Foliar Spraying of Salicylic Acid Enhances Growth, Yield, and Curcuminoid Biosynthesis Gene Expression as Well as Curcuminoid Accumulation in Curcuma longa
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Growth Conditions
2.2. Chemical Analysis
2.2.1. Measurement of Photosynthetic Pigments
2.2.2. Mineral Composition
2.3. GC/MS Analysis
2.4. Determination of Curcumin, Bisdemethoxycurcumin, and Demethoxycurcumin Contents by High-Performance Liquid Chromatography (HPLC)
2.4.1. Instrumentation
2.4.2. Preparation of Curcumin, Bisdemethoxycurcumin, and Demethoxycurcumin Standard Curves
2.4.3. Preparation of C. longa Rhizome Methanolic Extracts
2.4.4. HPLC Analysis Conditions
2.5. Analysis of CURS1, -2, and -3 and DCS Gene Expression by Real-Time Reverse Transcriptase Polymerase Chain Reaction (Real-Time RT-PCR)
2.5.1. Total RNA Preparation and cDNA Synthesis
2.5.2. Real Time RT-PCR and Curcuminoid Gene Expression Analysis
2.6. Statistical Analysis
3. Results
3.1. Effects of SA on Plant Development and Yield
3.2. Photosynthetic Pigments and Mineral Contents
3.3. Gas Chromatography–Mass Spectrometry (GC–MS) Analysis of Ethanolic Extracts
3.4. HPLC Results
3.5. Effect of SA on the Expression of Curcuminoid Biosynthesis Genes
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Setzer, W.N.; Duong, L.; Poudel, A.; Mentreddy, S.R. Variation in the chemical composition of five varieties of Curcuma longa rhizome essential oils cultivated in north alabama. Foods 2021, 10, 212. [Google Scholar] [CrossRef] [PubMed]
- Lakshmi, S.; Padmaja, G.; Remani, P. Antitumour Effects of Isocurcumenol Isolated from Curcuma zedoaria Rhizomes on Human and Murine Cancer Cells. Int. J. Med. Chem. 2011, 2011, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Itokawa, H.; Shi, Q.; Akiyama, T.; Morris-Natschke, S.L.; Lee, K.H. Recent advances in the investigation of curcuminoids. Chin. Med. 2008, 3, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prasad, S.; Tyagi, A.K.; Aggarwal, B.B. Recent developments in delivery, bioavailability, absorption and metabolism of curcumin: The golden pigment from golden spice. Cancer Res. Treat. 2014, 46, 2–18. [Google Scholar] [CrossRef] [Green Version]
- de Oliveira Filho, J.G.; de Almeida, M.J.; Sousa, T.L.; dos Santos, D.C.; Egea, M.B. Bioactive Compounds of Turmeric (Curcuma longa L.). Ref. Ser. Phytochem. 2021, 297–318. [Google Scholar]
- Urošević, M.; Nikolić, L.; Gajić, I.; Nikolić, V.; Dinić, A.; Miljković, V. Curcumin: Biological Activities and Modern Pharmaceutical Forms. Antibiotics 2022, 11, 135. [Google Scholar] [CrossRef]
- Oglah, M.K.; Mustafa, Y.F.; Bashir, M.K.; Jasim, M.H. Curcumin and its derivatives: A review of their biological activities. Syst. Rev. Pharm. 2020, 11, 472–481. [Google Scholar]
- Katsuyama, Y.; Kita, T.; Funa, N.; Horinouchi, S. Curcuminoid biosynthesis by two type III polyketide synthases in the herb Curcuma longa. J. Biol. Chem. 2009, 284, 11160–11170. [Google Scholar] [CrossRef] [Green Version]
- Katsuyama, Y.; Kita, T.; Horinouchi, S. Identification and characterization of multiple curcumin synthases from the herb Curcuma longa. FEBS Lett. 2009, 583, 2799–2803. [Google Scholar] [CrossRef] [Green Version]
- Sandeep, I.S.; Das, S.; Nasim, N.; Mishra, A.; Acharya, L.; Joshi, R.K.; Nayak, S.; Mohanty, S. Differential expression of CURS gene during various growth stages, climatic condition and soil nutrients in turmeric (Curcuma longa): Towards site specific cultivation for high curcumin yield. Plant Physiol. Biochem. 2017, 118, 348–355. [Google Scholar] [CrossRef]
- Ayer, D.K.; Modha, K.; Parekh, V.; Patel, R.; Vadodariya, G.; Ramtekey, V.; Bhuriya, A. Associating gene expressions with curcuminoid biosynthesis in turmeric. J. Genet. Eng. Biotechnol. 2020, 18, 83. [Google Scholar] [CrossRef]
