1. Introduction
Symbiotic relationships between fungi and plants range from mutualism to parasitism. The latter mode is a focus of research in agricultural biotechnology, because fungal diseases significantly reduce crop yields at both the pre-harvest and the post-harvest stages. The main strategy used by plants as antifungal response is the suppression of fungal growth through destruction of the cell wall, which ultimately leads to fungal cell death [
1,
2,
3].
In most fungi, the inner cell wall has a relatively conserved structure with an alkali-insoluble core consisting of branched covalently linked β-(1,3) and β-(1,6) glucans, chitin (a polymer of
N-acetyl-
D-glucosamine), and chitosan (a linear chitin derivative composed of acetylated (chitin) and deacetylated (β-(1→4)-linked
D-glucosamine) [
1]. While chitin is the second most abundant natural polysaccharide, chitosan is found only in fungi that express deacetylases [
2]. During fungal invasion, plants perceive chitin/chitosan as a conserved microbe/pathogen-associated molecular pattern (MAMP/PAMP) and activate defense mechanisms through generation of reactive oxygen species (ROS), induction of salicylic acid (SA) and jasmonic acid (JA) signaling, and subsequent mobilization of pathogenesis-related (PR) proteins [
1,
3]. Among the 17 currently known PR families, 13 are implicated in responses to fungal attacks [
4], including CAP-domain proteins (PR1), β-1,3-glucanases (PR2), chitinases (PR3), Barwin-domain proteins (PR4), thaumatin-like proteins (PR5), and defensins (PR12) [
5,
6,
7,
8,
9,
10]. PR1 proteins are involved in plant cell-wall thickening to prevent the spread of pathogens in the apoplast [
7], and PR12 defensins reduce fungal growth and hyphal elongation [
8]. PR2–PR5 families comprise enzymes with β-1,3-glucanase, chitinase, and chitosanase activities, which exert antifungal effects through destruction of the fungal cell wall [
5,
6,
9,
10]. Plant chitinases, which hydrolyze chitin/chitosan by cleaving β-1,4-glycosidic bonds that link acetylated
D-glucosamine monomers, are considered the most important enzymes in plant self-defense against fungi; the activation of chitinase genes depends on the acetylation degree of chitin/chitosan and polymerization [
11]. Deacetylated chitosan components are hydrolyzed by chitosanases [
12].
Because chitin and chitosan can elicit plant immune responses and are biodegradable, nontoxic, and non-allergenic, they are widely used in crop protective biotechnology to increase plant resistance to pathogens and reduce the application of bactericides and fungicides [
13]. Structural modifications impart novel physicochemical properties and biological activities to chitin/chitosan derivatives [
13]. Several studies have reported the positive effects of chitosan and its derivatives on plant growth and development, including enhancement of photosynthetic and antioxidant activities, and improvement of crop quality and yield [
14,
15,
16]. It has also been shown that foliar application of chitosan decreases plant transpiration and water use, while maintaining biomass production [
13].
Chitosan-triggered plant immunity may be regulated at the post-transcriptional and post-translational levels, from signal recognition and induction of defense genes to synthesis of defense-related proteins and other metabolites [
17]. Thus, chitosan has been reported to reduce the size of stomatal aperture used by pathogens to penetrate the plant cell [
17], activate defense-related genes (such as those encoding chitinases, β-1,3-glucanases, and lipoxygenases), and stimulate ROS production and energy assimilation [
13,
17] without induction of hypersensitivity leading to cell death [
17].
The protective activity of chitosan against fungal infection is based not only on the stimulation of plant immune response but also on negative effects on fungi, which, depending on the fluidity of the plasma membrane, can be chitosan-sensitive or -resistant [
18]. In the former, cell membranes are enriched in polyunsaturated fatty acids, whereas, in the latter, membranes mostly contain saturated fatty acids and, therefore, present a barrier to chitosan [
19]. It is thought that, in the membranes of chitosan-sensitive fungi, chitosan binds to negatively charged phospholipids and induces membrane permeabilization, which results in increased intracellular ROS production and oxidative stress [
18,
19]. Chitosan resistance is considered to be evolved in nematophagous and entomopathogenic fungi, which naturally encounter chitosan during infection of arthropods and nematodes [
19].
The most economically significant fungal pathogens in terms of negative impact on the yield of over 100 crop plants in all climate zones are soil-born hemibiotrophic ascomycetes of the
Fusarium genus causing
Fusarium basal rot (FBR),
Fusarium wilt (FW),
Fusarium head blight (FHB), and other diseases [
17,
20,
21,
22]. On the basis of their phylogeny and plasma membrane fluidity,
Fusarium spp. are classified as chitosan-sensitive fungi [
19]. Thus, chitosan treatment suppresses
F. graminearum growth in wheat [
23] and triggers immunity to
F. oxysporum in chickpea [
17]. The mechanisms of chitosan-induced protection against
Fusarium invasion include transcriptional upregulation of PR1–5, PR8, and PR11–13 family members and other defense-related enzymes such as phenylalanine ammonia-lyase (PAL) [
5,
6,
10,
17,
18]. Furthermore, it has been reported that, in plants, chitosan pretreatment stimulates the expression of extracellular matrix structural proteins, lysin motif domain-containing glycosylphosphatidylinositol-anchored protein 2 (LYM2), and receptor-like kinases, as well as the production of immunity-related metabolites (sugars, sugar and fatty alcohols, and organic acids) associated with ROS generation and regulation of stomatal function [
17].
