Green Version]
- Cao, K.; Ding, T.; Mao, D.; Zhu, G.; Fang, W.; Chen, C.; Wang, L. Transcriptome analysis reveals novel genes involved in anthocyanin biosynthesis in the flesh of peach. Plant Physiol. Biochem. 2018, 123, 94–102. [Google Scholar] [CrossRef]
- Kong, J.M.; Chia, L.S.; Goh, N.K.; Chia, T.F.; Brouillard, R. Analysis and biological activities of anthocyanins. Phytochemistry 2003, 64, 923–933. [Google Scholar] [CrossRef]
- Zhao, Y.; Dong, W.; Wang, K.; Zhang, B.; Allan, A.C.; Lin-Wang, K.; Xu, C. Differential sensitivity of fruit pigmentation to ultraviolet light between two peach cultivars. Front. Plant Sci. 2017, 8, 1552. [Google Scholar] [CrossRef]
- Gonçalves, A.C.; Nunes, A.R.; Falcão, A.; Alves, G.; Silva, L.R. Dietary effects of anthocyanins in human health: A comprehensive review. Pharmaceuticals 2021, 14, 690. [Google Scholar] [CrossRef]
- Orazem, P.; Stampar, F.; Hudina, M. Fruit quality of Redhaven and Royal Glory peach cultivars on seven different rootstocks. J. Agric. Food Chem. 2011, 59, 9394–9401. [Google Scholar] [CrossRef]
- Lin-Wang, K.; Bolitho, K.; Grafton, K.; Kortstee, A.; Karunairetnam, S.; McGhie, T.K.; Allan, A.C. An R2R3 MYB transcription factor associated with regulation of the anthocyanin biosynthetic pathway in Rosaceae. BMC Plant Biol. 2010, 10, 50. [Google Scholar] [CrossRef] [Green Version]
- Grotewold, E. The genetics and biochemistry of floral pigments. Annu. Rev. Plant Biol. 2006, 57, 761–780. [Google Scholar] [CrossRef]
- Dare, A.P.; Schaffer, R.J.; Lin-Wang, K.; Allan, A.C.; Hellens, R.P. Identification of a cis-regulatory element by transient analysis of co-ordinately regulated genes. Plant Methods 2008, 4, 17. [Google Scholar] [CrossRef] [Green Version]
- Jiang, S.; Chen, M.; He, N.; Chen, X.; Wang, N.; Sun, Q.; Chen, X. MdGSTF6, activated by MdMYB1, plays an essential role in anthocyanin accumulation in apple. Hortic. Res. 2019, 6, 40. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niu, S.S.; Xu, C.J.; Zhang, W.S.; Zhang, B.; Li, X.; Lin-Wang, K.; Chen, K.S. Coordinated regulation of anthocyanin biosynthesis in Chinese bayberry (Myrica rubra) fruit by a R2R3 MYB transcription factor. Planta 2010, 231, 887–899. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, S.; Goto-Yamamoto, N.; Hirochika, H. Retrotransposon-induced mutations in grape skin color. Science 2004, 304, 982. [Google Scholar] [CrossRef]
- Feng, S.; Wang, Y.; Yang, S.; Xu, Y.; Chen, X. Anthocyanin biosynthesis in pears is regulated by a R2R3-MYB transcription factor PyMYB10. Planta 2010, 232, 245–255. [Google Scholar] [CrossRef]
- Chagné, D.; Lin-Wang, K.; Espley, R.V.; Volz, R.K.; How, N.M.; Rouse, S.; Allan, A.C. An ancient duplication of apple MYB transcription factors is responsible for novel red fruit-flesh phenotypes. Plant Physiol. 2013, 161, 225–239. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Butelli, E.; Licciardello, C.; Zhang, Y.; Liu, J.; Mackay, S.; Bailey, P.; Reforgiato-Recupero, G.; Martin, C. Retrotransposons control fruit-specific, cold-dependent accumulation of anthocyanins in blood oranges. Plant Cell 2012, 24, 1242–1255. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.Y.; Mao, K.; Zhao, C.; Zhao, X.Y.; Zhang, H.L.; Shu, H.R.; Hao, Y.J. MdCOP1 ubiquitin E3 ligases interact with MdMYB1 to regulate light-induced anthocyanin biosynthesis and red fruit coloration in apple. Plant Physiol. 2012, 160, 1011–1022. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Meng, D.; Wang, A.; Li, T.; Jiang, S.; Cong, P.; Li, T. The methylation of the PcMYB10 promoter is associated with green-skinned sport in Max Red Bartlett pear. Plant Physiol. 2013, 162, 885–896. [Google Scholar] [CrossRef] [Green Version]