- Hayat, Q.; Hayat, S.; Alyemeni, M.N.; Ahmad, A. Salicylic acid mediated changes in growth, photosynthesis, nitrogen metabolism and antioxidant defense system in Cicer arietinum L. Plant Soil Environ. 2012, 58, 417–423. [Google Scholar] [CrossRef] [Green Version]
- Pedroso, R.C.N.; Branquinho, N.A.D.Á.; Hara, A.C.B.D.A.M.; Silva, F.G.; Kellner Filho, L.C.; Silva, M.L.A.; Cunha, W.R.; Pauletti, P.M.; Januario, A.H. Effect of salicylic acid and silver nitrate on rutin production by hyptis marrubioides cultured in vitro. Cienc. Rural. 2019, 49. [Google Scholar] [CrossRef] [Green Version]
- Pedro, H.G.; Ana, C.A.U.P. Growth promotion and elicitor activity of salicylic acid in Achillea millefolium L. Afr. J. Biotechnol. 2016, 15, 657–665. [Google Scholar] [CrossRef] [Green Version]
- Hashemi, S.M.; Naghavi, M.R.; Ghorbani, M.; Priyanatha, C.; Zandi, P. Effects of abiotic elicitors on expression and accumulation of three candidate benzophenanthridine alkaloids in cultured greater celandine cells. Molecules 2021, 26, 1395. [Google Scholar] [CrossRef] [PubMed]
- Kandoudi, W.; Radácsi, P.; Gosztola, B.; Németh, É.Z. Elicitation of medicinal plants in vivo—Is it a realistic tool? The effect of methyl jasmonate and salicylic acid on lamiaceae species. Horticulturae 2022, 8, 5. [Google Scholar] [CrossRef]
- Lan, T.T.P.; Huy, N.D.; Luong, N.N.; Quang, H.T.; Tan, T.H.; Thu, L.T.A.; Loc, N.H. Effect of salicylic acid and yeast extract on curcuminoids biosynthesis gene expression and curcumin accumulation in cells of Curcuma zedoaria. J. Plant Biotechnol. 2019, 46, 172–179. [Google Scholar] [CrossRef] [Green Version]
- Maruri-López, I.; Aviles-Baltazar, N.Y.; Buchala, A.; Serrano, M. Intra and extracellular journey of the phytohormone salicylic acid. Front. Plant Sci. 2019, 10, 423. [Google Scholar] [CrossRef]
- Elwan, M.W.M.; El-Hamahmy, M.A.M. Improved productivity and quality associated with salicylic acid application in greenhouse pepper. Sci. Hortic. 2009, 122, 521–526. [Google Scholar] [CrossRef]
- Alyemeni, M.N.; Hayat, Q.; Wijaya, L.; Hayat, S. Effect of salicylic acid on the growth, photosynthetic efficiency and enzyme activities of leguminous plant under cadmium stress. Not. Bot. Hortic. Agrobot. 2014, 42, 440–445. [Google Scholar] [CrossRef] [Green Version]
- Association of Official Analytical Chemists. Official Method of Analysis of the Association of Officail Chemist. In Official Methods of Analysis, 14th ed.; Howitz, Ed.; Association of Official Analytical Chemists: Washington, DC, USA, 1984. [Google Scholar]
- Piper, C.S. Soil and Plant Analysis; Hans Publishers: Delhi, India, 1942. [Google Scholar]
- Jackson, M.L. Soil Chemica Analysis; Prentice Hall: New Delhi, India, 1967. [Google Scholar]
- Murphy, J.; Riley, J.P. Modified single-solution method for the determination of phosphorus in natural water. Anal. Chim. Acta 1962, 27, 31–36. [Google Scholar] [CrossRef]
- Mazumdar, B.; Majumder, K.; Mazumdar, B.C.; Majumder, K. Methods of Physiochemical Analysis of Fruits; Daya Publications: Delhi, India, 2003. [Google Scholar]
- Page, A.L. Methods of Soil Analysis—Part 2: Chemical and Microbiological Properties, 2nd ed.; American Society of Agronomy, Inc.; Soil Science Society of America, Inc.: Madison, WI, USA, 1982; Volume 9, pp. 421–422. [Google Scholar]
- El Sherif, F.; Albotnoor, N.; Yap, Y.K.; Meligy, A.; Khattab, S. Enhanced bioactive compounds composition in Lavandula officinalis in-vitro plantlets using NaCl and Moringa oleifera, Aloe vera and Spirulina platensis extracts. Ind. Crops Prod. 2020, 157, 112890. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- StatSoft STATISTICA for Windows, Version 6; StatSoft Inc.: Tulsa, OK, USA, 2001.