Among the crops sensitive to
Fusarium spp., garlic (
Allium sativum L.) is one of the 20 most important vegetables and the second most important bulbous crop in the world [
24]. Garlic is mostly infected with
F. oxysporum (f. sp.
cepae) and
F. proliferatum, but is also susceptible to
F. acutatum,
F. anthophilium,
F. verticillioides,
F. solani, and
F. acuminatum attacks; characteristic symptoms include leaf discoloration and wilting (FW) or dry brown necrotic spots, white mycelium, and water-soaked signs at the clove surface (FBR) [
25,
26,
27,
28]. Previous studies of FBR-resistant and -susceptible
A. sativum cultivars have indicated that PR1–5 proteins, including chitinases and endo-β-1,3-glucanases, are involved in the immune response of garlic to
Fusarium infection [
5,
6].
The aim of this study was to investigate the effects of chitosan on the activity of immunity-related enzymes, chitinases and endo-β-1,3-glucanases, in garlic. Bulbs were treated with chitosan derivatives of low and medium molecular weight (MW) before and after infection with F. proliferatum and analyzed for the activity of chitinases and endo-β-1,3-glucanases and the expression of respective genes. Our results provide useful insights into the effect of chitosan on A. sativum, which can be used in the development of measures to increase the resistance of garlic to Fusarium infection.
2. Materials and Methods
2.1. Plants and Fungi
Bulbs of FBR-resistant winter garlic cultivar (cv.) Sarmat were kindly provided by the Federal Scientific Vegetable Center (FSVC, Moscow region, Russia); the number of cloves in the bulb was 7–11.
The
F. proliferatum strain, kindly provided by the Group of Experimental Mycology (Research Center of Biotechnology of the RAS, Moscow, Russia), was previously isolated from field-grown FBR-sensitive cv. Strelets bulbs [
29]. The pathogenicity test showed that the first signs of the disease appear on the surface of the treated cloves after 3–5 days of infection [
5,
6,
29]. The used strain FPSt2021 was stored in the collection of the group (as a stock culture in potato-dextrose (PD) medium supplemented with 30% glycerol at −80 °C). For the experiment, the strain was sown from the stock on a PD; freshly grown colonies were transferred to PD agar and used for further experiments.
2.2. Chitosan Hydrolysates
Crab shell chitosan (MW 1000 kDa, deacetylation degree (DD) 85%) was obtained from Bioprogress (Shchelkovo, Russia). Laminarin, dinitrosalicylic acid (DNS), iodonitrotetrazolium chloride, 1-methoxy-phenazine-methosulfate, and other chemicals of analytical grade were obtained from Sigma (St. Louis, MO, USA).
Chitosan hydrolysates (Chs) were prepared by chemical depolymerization of crab shell chitosan using nitric acid as described previously [
30] with some modifications. Briefly, 10 g of chitosan was dispersed in 200 mL of 6.5% (Ch1) or 1.95% (Ch2) nitric acid, incubated for 7 h at 70 °C with stirring, cooled to room temperature, and kept without stirring for 16 h at 23 °C. Then, the pH was adjusted to 5.0–5.2 with 25% ammonium hydroxide, and the mixture was diluted by distilled water to a final volume of 400 mL (
Table 1).
The MW of Ch1 and Ch2 was determined by high-performance gel permeation chromatography in an S 2100 Sykam chromatograph (Sykam, Germany) using a separation column (8 mm × 300 mm; PSS NOVEMA Max analytical 1000 A) and a pre-column (8.0 mm × 50 mm) [
31]. Pullulans were used as calibration standards.
The DD of Chs was determined by proton nuclear magnetic resonance (1H-NMR). Samples were prepared in deuterated water, and proton spectra were recorded on a Bruker AMX 400 spectrometer (Brucker, Watertown, MA, USA); 4,4-dimethyl-4-silapentane-sulfonic acid was used as a standard.
2.3. Evaluation of Chitosan Effects on F. proliferatum In Vitro
2.3.1. Effect of Chitosan on F. proliferatum Metabolic Activity (MA)
Twofold serial dilutions of Chs (from 4.000 to 0.625 mg/mL) were prepared in PD medium as previously described [
32] and distributed into wells of 96-well flat-bottom plates (SPL Life Sciences, Gyeonggi-do, Korea) at 100 µL/well.