- Telias, A.; Lin-Wang, K.; Stevenson, D.E.; Cooney, J.M.; Hellens, R.P.; Allan, A.C.; Bradeen, J.M. Apple skin patterning is associated with differential expression of MYB10. BMC Plant Biol. 2011, 11, 93. [Google Scholar] [CrossRef] [Green Version]
- Ravaglia, D.; Espley, R.V.; Henry-Kirk, R.A.; Andreotti, C.; Ziosi, V.; Hellens, R.P.; Allan, A.C. Transcriptional regulation of flavonoid biosynthesis in nectarine (Prunus persica) by a set of R2R3 MYB transcription factors. BMC Plant Biol. 2013, 13, 68. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, Y.C.; Zhang, B.; Allan, A.C.; Lin-Wang, K.; Zhao, Y.; Wang, K.; Chen, K.S.; Xu, C.J. DNA demethylation is involved in the regulation of temperature-dependent anthocyanin accumulation in peach. Plant J. 2020, 102, 965–976. [Google Scholar] [CrossRef] [PubMed]
- Tsuda, T.; Yamaguchi, M.; Honda, C.; Moriguchi, T. Expression of anthocyanin biosynthesis genes in the skin of peach and nectarine fruit. J. Am. Soc. Hortic. Sci. 2004, 129, 857–862. [Google Scholar] [CrossRef] [Green Version]
- Dardick, C.; Callahan, A.M. Evolution of the fruit endocarp: Molecular mechanisms underlying adaptations in seed protection and dispersal strategies. Front. Plant Sci. 2014, 5, 284. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Zhou, H.; Lin-Wang, K.; Vimolmangkang, S.; Espley, R.V.; Wang, L.; Han, Y. Transcriptome analysis and transient transformation suggest an ancient duplicated MYB transcription factor as a candidate gene for leaf red coloration in peach. BMC Plant Biol. 2014, 14, 388. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uematsu, C.; Katayama, H.; Makino, I.; Inagaki, A.; Arakawa, O.; Martin, C. Peace, a MYB-like transcription factor, regulates petal pigmentation in flowering peach ‘Genpei’bearing variegated and fully pigmented flowers. J. Exp. Bot. 2014, 65, 1081–1094. [Google Scholar] [CrossRef] [Green Version]
- Rahim, M.A.; Busatto, N.; Trainotti, L. Regulation of anthocyanin biosynthesis in peach fruits. Planta 2014, 240, 913–929. [Google Scholar] [CrossRef]
- Wang, G.; Chen, B.; Du, H.; Zhang, F.; Zhang, H.; Wang, Y.; Zhang, X. Genetic mapping of anthocyanin accumulation-related genes in pepper fruits using a combination of SLAF-seq and BSA. PLoS ONE 2018, 13, e0204690. [Google Scholar] [CrossRef]
- Guo, J.; Cao, K.; Li, Y.; Yao, J.L.; Deng, C.; Wang, Q.; Zhu, G.; Fang, W.; Chen, C.; Wang, X.; et al. Comparative transcriptome and microscopy analyses provide insights into flat shape formation in peach (Prunus persica). Front. Plant Sci. 2018, 8, 2215. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [Green Version]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef] [PubMed]