- Bagautdinova, Z.Z.; Omelyanchuk, N.; Tyapkin, A.V.; Kovrizhnykh, V.V.; Lavrekha, V.V.; Zemlyanskaya, E.V. Salicylic Acid in Root Growth and Development. Int. J. Mol. Sci. 2022, 23, 2228. [Google Scholar] [CrossRef]
- Youssef, S.M.; López-Orenes, A.; Ferrer, M.A.; Calderón, A.A. Salicylic-Acid-Regulated Antioxidant Capacity Contributes to Growth Improvement of Okra (Abelmoschus esculentus cv. Red Balady). Agronomy 2022, 12, 168. [Google Scholar] [CrossRef]
- Mohammad, F.; Wajid, M.A.; Bhat, M.A. Effect of Salicylic Acid Sprays on the Performance of Fenugreek Grown with Graded Levels of Salinity. Haya Saudi J. Life Sci. 2019, 4, 346–354. [Google Scholar] [CrossRef]
- Gonçalves, F.C.D.M.; Parreiras, N.D.S.; Campos, F.G.; Mantoan, L.P.B.; Boaro, C.S.F. Exogenous salicylic acid modifies gas exchange and biomass production of Mentha x piperita L. Aust. J. Crop Sci. 2020, 14, 98–107. [Google Scholar] [CrossRef]
- Dehnavi, A.R.; Zahedi, M.; Ludwiczak, A.; Piernik, A. Foliar Application of Salicylic Acid Improves Salt Tolerance of Sorghum (Sorghum bicolor (L.) Moench). Plants 2022, 11, 368. [Google Scholar] [CrossRef]
- Liu, R.; Bao, Z.X.; Zhao, P.J.; Li, G.H. Advances in the study of metabolomics and metabolites in some species interactions. Molecules 2021, 26, 3311. [Google Scholar] [CrossRef]
- Bekinbo, M.; Tariah, F.S.A.; Dapper, D. Comparative GC-MS determination of bioactive constituents of the methanolic extracts of Curcuma longa rhizome and Spondias mombin leaves Bekinbo MT, Amah-Tariah FS and Dapper DV. J. Med. Plants Stud. 2020, 8, 1–6. Available online: http://www.plantsjournal.com (accessed on 22 April 2022).
- Arivoli, S.; Tennyson, S.; Divya, S.; Rani, S.; Marin, G. GC-MS analysis of bioactive compounds of Curcuma longa Linnaeus (Zingiberaceae) rhizome extract. J. Pharmacogn. Phytochem. 2019, 8, 49–52. Available online: http://www.phytojournal.com (accessed on 22 April 2022).