To obtain conidia, fresh
F. proliferatum mycelium grown for 7 days on potato-dextrose agar (PDA) was scraped with a glass rod, suspended in 10 mL of liquid PD medium, filtered through sterile cotton to remove residual mycelium, and distributed evenly among the wells (100 µL per well) to a final concentration 1.25 × 10
5 conidia/mL. Plates were incubated for 24 h at 25 °C in the dark, and the effect of Chs was analyzed using a modified tetrazolium method, in which the production of purple formazan crystals due to NADPH-dependent oxidoreductase activity reflected cellular MA and cell viability. Briefly, 10 µL of iodonitrotetrazolium chloride solution (5 mg/mL in 0.1 M PBS, pH 7.4), containing 1-methoxy-phenazine-methosulfate (4 mg/mL), was added to each well, and plates were incubated for 4 h at 37 °C. Then, supernatant was carefully removed, and insoluble formazan crystals were dissolved in 150 µL of dimethyl sulfoxide for 16 h at 37 °C with stirring (100 rpm). Optical density (OD) was measured at 540 nm in a microplate photometer (Multiskan
TM FC Microplate Photometer, Thermo Fisher Scientific Inc., Waltham, MA, USA), and fungal MA was calculated as
where
ODt and
ODc are mean OD values in the test and control wells, respectively.
The results were based on three biological replicates with three technical replicates for each tested concentration. Biological replicates were wells with a culture for a specific concentration of chitosan; technical replicates were the Ch effect analysis in each well. The Ch concentration causing 50% inhibition of fungal MA (EC50) was considered as reference.
2.3.2. Effect of Chitosan on the Growth of F. proliferatum
The antifungal activity (AA) of Chs was evaluated in vitro as previously described [
33]. An agar disc (11 mm in diameter) with 1 day F. proliferatum culture was placed on the surface of 90 mm Petri dishes containing PD agar supplemented or not with Chs (1, 2, 4, or 6 mg/mL) and incubated at 25 °C in the dark. The mycelial growth was evaluated by measuring diameters of fungal growth areas after 3, 7, and 14 days, and AA was calculated as the percentage growth inhibition:
where
Dt and
Dc are the diameters (mm) of fungal culture growth areas in the test and control plates, respectively.
The data were expressed as the arithmetic mean value of three independent replicates for each concentration.
2.4. Chitosan Treatment and F. proliferatum Infection of Garlic Cloves
Cloves of cv. Sarmat were sterilized by soaking in 10% NaCl with 2.5% NaHCO3 for 30 min, rinsed with distilled water, incubated in 70% ethanol for 3 min, and rinsed with sterile water three times.
2.4.1. Pretreatment with Chitosan and F. proliferatum Infection
Twenty-five cloves were divided into five equal sets and soaked for 24 h in distilled water, two control solutions (S1 and S2), and Ch1 and Ch2 solutions (2 mg/mL), respectively. Then, cloves were placed on wet filter paper in Petri dishes and incubated at room temperature (23–25 °C) in the dark. After 6 days, active root growth was observed, and the roots of one clove from each set were used to analyze chitinase and β-1,3-glucanase activities and the expression of chitinase and β-1,3-glucanase genes.
Two cloves from each set were infected by soaking in
F. proliferatum conidial suspension (~10
6 conidia per mL) for 5 min according to the procedure described in [
5]. The inoculated cloves were transferred to fresh Petri dishes with wet filter paper and incubated at room temperature in the dark; the remaining two cloves of each set were used as control. Test and control roots were collected at 24 and 96 h post inoculation (hpi), immediately frozen in liquid nitrogen, and stored at −80 °C until gene expression and enzyme activity analyses; the timepoints were chosen on the basis of reports that certain
PR genes show peak expression 1–3 days after inoculation with hemibiotrophic pathogens [
34,
35].
The experiment was carried out in three biological replicates; in total, 75 cloves were processed.
2.4.2. F. proliferatum Infection and Post-Treatment with Chitosan
Cloves were incubated on wet filter paper in Petri dishes at room temperature (23–25 °C) in the dark for 72 h to initiate active rooting. Twenty cloves were divided into two sets (10 cloves per each): one was infected by soaking in F. proliferatum conidial suspension (~106 conidia per mL) for 5 min and transferred to fresh Petri dishes, whereas the other was used as control. After incubation for 24 h at room temperature in the dark, each set was divided into five groups (two cloves each), which were treated by soaking in distilled water, S1, S2, and Ch1 and Ch2 (2 mg/mL), respectively, for 24 h. Then, cloves were transferred to fresh Petri dishes with wet filter paper, incubated at room temperature (23–25 °C) in the dark for 24 and 72 h, frozen in liquid nitrogen, and stored at −80 °C until analyses of gene expression and enzymatic activity.
The experiment was carried out in three biological replicates; a total of 60 cloves were analyzed.
2.5. RNA Extraction and Quantitative Real-Time PCR (qRT-PCR)
Root samples (~0.2–0.5 g) were ground to powder in liquid nitrogen and used for total RNA extraction (RNeasy Plant Mini Kit, QIAGEN, Hilden, Germany). After removal of genomic DNA with an RNase-free DNase set (QIAGEN) and analysis by gel electrophoresis, RNA samples were used for first-strand cDNA synthesis (GoScript Reverse Transcription System; Promega, Madison, WI, USA) with an oligo-dT primer. RNA and cDNA concentrations were quantified by fluorimetry (Qubit® Fluorometer, Thermo Fisher Scientific). qRT-PCR was performed in a CFX96 Real-Time PCR Detection System (Bio-Rad Laboratories, Hercules, CA, USA) with 3.0 ng of cDNA, gene-specific primers, and SYBR Green RT-PCR mixture (Syntol, Moscow, Russia) at the following cycling conditions: initial denaturation at 95 °C for 5 min, 40 cycles of denaturation at 95 °C for 15 s, and annealing/extension at 60 °C for 40 s.