- Frazee, A.C.; Pertea, G.; Jaffe, A.E.; Langmead, B.; Salzberg, S.L.; Leek, J.T. Ballgown bridges the gap between transcriptome assembly and expression analysis. Nat. Biotechnol. 2015, 33, 243–246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B (Methodol.) 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Cao, K.; Zhou, Z.; Wang, Q.; Guo, J.; Zhao, P.; Zhu, G.; Fang, W.; Chen, C.; Wang, X.; Wang, X.; et al. Genome-wide association study of 12 agronomic traits in peach. Nat. Commun. 2016, 7, 13246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, H.; Cao, K.; Zhu, G.; Fang, W.; Chen, C.; Wang, X.; Wang, L. Genome-wide Association Analysis of Red Flesh Character Based on Resequencing Approach in Peach. J. Am. Soc. Hortic. Sci. 2019, 144, 209–216. [Google Scholar] [CrossRef] [Green Version]
- Cheng, G.W.; Breen, P.J. Activity of phenylalanine ammonia-lyase (PAL) and concentrations of anthocyanins and phenolics in developing strawberry fruit. J. Am. Soc. Hortic. Sci. 1991, 116, 865–869. [Google Scholar] [CrossRef]
- Wang, Y.; Zhou, L.J.; Wang, Y.; Geng, Z.; Ding, B.; Jiang, J.; Chen, S.; Chen, F. An R2R3-MYB transcription factor CmMYB21 represses anthocyanin biosynthesis in color fading petals of chrysanthemum. Sci. Hortic. 2022, 293, 110674. [Google Scholar] [CrossRef]
- Guo, J.; Cao, K.; Deng, C.; Li, Y.; Zhu, G.; Fang, W.; Chen, C.; Wang, X.; Wu, J.; Guan, L.; et al. An integrated peach genome structural variation map uncovers genes associated with fruit traits. Genome Biol. 2020, 21, 1–19. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, W.; Dou, Y.; Zhang, J.; Jiang, G.; Miao, L.; Zhang, Z. Transcript quantification by RNA-seq reveals differentially expressed genes in the red and yellow fruits of Fragaria vesca. PLoS ONE 2015, 10, e0144356. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Wang, L.; Zeng, X.; Chen, R.; Yang, S.; Pan, S. Comparative transcriptome analysis reveals fruit discoloration mechanisms in postharvest strawberries in response to high ambient temperature. Food Chem. X 2019, 2, 100025. [Google Scholar] [CrossRef]
- Chen, P.N.; Chu, S.C.; Chiou, H.L.; Kuo, W.H.; Chiang, C.L.; Hsieh, Y.S. Mulberry anthocyanins, cyanidin 3-rutinoside and cyanidin 3-glucoside, exhibited an inhibitory effect on the migration and invasion of a human lung cancer cell line. Cancer Lett. 2006, 235, 248–259. [Google Scholar] [CrossRef] [PubMed]
- Tuan, P.A.; Bai, S.; Yaegaki, H.; Tamura, T.; Hihara, S.; Moriguchi, T.; Oda, K. The crucial role of PpMYB10. 1 in anthocyanin accumulation in peach and relationships between its allelic type and skin color phenotype. BMC Plant Biol. 2015, 15, 280. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, H.; Peng, Q.; Zhao, J.; Owiti, A.; Ren, F.; Liao, L.; Han, Y. Multiple R2R3-MYB transcription factors involved in the regulation of anthocyanin accumulation in peach fruit. Front. Plant Sci. 2016, 7, 1557. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Cai, Z.X.; Shen, Z.J.; Ma, R.J.; Yu, M.L. Proanthocyanidin monomers and cyanidin 3-o-glucoside accumulation in blood-flesh peach (Prunus persica (l.) Batsch) fruit. Arch. Biol. Sci. 2017, 69, 611–617. [Google Scholar] [CrossRef]
- Zhao, Y.; Dong, W.; Zhu, Y.; Allan, A.C.; Lin-Wang, K.; Xu, C. PpGST1, an anthocyanin-related glutathione S-transferase gene, is essential for fruit coloration in peach. Plant Biotechnol. J. 2020, 18, 1284–1295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Tikunov, Y.; Schouten, R.E.; Marcelis, L.F.; Visser, R.G.; Bovy, A. Anthocyanin biosynthesis and degradation mechanisms in Solanaceous vegetables: A review. Front. Chem. 2018, 6, 52. [Google Scholar] [CrossRef]