- Kulyal, P.; Acharya, S.; Ankari, A.B.; Kokkiripati, P.K.; Tetali, S.D.; Raghavendra, A.S. Variable Secondary Metabolite Profiles Across Cultivars of Curcuma longa L. and C. aromatica Salisb. Front. Pharmacol. 2021, 12, 659546. [Google Scholar] [CrossRef] [PubMed]
- Amalraj, A.; Pius, A.; Gopi, S.; Gopi, S. Biological activities of curcuminoids, other biomolecules from turmeric and their derivatives—A review. J. Tradit. Complement. Med. 2017, 7, 205–233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, W.T.; Arasu, M.V.; Al-Dhabi, N.A.; Yeo, S.K.; Jeon, J.; Park, J.S.; Lee, S.Y.; Park, S.U. Yeast extract and silver nitrate induce the expression of phenylpropanoid biosynthetic genes and induce the accumulation of rosmarinic acid in agastache rugosa cell culture. Molecules 2016, 21, 426. [Google Scholar] [CrossRef]
- Yap, Y.K.; El-Sherif, F.; Habib, E.S.; Khattab, S. Moringa oleifera leaf extract enhanced growth, yield and silybin content while mitigating salt-induced adverse effects on the growth of silybum marianum. Agronomy 2021, 11, 2500. [Google Scholar] [CrossRef]
| Characteristic | Value |
|---|---|
| Texture | Sandy |
| Sand % | 91.51 |
| Silt % | 5.74 |
| Clay % | 2.75 |
| Saturation % | 23 |
| pH | 7.5 |
| Electrical conductivity (EC) (dS/m) | 2.2 |
| Organic matter (OM) % | 0.05 |
| Total N % | 0.014 |
| Available P ppm | 3.9 |
| Available K ppm | 110 |
| Salinity Level (ppm) | Cations (meq/L) | Anions (meq/L) | Sodium Adsorption Ratio (SAR) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Ca2+ | Mg2+ | Na+ | K+ | CO32− | HCO3− | SO42− | Cl− | ||
| 864 | 5.72 | 2.02 | 7.27 | 0.38 | 0.28 | 2.68 | 4.03 | 8.4 | 3.43 |
| Gene | Primers Sequence | Amplicon Length (bp) | GenBank Accession Number | References |
|---|---|---|---|---|
| Diketide-CoA synthase (DCS) | 5′-GTGCTGTTCATCCTGGACGAG-3′ (forward primer) | 21 | AB495006.1 | [8] |
| 5′-CAACAGCACGCCCCAGTCGA-3′ (reverse primer) | 20 | |||
| Curcumin synthase 1 (CURS1) | 5′-CATCATTGACGCCATCGAAGC-3′ (forward primer) | 21 | AB495007.1 | [8] |
| 5′-TCAGCTCATCCATCACGAAGTACAC-3′ (reverse primer) | 25 | |||
| Curcumin synthase 2 (CURS2) | 5′-TCGGGATCAAGGACTGGAACAAC-3′ (forward primer) | 23 | AB506762.1 | [8] |