Gene-specific primer pairs were as follows (accession numbers are given according to [
36]): 5′–GCTAGAAACCATATCGTTGCCT–3′/5′–GCATACCGTAGCATACTCCGA–3′ (
AsPR2a; Asa2G01057.1), 5′–GGTCGCATTTCTCCTAGGCAT–3′/5′–GCGTCGCCTGCTGATGGAA–3′ (
AsPR2b; Asa2G01195.1), 5′–GTACCACTGGGGATACCGAT–3′/5′–CCCCATGAATATGGTCCATCG–3′ (
AsCHI33; Asa6G07427.1), 5′–GGAACCACTGGAGACATCAATG–3′/5′–GCCTTGTTCTTGCTTGAAGCAG–3′ (
AsCHI28; Asa6G07412.1), 5′–CCGCTTTCTTCGCACAGACTT–3′/5′–TCCCCTGCTCTTCCACAAAG–3′ (
AsCHI1; Asa1G02082.1), and 5′–CTTTTCTTGGCCATGTTGGTGC–3′/5′–TCAGCACAATAGGACTGGCTC–3′ (
AsCHI34; Asa7G05194.1).
The gene expression data were normalized using two reference garlic genes,
GAPDH [
37] and
UBQ [
38], and statistically analyzed with Graph Pad Prism version 8 (GraphPad Software Inc., San Diego, CA, USA;
https://www.graphpad.com/scientific-software/prism/ accessed on 1 February 2021). The results were expressed as the mean ± standard deviation (SE) from three technical replicates of three biological replicates for each combination of cDNA and primer pairs. The unequal variance (Welch’s)
t-test was applied to assess differences in gene expression;
p < 0.01 was considered to indicate statistical significance. Sample calibrators were garlic samples treated with water against all other samples (1) and samples treated with solution S1 or S2 in relation to Ch1 or Ch2 treatment (2).
2.6. Chitinase and β-1,3-Glucanase Activity
Garlic roots were ground in liquid nitrogen, and 0.2 g was transferred to a 1.5 mL centrifuge tube, mixed with 0.2 mL of prechilled 0.05 M sodium phosphate buffer (pH 6.0), and centrifuged at 14,000×
g for 20 min at room temperature; the supernatant was used for enzyme activity assays [
39].
2.6.1. Chitinase Activity Assay
Total chitinase activity was measured using colloidal chitin as a substrate, which was prepared as described [
40] with some modifications. Briefly, 10 g of sieved crab shell flakes were thoroughly mixed with 50 mL of 85% phosphoric acid and incubated for 20 h at room temperature. The resulting suspension was filtered under vacuum through a glass porous filter No. 1, and the filtrate was diluted 20 times with distilled water. After vigorous stirring, the precipitated chitin was washed with water by decanting until pH 5.5–6.0 and freeze-dried. The obtained chitin powder (5 g) was suspended in 500 mL of sterile distilled water and used as a substrate to determine chitinolytic activity as previously described [
41] with some modifications. Briefly, 10 µL of colloidal chitin (10 mg/mL) was mixed with 10 µL of 0.05 M sodium-phosphate buffer (pH 6.0) and 10 µL of root extract supernatant (1 mg/mL), and incubated for 60 min at 50 °C. The reaction was terminated by heating in a boiling water bath for 5 min; then, the mixture was centrifuged at 5000×
g for 5 min at room temperature, and 10 µL of the supernatant was mixed with 90 µL of sterile water and 150 μL of DNS and incubated in boiling water for 20 min. After chilling, the mixture was centrifuged at 5000× g for 5 min at room temperature, and the supernatant was used for analysis of reducing sugar content calculated according to the OD at 540 nm. One unit of chitinase activity was defined as the amount of enzyme required to release 1 µg of reducing sugar per 1 min per 1 mg of raw tissue.
2.6.2. β-1,3-Glucanase Activity Assay
Total β-1,3-glucanase activity was determined with laminarin and
D-glucose as the substrate and standard, respectively, as described previously [
42] with some modifications. Briefly, 5 µL of root extract supernatant (1 mg/mL) was mixed with 5 µL of 0.05 M sodium phosphate buffer (pH 6.0) and 10 µL of laminarin (2 mg/mL), and incubated for 60 min at 50 °C. The reaction was terminated by heating in boiling water for 5 min; the mixture was then centrifuged at 5000× g for 5 min at the room temperature, and 10 µL was mixed with 90 µL of sterile water and 150 μL of DNS, before incubating in boiling water for 20 min. After chilling, the mixture was centrifuged at 5000× g for 5 min at room temperature, and the supernatant was analyzed for reducing sugar content calculated according to the OD at 540 nm. One unit of β-glucanase activity was defined as the amount of enzyme required to release 1 µg of reducing sugar per 1 min per 1 mg of raw tissue.
3. Results
3.1. Chitosan Hydrolysates and Their Effect on the Growth of F. proliferatum
The characteristics of Ch1 and Ch2 samples with low and medium MW, respectively, are shown in
Table 1.