- Khan, I.A.; Cao, K.; Guo, J.; Li, Y.; Wang, Q.; Yang, X.; Wang, L. Identification of key gene networks controlling anthocyanin biosynthesis in peach flower. Plant Sci. 2022, 316, 111151. [Google Scholar] [CrossRef]
- Moyano, E.; Portero-Robles, I.; Medina-Escobar, N.; Valpuesta, V.; Munoz-Blanco, J.; Luis Caballero, J. A fruit-specific putative dihydroflavonol 4-reductase gene is differentially expressed in strawberry during the ripening process. Plant Physiol. 1998, 117, 711–716. [Google Scholar] [CrossRef] [Green Version]
- Piero, A.R.L.; Puglisi, I.; Petrone, G. Gene characterization, analysis of expression and in vitro synthesis of dihydroflavonol 4-reductase from [Citrus sinensis (L.) Osbeck]. Phytochemistry 2006, 67, 684–695. [Google Scholar] [CrossRef]
- Tian, J.; Han, Z.Y.; Zhang, J.; Hu, Y.; Song, T.; Yao, Y. The balance of expression of dihydroflavonol 4-reductase and flavonol synthase regulates flavonoid biosynthesis and red foliage coloration in crabapples. Sci. Rep. 2015, 5, 12228. [Google Scholar] [CrossRef]
- Luo, P.; Ning, G.; Wang, Z.; Shen, Y.; Jin, H.; Li, P.; Bao, M. Disequilibrium of flavonol synthase and dihydroflavonol-4-reductase expression associated tightly to white vs. red color fruit formation in plants. Front. Plant Sci. 2016, 6, 1257. [Google Scholar] [CrossRef] [PubMed] [Green Version]





| Sample | Raw Reads | Clean Reads | Clean Reads Rate (%) | Q30 (%) |
|---|---|---|---|---|
| HB-SI | 51,170,556 | 47,624,064 | 93.07 | 94.05 |
| HB-SII | 50,971,600 | 46,185,792 | 90.61 | 94.3 |
| HB-SIII | 47,834,472 | 42,998,916 | 89.89 | 94.2 |
| AH-SI | 46,768,718 | 42,826,842 | 91.57 | 93.76 |
| AH-SII | 48,688,728 | 45,267,666 | 92.97 | 94.4 |
| AH-SIII | 50,232,674 | 46,087,396 | 91.75 | 94.14 |
| Groups | Total DEGs | Up-Regulated | Down-Regulated |
|---|---|---|---|
| HB SI_V_AS SI | 38 | 269 | 112 |
| HB SII_V_AS SII | 219 | 148 | 71 |
| HB SIII_V_AS SIII | 185 | 125 | 60 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khan, I.A.; Rahman, M.U.; Sakhi, S.; Nawaz, G.; Khan, A.A.; Ahmad, T.; Adnan, M.; Khan, S.M. PpMYB39 Activates PpDFR to Modulate Anthocyanin Biosynthesis during Peach Fruit Maturation. Horticulturae 2022, 8, 332. https://doi.org/10.3390/horticulturae8040332
Khan IA, Rahman MU, Sakhi S, Nawaz G, Khan AA, Ahmad T, Adnan M, Khan SM. PpMYB39 Activates PpDFR to Modulate Anthocyanin Biosynthesis during Peach Fruit Maturation. Horticulturae. 2022; 8(4):332. https://doi.org/10.3390/horticulturae8040332
Chicago/Turabian StyleKhan, Irshad Ahmad, Masood Ur Rahman, Shazia Sakhi, Ghazala Nawaz, Aftab Ahmad Khan, Tanveer Ahmad, Mohammad Adnan, and Shah Masaud Khan. 2022. "PpMYB39 Activates PpDFR to Modulate Anthocyanin Biosynthesis during Peach Fruit Maturation" Horticulturae 8, no. 4: 332. https://doi.org/10.3390/horticulturae8040332
APA StyleKhan, I. A., Rahman, M. U., Sakhi, S., Nawaz, G., Khan, A. A., Ahmad, T., Adnan, M., & Khan, S. M. (2022). PpMYB39 Activates PpDFR to Modulate Anthocyanin Biosynthesis during Peach Fruit Maturation. Horticulturae, 8(4), 332. https://doi.org/10.3390/horticulturae8040332