| 5′-TGTTGCCGAACTCGGAGAAGAC-3′ (reverse primer) | 22 | |||
| Curcumin synthase 3 (CURS3) | 5′-TGGAGCCCTCCTTCGACGACC-3′ (forward primer) | 21 | AB506763.1 | [8] |
| 5′-CCCATTCCTTGATCGCCTTTTCC-3′ (reverse primer) | 23 | |||
| Actin | 5′-GGATATGCTCTTCCTCATGCT-3′ (forward primer) | 21 | CP002686.1 AK118354.1 AY087740.1 | [8] |
| 5′-TCTGCTGTGGTGGTGAATGA-3′ (reverse primer) | 20 |
| SA (M) Treatments | Plant Height (cm) | No. of Leaves (n) | No. of Roots (n) | Leaves’ Dry Weight (g) | Roots’ Dry Weight (g) | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Season (2020) | Season (2021) | Season (2020) | Season (2021) | Season (2020) | Season (2021) | Season (2020) | Season (2021) | Season (2020) | Season (2021) | |
| Control | 150.33 b * | 130.25 c | 11.67 ab | 8.75 ab | 29.0 b | 24.75 b | 26.9 b | 23.53 b | 3.33 a | 3.5 a |
| 10−4 | 167.75 a | 139.75 b | 14.25 a | 10.25 a | 33.75 ab | 24.0 b | 52.0 a | 29.15 ab | 3.5 a | 3.67 a |
| 10−3 | 176.25 a | 166.75 a | 10.5 b | 8.33 b | 40.5 a | 30.0 a | 59.33 a | 35.5 a | 2.25 b | 3.0 b |
| SA (M) Treatments | No. of Rhizomes (n) | Rhizome Dry Weight (g) | Rhizome Diameter (mm) | |||
|---|---|---|---|---|---|---|
| Season (2020) | Season (2021) | Season (2020) | Season (2021) | Season (2020) | Season (2021) | |
| Control | 16.67 c * | 10.70 b | 13.6 c | 7.23 b | 16.3 b | 14.47 c |
| 10−4 | 22.75 b | 16.5 a | 39.75 b | 30.0 ab | 18.18 a | 17.81 b |
| 10−3 | 34.33 a | 19.0 a | 78.87 a | 61.23 a | 18.25 a | 20.15 a |
| SA (M) Treatments | Chl a (mg/100 g F.W.) | Chl b (mg/100 g F.W.) | Carotenoids (mg/100 g F.W.) | |||
|---|---|---|---|---|---|---|
| Season (2020) | Season (2021) | Season (2020) | Season (2021) | Season (2020) | Season (2021) | |
| Control | 75.37 b * | 107.59 a | 26.95 b | 39.02 b | 83.58 b | 117.31 b |
| 10−4 | 92.43 ab | 111.74 a | 37.85 a | 39.96 b | 99.69 ab | 122.86 ab |
| 10−3 | 106.24 a | 126.33 a | 35.02 ab | 43.09 a | 118.09 a | 146.12 a |
| SA (M) Treatments | N% | P% | K% | |||
|---|---|---|---|---|---|---|
| Season (2020) | Season (2021) | Season (2020) | Season (2021) | Season (2020) | Season (2021) | |
| Control | 3.102 a * | 1.688 b | 1.34 a | 2.085 b | 1.608 a | 1.36 a |