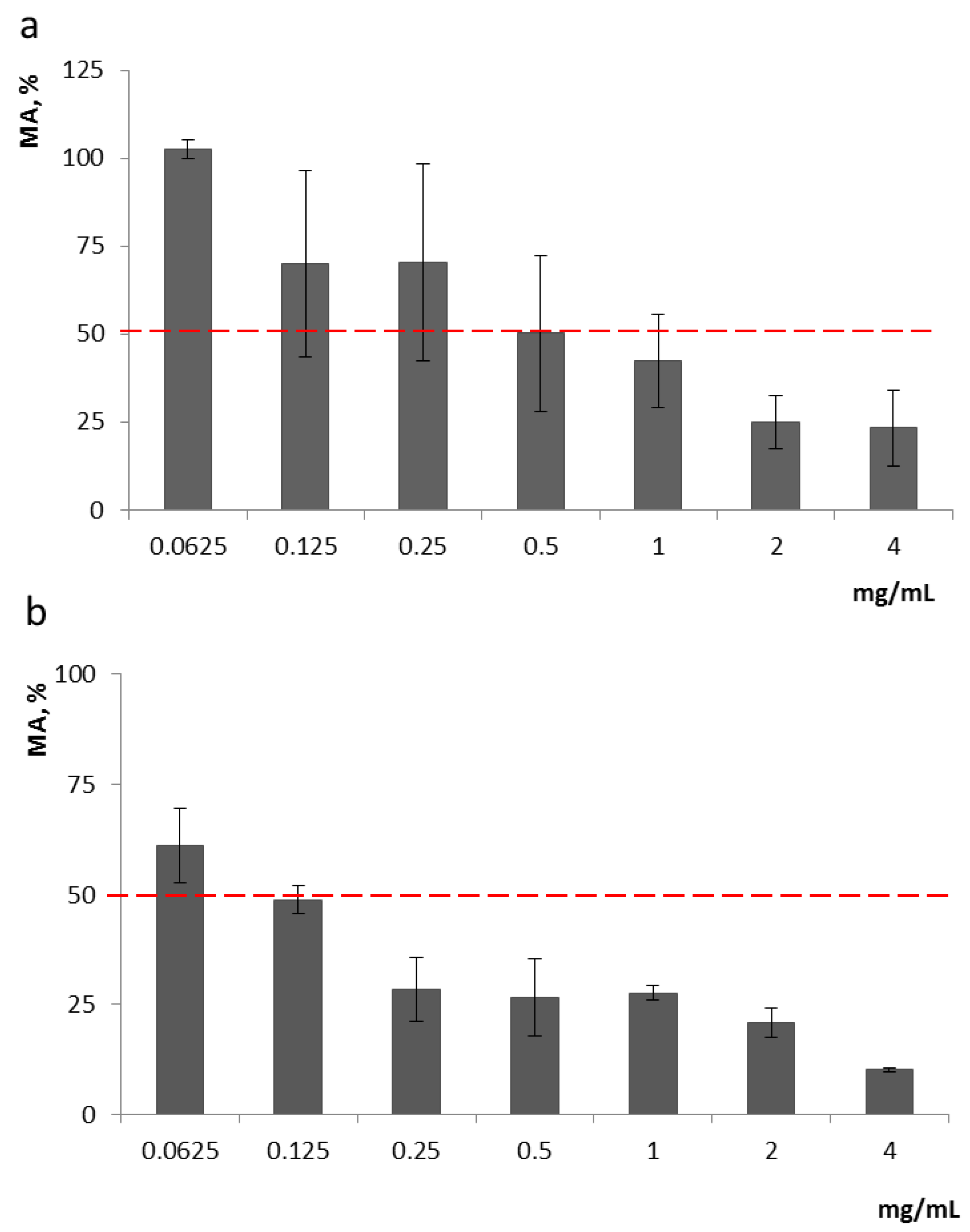
Analysis of Ch effects on the growth of
F. proliferatum revealed that Ch1 and Ch2 started to cause at least 50% inhibition of
F. proliferatum MA (metabolic activity/cell viability) at 0.5 mg/mL and 0.125 mg/mL, respectively, and they were also effective at lower concentrations (0.125–0.250 mg/mL and 0.0625–0.125 mg/mL, respectively), inhibiting fungal growth by 30% (
Figure 1).
Next, we analyzed the radial growth rate of
F. proliferatum on the medium supplemented with various concentrations of Ch1 and Ch2 (
Figure 2). During radial growth on control PDA,
F. proliferatum mycelium acquired a deep purple color, and the pigment was released into medium, whereas on Ch1- or Ch2-containing media, pigmentation was weak and observed only in the central disc colonies, which were initially grown on Ch-free PDA (
Figure 2).
On days 3 and 7, the diameters of fungal growth areas on plates with Ch1 and Ch2 were 1.5–4.6 cm and 1.9–8.3 cm, respectively, which corresponded to growth inhibition indices of 92–59% and 80–13%, respectively (
Table 2). The data indicated that Ch1 at concentrations 1–6 mg/mL could more effectively suppress the growth of
F. proliferatum than Ch2. However, the inhibitory effect of Ch2 at 4–6 mg/mL was longer lasting than that of Ch1 and could be seen on day 14 of
F. proliferatum culture.
3.2. Analysis of Chitosan Pretreatment Effects on Garlic Infected with F. proliferatum
According to the growth inhibition results, the Ch concentration of 2 mg/mL, which produced a moderate antifungal effect, was used in further experiments.
Cloves pretreated with Ch1 and Ch2 and respective control solutions were infected or not with
F. proliferatum, and the developed roots were analyzed for enzymatic activity and gene expression. The results indicated that control solutions (S1 and S2) and Ch1 and Ch2 inhibited total chitinase and β-1,3-glucanase activities compared with water-treated control at 144 h post soaking (hps) (
Figure 3a,b). The inhibition caused by Ch1/2 was approximately two times stronger compared to control solutions, except for β-1,3-glucanase activity in Ch1 and S1 samples (
Figure 3a). At 168 hps, chitinase and β-1,3-glucanase activities were recovered to the control (W) level (except for Ch2-treated samples), whereas, at 240 hps, the enzymatic activities decreased in S1- and Ch1-treated roots to about 50% of control (
Figure 3a,b).
In our previous study [
5,
6], we observed differential expression of genes encoding endo-β-1,3-glucanases (AsPR2a and AsPR2b) and chitinases of class I (AsCHI33 and AsCHI1), class II (AsCHI34), and IV (AsCHI1) in garlic bulbs infected with
Fusarium. Here, we analyzed the expression of these genes in the roots. At 144 hps with Ch1, the expression of AsPR2b, AsCHI33, and AsCHI28 genes was downregulated, whereas that of AsPR2a, AsCHI1, and AsCHI34 was not affected. Treatment with Ch2 significantly upregulated the expression of AsPR2a, AsPR2b, AsCHI33, and AsCHI28, downregulated that of AsCHI34, and did not affect that of AsCHI1 (
Figure 3c–h). At 168 hps, the transcription of AsPR2a was downregulated by Ch2, whereas that of AsPR2b, AsCHI33, and AsCHI28 was upregulated by both Ch1 and Ch2, and that of AsCHI34 was upregulated by Ch2; the expression of AsCHI1 was unchanged. At 240 hps, Ch2 induced the expression of AsPR2a, AsPR2b, and AsCHI28 and reduced that of AsCHI1, whereas Ch1 induced the expression of AsCHI34 and reduced that of AsCHI28. The effects of respective control solutions on gene expression compared with Chs were observed for AsPR2a, AsCHI1, and AsCHI34 (S2) and AsPR2a, AsCHI28, and AsCHI34 (S1), indicating the complex influence of chitosan and control solutions on the enzymatic activity and gene expression patterns (
Figure 3c–h).
Thus, treatment with Ch1/S1 significantly inhibited both β-1,3-glucanase and chitinase activities at 144 and 240 hps, whereas that with Ch2/S2 inhibited chitinase activity at 144, 168, and 240 hps and β-1,3-glucanase activity at 144 and 168 hps (
Figure 3a,b). At the same time, the expression of individual chitinase and endo-β-1,3-glucanase genes was differentially regulated by Ch1/S1 and Ch2/S2 (
Figure 3c–h).
After inoculation with
F. proliferatum, disease symptoms were observed only at 96 hpi (240 hps) on the surface of cloves pretreated with water (dry brown necrotic and/or water-soaked spots) and S1/2 (weaker signs), whereas pretreatment with Ch1/2 prevented the appearance of symptoms.
Figure 4a,b shows enzymatic activities at 24 and 96 hpi (168 and 240 hps) in the roots of cloves infected with
F. proliferatum. At 24 hpi, chitinase and β-1,3-glucanase activities were significantly decreased in S1-, Ch1-, and Ch2-pretreated samples and increased in S2-pretreated samples. At 96 hpi, the activity of chitinase significantly increased in S2, Ch1, and Ch2 roots and that of β-1,3-glucanase decreased in S1 and S2 roots, whereas no changes were observed in Ch1 and Ch2 samples compared to water-treated control.
Analysis of gene expression indicated that, at 24 hpi, AsPR2a, AsCHI33, AsCHI34, and AsCHI28 were upregulated and AsPR2b was downregulated in Ch1-treated roots, whereas AsPR2a, AsPR2b, AsCHI33, and AsCHI28 were upregulated and AsCHI34 was downregulated in Ch2-treated roots. At 96 hpi, all genes (except AsCHI1) were downregulated in Ch1-treated samples, whereas AsPR2a was upregulated and AsCHI34 was downregulated in Ch2-treated samples. At 24 and/or 96 hpi, the expression of several genes was increased in the roots treated with S1 (AsPR2a, AsCHI33, and AsCHI28) and S2 (AsPR2b, AsCHI33, AsCHI34, and AsCHI28), whereas that of the others (AsCHI34 and AsCHI28 in S1 samples and AsPR2a and AsPR2b in S2 samples) was decreased.
Thus, we observed an initial inhibitory and then stimulatory effect of Ch1 and Ch2 pretreatments on the activity of chitinases and β-1,3-glucanases in F. proliferatum-infected garlic, which was probably due to the transcriptional regulation of chitinase and β-1,3-glucanase genes by chitosan.