| 10−4 | 3.295 a | 1.736 b | 1.427 a | 3.82 a | 1.648 a | 1.4 a |
| 10−3 | 3.363 a | 2.067 a | 1.54 a | 3.143 a | 1.708 a | 1.45 a |
| SA Treatment (M) | Peak | Essential Oil Compounds | RT, min | Area, % | Molecular Weight | Molecular Formula |
|---|---|---|---|---|---|---|
| Season (2020) | ||||||
| Control | 1 | Alpha-curcumene | 14.47 | 4.65 | 202.33 | C15H22 |
| 2 | (−)-Zingiberene | 14.64 | 25.03 | 204.35 | C15H24 | |
| 3 | Beta-bisabolene | 14.81 | 3.66 | 204.35 | C15H24 | |
| 4 | (−)-Beta-sesquiphellandrene | 15.02 | 21.54 | 204.35 | C15H24 | |
| 5 | 4-(2,2-Dimethyl-6-methylenecyclohexyl)butanal | 16.12 | 1.5 | 194.3132 | C13H22O | |
| 6 | Ar-turmerone | 16.68 | 4.82 | 216.32 | C15H20O | |
| 7 | Curlone | 16.74 | 21.8 | 218.3346 | C15H22O | |
| 8 | Cyclopentane | 17.13 | 12.23 | 70.13 | C5H10 | |
| 9 | Tricyclo | 17.47 | 2.22 | 204.35 | C15H24 | |
| 10 | Cyclohexane, 1-methyl-2,4-bis(1-methylethenyl)- | 18.06 | 2.55 | 204.35 | C15H24 | |
| 10−4 | 1 | Alpha-curcumene | 14.47 | 4.89 | 202.33 | C15H22 |
| 2 | (−)-Zingiberene | 14.64 | 32.7 | 204.35 | C15H24 | |
| 3 | Beta-bisabolene | 14.81 | 4.37 | 204.35 | C15H24 | |
| 4 | (−)-Beta-sesquiphellandrene | 15.02 | 23.32 | 204.35 | C15H24 | |
| 5 | Caryophyllene | 15.55 | 0.78 | 204.357 | C15H24 | |
| 6 | 4-(2,2-Dimethyl-6-methylenecyclohexyl)butanal | 16.12 | 0.95 | 194.31 | C13H22O | |
| 7 | Ar-turmerone | 16.68 | 4.15 | 216.32 | C15H20O | |
| 8 | Curlone | 16.74 | 17.73 | 218.3346 | C15H22O | |
| 9 | Beta-turmerone | 17.13 | 9.46 | 218.33 | C15H22O | |
| 10 | Cyclohexane, 1-methyl-2,4-bis(1-methylethenyl)- | 18.06 | 1.65 | 204.35 | C15H24 | |
| 10−3 | 1 | Alpha-curcumene | 14.47 | 4.55 | 174.282 | C15H22 |
| 2 | (−)-Zingiberene | 14.64 | 30.86 | 204.35 | C15H24 | |
| 3 | Beta-bisabolene | 14.81 | 4.1 | 204.35 | C15H24 | |
| 4 | Beta-sesquiphellandrene | 15.02 | 22.86 | 204.35 | C15H24 | |
| 5 | Caryophyllene | 15.55 | 0.65 | 204.357 | C15H24 | |
| 6 | Butanal | 16.12 | 1.1 | 74.12 | C4H8O | |
| 7 | Ar-turmerone | 16.68 | 5.13 | 216.32 | C15H20O | |
| 8 | Curlone | 16.74 | 18.92 | 218.33 | C15H22O | |
| 9 | Beta-turmerone | 17.13 | 10.14 | 218.33 | C15H22O | |
| 10 | Cyclohexane | 18.07 | 1.69 | 84.16 | C6H12 | |