3.3. Analysis of Chitosan Post-Treatment Effects on Garlic Infected with F. proliferatum
Next, sterilized garlic cloves with initiated roots were inoculated or not with F. proliferatum and soaked in water, S1, S2, Ch1, or Ch2. Disease symptoms (dry brown necrotic and/or water-soaked spots) were observed at 72 hps on the surface of all infected cloves, regardless of the treatment type. The roots were analyzed for enzymatic activity and gene expression at 24 and 72 hps.
In uninfected S1, S2, Ch1, and Ch2 samples, chitinase and β-1,3-glucanase activity profiles were overall similar at 24 and 72 hps (
Figure 5a,b). Analysis of gene expression revealed that, at 24 hps, the transcription of AsPR2a, AsPR2b, and AsCHI28 was upregulated, whereas that of AsCHI34 and AsCHI1 was downregulated by Ch1 and Ch2, and that of AsCHI33 was upregulated by Ch1 and not affected by Ch2 (
Figure 5c–h). At 72 hps, the transcription of AsPR2a, AsCHI28, AsCHI33, and AsCHI1 was upregulated and that of AsPR2b and AsCHI34 was unchanged by Ch1, whereas the transcription of AsPR2a and AsPR2b was downregulated and that of AsCHI28, AsCHI33, AsCHI1, and AsCHI34 was upregulated by Ch2. The treatment with S1 upregulated AsPR2a at both timepoints, AsPR2b at 24 hps, and AsCHI28 and AsCHI33 at 72 hps. S2 upregulated AsPR2a at both timepoints, AsPR2b, AsCHI33, and AsCHI28 at 24 hps, and AsCHI1 and AsCHI34 at 72 hps, while it downregulated AsPR2b at 72 hps and AsCHI1 and AsCHI34 at 24 hps (
Figure 5c–h).
Thus, at 24 hps, Ch1 and Ch2 exerted only slight effects on the chitinase and β-1,3-glucanase activities, whereas, at 72 hps, Ch1 induced and Ch2 reduced their activity. The most pronounced effects on gene expression were observed for Ch2, which upregulated AsCHI33, AsCHI1, and AsCHI34, and for S1 and S2, which upregulated AsPR2a and AsPR2b.
Figure 6a,b show the enzymatic activities at 24 and 72 hps in garlic roots infected with
F. proliferatum and then treated with water, S1, S2, Ch1, or Ch2. In Ch1- and Ch2-treated roots, the activity of both chitinase and β-1,3-glucanase was significantly increased at 24 hps; at 72 hps, the former was decreased, whereas the latter was either unchanged (Ch1) or increased (Ch2) compared to control (water). S1 stimulated the activity of β-1,3-glucanase and inhibited that of chitinase at 72 hps.
The mRNA expression of chitinase and β-1,3-glucanase genes at 24 and 72 hps is shown in
Figure 6c–h. At 24 hps, the transcription of all the genes (except
AsCHI28) was downregulated in Ch1-treated roots, whereas that of
AsPR2a,
AsPR2b,
AsCHI33, and
AsCHI28 was upregulated and that of
AsCHI1 and
AsCHI34 was downregulated in Ch2-treated roots compared to control (water treatment). At 72 hps, the transcription of
AsCHI1 and
AsCHI28 was upregulated and that of
AsPR2a was downregulated in Ch1-treated samples, whereas the transcription of
AsPR2a and
AsPR2b was downregulated in Ch2-treated samples. The upregulation of gene expression was observed in S1-treated roots (
AsCHI34 at 72 hps and
AsCHI1 at 24 and 72 hps) and S2-treated roots (
AsPR2b,
AsCHI33, and
AsCHI28 at 24 and 72 hps;
AsCHI34 and
AsCHI1 at 72 hps). Transcriptional downregulation was observed for
AsCHI34 at 24 hps and for
AsPR2a,
AsPR2b,
AsCHI33, and
AsCHI28 at both timepoints in S1-treated samples, while it was observed for
AsPR2a at both timepoints in S2-treated samples (
Figure 6c–h).
Thus, both Ch1 and Ch2 initially caused the induction of chitinase and β-1,3-glucanase activities in F. proliferatum-infected roots, which was sustained at the later stage only for β-1,3-glucanase activity in Ch2-treated roots.
4. Discussion
Chitosan is considered to be one of the key signaling molecules in plant cells and, as such, is used as an elicitor of plant defense responses. It has been established that the effects of chitosan depend on its structural characteristics and concentrations used for seed/plant treatment [
17]. In plants, both fungal infection and external chitosan application cause receptor-mediated activation of genes associated with defense response, such as those encoding endo-β-1,3-glucanases and chitinases of the GH19 family (classes I, II, IV, VI, and VII) [
43,
44]. These enzymes can directly hydrolyze fungal cell-wall polysaccharides, thus promoting cell lysis, and/or activate plant immune response through generation of oligosaccharide elicitors from chitin/chitosan and β-1,3-/β-1,6-glucans, which are recognized by plants as molecular signals for the induction of downstream defense-related pathways [
43,
44,
45,
46,
47]. Such versatile antifungal activity makes chitinases and endo-β-1,3-glucanases promising targets in agricultural plant breeding programs, such as those aimed at prevention of pre- and post-harvest crop losses of garlic (
A. sativum L.) due to
Fusarium infection [
48].