| Season (2021) | ||||||
| Control | 1 | Alpha-curcumene | 14.47 | 4.17 | 174.282 | C15H22 |
| 2 | (−)-Zingiberene | 14.64 | 34.58 | 204.35 | C15H24 | |
| 3 | Beta-bisabolene | 14.81 | 4.15 | 204.35 | C15H24 | |
| 4 | Beta-sesquiphellandrene | 15.02 | 24.63 | 204.35 | C15H24 | |
| 5 | Caryophyllene | 15.55 | 9.57 | 204.357 | C15H24 | |
| 6 | 4-(2,2-Dimethyl-6-methylenecyclohexyl)butanal | 16.12 | 1.5 | 194.31 | C13H22O | |
| 7 | Ar-turmerone | 16.68 | 1.82 | 216.32 | C15H20O | |
| 8 | Curlone | 16.74 | 17.98 | 218.33 | C15H22O | |
| 9 | Cyclopentane | 17.13 | 2.91 | 70.13 | C5H10 | |
| 10 | Cyclohexane | 18.07 | 1.76 | 84.16 | C6H12 | |
| 10−4 | 1 | Alpha-curcumene | 14.47 | 4.77 | 174.282 | C15H22 |
| 2 | (−)-Zingiberene | 14.64 | 32.58 | 204.35 | C15H24 | |
| 3 | Beta-bisabolene | 14.81 | 4.25 | 204.35 | C15H24 | |
| 4 | Beta-sesquiphellandrene | 15.02 | 23.32 | 204.35 | C15H24 | |
| 5 | Caryophyllene | 15.55 | 0.83 | 204.357 | C15H24 | |
| 6 | Butanal | 16.124 | 1.02 | 74.12 | C4H8O | |
| 7 | Ar-turmerone | 16.68 | 3.71 | 216.32 | C15H20O | |
| 8 | Curlone | 16.74 | 18.18 | 218.33 | C15H22O | |
| 9 | Cyclopentane | 17.13 | 9.63 | 70.13 | C9H13N | |
| 10 | Cyclohexane | 18.07 | 1.71 | 84.16 | C6H12 | |
| 10−3 | 1 | Alpha-curcumene | 14.47 | 4.51 | 174.282 | C15H22 |
| 2 | (−)-Zingiberene | 14.64 | 32.53 | 204.35 | C15H24 | |
| 3 | Beta-bisabolene | 14.81 | 4.18 | 204.35 | C15H24 | |
| 4 | Beta-sesquiphellandrene | 15.02 | 22.98 | 204.35 | C15H24 | |
| 5 | Cycloheptane | 15.55 | 0.77 | 204.35 | C15H24 | |
| 6 | Butanal | 16.12 | 1.12 | 74.12 | C4H8O | |
| 7 | Ar-turmerone | 16.68 | 4.15 | 216.32 | C15H20O | |
| 8 | Curlone | 16.74 | 18.3 | 218.33 | C15H22O | |
| 9 | Cyclopentane | 17.13 | 9.77 | 70.13 | C9H13N | |
| 10 | Cyclohexane | 18.07 | 1.69 | 84.16 | C6H12 |
| Phytochemical | Composition (Area %) | |||||
|---|---|---|---|---|---|---|
| Control | SA (10−4 M) | SA (10−3 M) | ||||
| Season (2020) | Season (2021) | Season (2020) | Season (2021) | Season (2020) | Season (2021) | |
| Alpha-curcumene | 4.65 c * | 4.17 f | 4.89 a | 4.77 b | 4.55 d | 4.51 e |
| (−)-Zingiberene | 25.03 f | 34.58 a | 32.7 b | 32.58 c | 30.86 e | 32.53 d |
| Beta-sesquiphellandrene | 21.54 d | 24.63 a | 23.32 b | 23.32 b | 22.86 c | 22.98 c |
| Ar-turmerone | 4.82 b | 1.82 e | 4.15 c | 3.71 d | 5.13 a | 4.15 c |
| Curlone | 21.8 a | 17.98 d | 17.73 e | 18.18 c | 18.92 b | 18.3 c |
| Beta-turmerone | ND | ND | 9.46 a | ND | 10.14 a | ND |
| Caryophyllene | ND | 9.57 a | 0.78 b | 0.83 b | 0.65 b | ND |
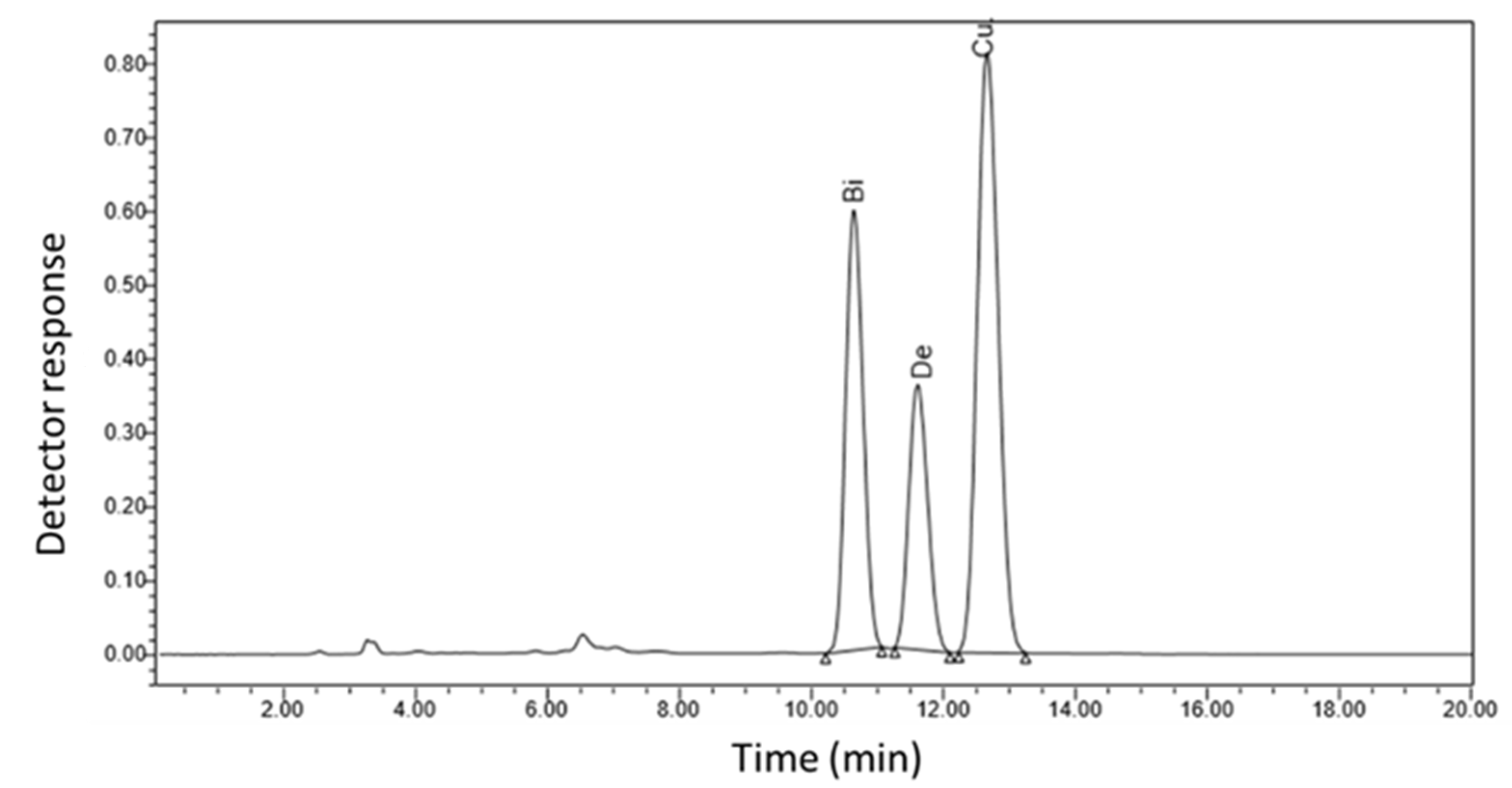
| SA Treatments (M) | Bisdemethoxycurcumin (μg/mL) | Demethoxycurcumin (μg/mL) | Curcumin (μg/mL) |
|---|---|---|---|
| Control | 70.76 c * | 40.29 c | 131.98 b |
| 10−4 | 109.03 b | 92.13 b | 338.31 a |
| 10−3 | 196.13 a | 104.09 a | 338.83 a |
| SA Treatments (M) | DCS | CURS3 | CURS2 | CURS1 |
|---|---|---|---|---|
| Control | 1 a * ± 0.158544 | 1 c ± 0.001449 | 1 c ± 0.063084 | 1 c ± 0.001827 |
| 10−4 | 1.145 a ± 0.047043 | 4.475 b ± 0.034282 | 3.238 b ± 0.22513 | 12.553 a ± 0.141362 |
| 10−3 | 2.129 a ± 0.011599 | 36.005 a ± 0.034282 | 23.412 a ± 0.182553 | 8.696 b ± 0.213558 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
El Sherif, F.; Alkuwayti, M.A.; Khattab, S. Foliar Spraying of Salicylic Acid Enhances Growth, Yield, and Curcuminoid Biosynthesis Gene Expression as Well as Curcuminoid Accumulation in Curcuma longa. Horticulturae 2022, 8, 417. https://doi.org/10.3390/horticulturae8050417
El Sherif F, Alkuwayti MA, Khattab S. Foliar Spraying of Salicylic Acid Enhances Growth, Yield, and Curcuminoid Biosynthesis Gene Expression as Well as Curcuminoid Accumulation in Curcuma longa. Horticulturae. 2022; 8(5):417. https://doi.org/10.3390/horticulturae8050417
Chicago/Turabian StyleEl Sherif, Fadia, Mayyadah Abdullah Alkuwayti, and Salah Khattab. 2022. "Foliar Spraying of Salicylic Acid Enhances Growth, Yield, and Curcuminoid Biosynthesis Gene Expression as Well as Curcuminoid Accumulation in Curcuma longa" Horticulturae 8, no. 5: 417. https://doi.org/10.3390/horticulturae8050417
APA StyleEl Sherif, F., Alkuwayti, M. A., & Khattab, S. (2022). Foliar Spraying of Salicylic Acid Enhances Growth, Yield, and Curcuminoid Biosynthesis Gene Expression as Well as Curcuminoid Accumulation in Curcuma longa. Horticulturae, 8(5), 417. https://doi.org/10.3390/horticulturae8050417