In our previous studies of PR genes in FBR-resistant and -susceptible garlic cultivars, we identified and characterized seven genes encoding class I chitinases, (
AsCHI1–7) and three genes encoding endo-β-1,3-glucanases (
AsPR2a–c) [
5,
6]. Other studies of plant response to
F. proliferatum infection have suggested that
AsCHI2 (syn.
AsCHI28),
AsCHI3,
AsCHI5,
AsCHI7 (syn.
AsCHI33),
AsPR2a,
AsPR2b, and
AsPR2c (accession numbers provided in [
5,
6,
36]) may define the protection of garlic against
Fusarium attacks [
5,
6].
Here, we investigated the antifungal activity of chitosan by analyzing its effects on
F. proliferatum growth and ability to infect garlic bulbs, as well as on garlic immune responses. It is known that the functional activity of chitosan is directly related to its MW, which can be low (<100 kDa), medium (100–1000 kDa), or high (>1000 kDa) [
49]. A previous study showed that chitosan of low (50–190 kDa) and medium (190–310 kDa) MW exerted the strongest protective effect on durum wheat seedlings under oxidative stress [
50]. Therefore, we prepared and analyzed chitosan hydrolysates of low (39 kDa) and medium (135 kDa) MW (
Table 1). Since only
F. proliferatum was used, the results obtained in our study can be used to interpret the interaction of this fungal species, first of all, with garlic plants during growth and/or storage.
The
Fusarium spp. growth is accompanied by the production of secondary metabolites defining yellow-to-red (carotenoids and polyketides) and black-violet (perithecial melanin) pigmentation [
51,
52]. Some of the pigments are mycotoxins (dark-red aurofusarin or orange-brown rubrofusarin) or participate in mycotoxin synthesis in
Fusarium [
51] and, consequently, are directly associated with
Fusarium virulence and pathogenicity or stress resistance mechanisms; thus, spore melanins as oxidizing agents provide protection against ionizing radiation [
53]. In view of these data, the suppression of
F. proliferatum growth and reduction in mycelium pigmentation by Ch1 and Ch2 observed in this study (
Figure 2) suggest that Chs inhibited fungal development and reduced virulence.
We analyzed the practical significance of Ch1/Ch2 antifungal effects in the pretreatment and post-treatment modes, by soaking garlic cloves in chitosan solutions (2 mg/mL) prior to or after
F. proliferatum infection. The concentration was chosen on the basis of moderate antifungal effects (
Figure 1 and
Figure 2,
Table 2) and a previous finding that medium MW chitosan at 2.5 mg/mL enhances the growth of cucumber seedlings [
54]. The protective effect, i.e., the absence of disease signs on the cloves, was observed only in the pretreatment mode, indicating that Ch1/Ch2 could prevent
Fusarium infection through mobilization of plant defense mechanisms rather than by inhibiting the growth of fungi through direct damage of fungal cells. We suggest an analogy between chitosan pretreatment in this study and inoculation of plants with a nonpathogenic fungal strain prior to infection in a previous study [
55]. Pathogenic and nonpathogenic
F. oxysporum strains cause hyper- and hypomethylation, respectively, of endo-β-1,3-glucanase and chitinase genes in flax (
Linum usitatissimum L.), and it is suggested that pretreatment with a nonpathogenic strain makes plants memorize the hypomethylation pattern and then respond more effectively to the attack of the pathogen [
55]. It remains to be determined whether Ch, similar to elicitors provided by nonpathogenic fungi, may cause genome-wide hypomethylation of endo-β-1,3-glucanase and chitinase genes, making garlic reaction to
F. proliferatum more effective.
Given that nitrogen, which is critical for plant growth, is absorbed mainly in the form of ammonium and nitrate compounds [
56], it was not surprising that control solutions (S1 and S2) containing ammonium nitrate affected chitinase and glucanase enzymatic activity and gene transcription, enhancing or weakening Ch1- and Ch2-elicited responses (
Figure 3,
Figure 4,
Figure 5 and
Figure 6). Such combined effects were taken into account when making conclusions about the stimulatory or inhibitory role of the used chitosan. Our data on chitinase and glucanase activities in infected garlic roots showed early inhibitory and late stimulatory effects of Ch1/Ch2 pretreatment, early stimulatory effects of Ch1/Ch2 post-treatment, and late inhibitory or stimulatory effects of post-treatment with Ch1 or Ch2, respectively (
Figure 4 and
Figure 6). Individual gene expression data did not show apparent correlation with chitosan exposure (
Figure 3,
Figure 4,
Figure 5 and
Figure 6). However, it can be deduced that the stimulatory effect of Ch2 on gene expression was stronger than that of Ch1 in the pretreatment mode, whereas the opposite trend was observed in the post-treatment mode (
Figure 4 and
Figure 6). These results suggest that pretreatment with medium MW Ch2 can be more effective to prevent fungal infection than that with low MW Ch1.