Genome-Wide Analysis of the Trehalose-6-Phosphate Synthase Gene Family in Rose (Rosa chinensis) and Differential Expression under Heat Stress
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Heat Stress Experiment
2.2. Trehalose Content Detection in R. chinensis
2.3. Identification of Rose TPS Genes
2.4. Multiple Sequence Alignment and Phylogenetic Analysis
2.5. Selective Pressure Analysis
2.6. Protein Physicochemical Properties and Subcellular Localization Analysis
2.7. TPS Gene Structure and Cis-Acting Element Analyses
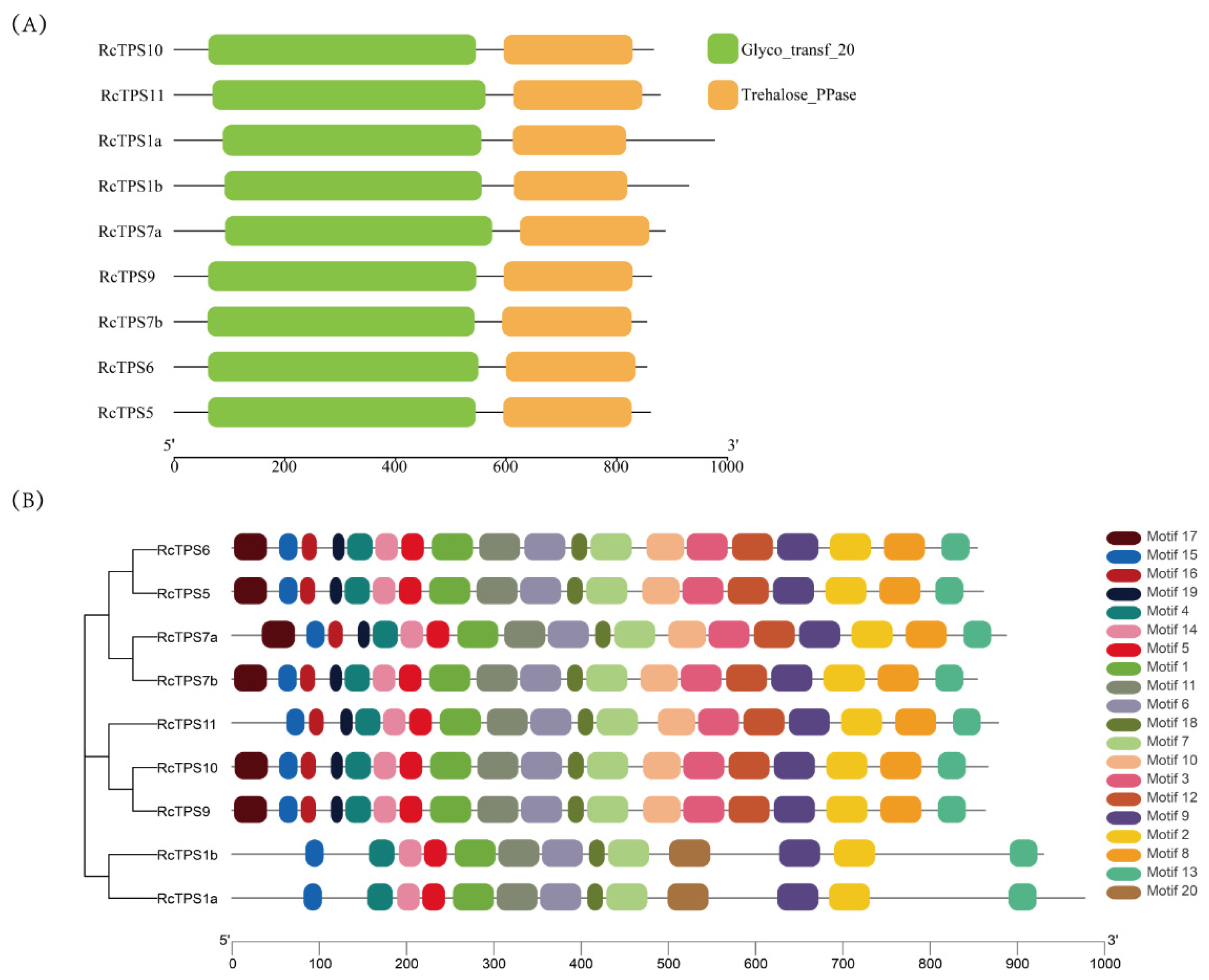
2.8. Protein Motif and Conserved Domain Analysis
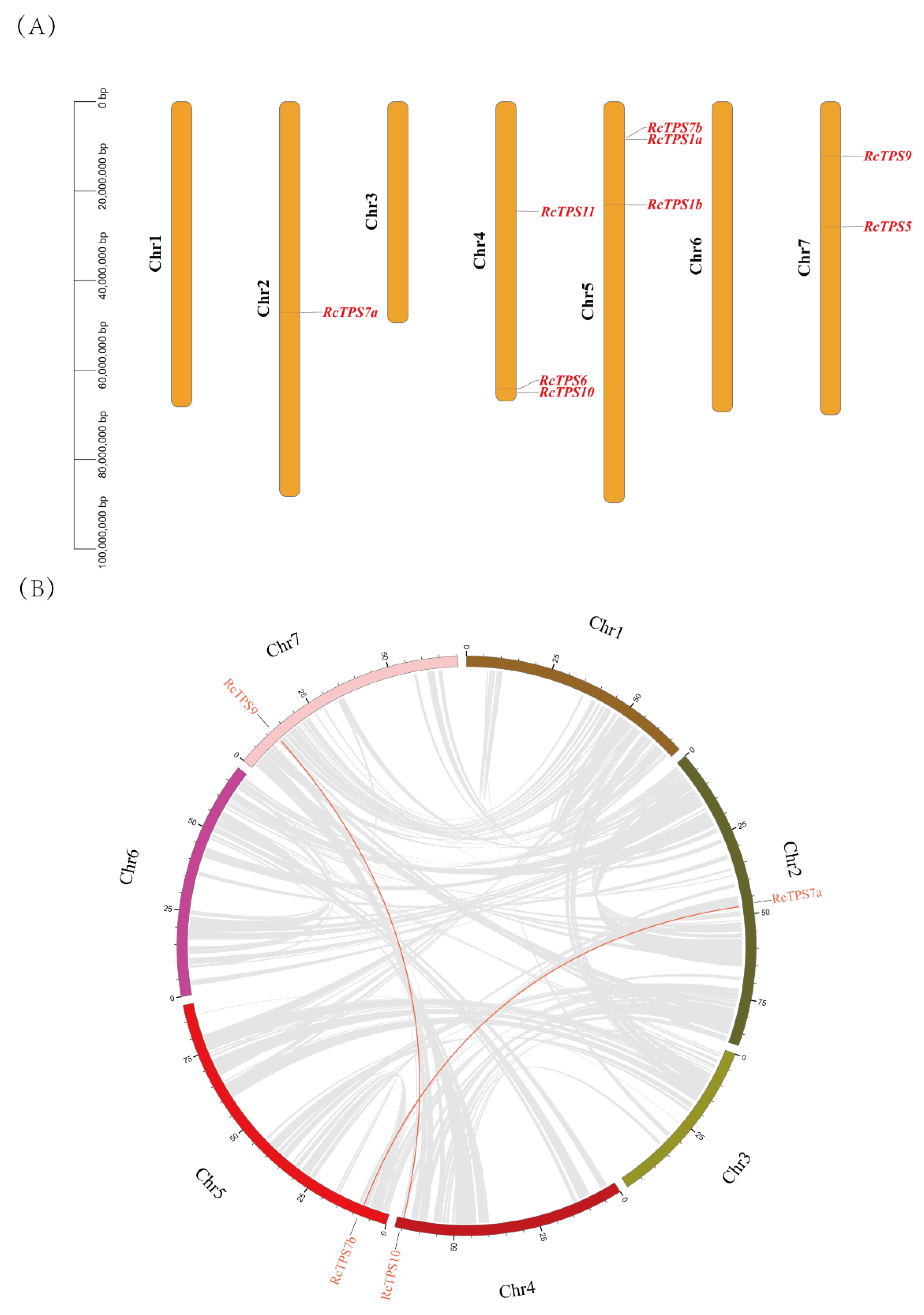
2.9. Gene Chromosomal Location and Collinearity Analysis
2.10. Real-Time Quantitative PCR
3. Results
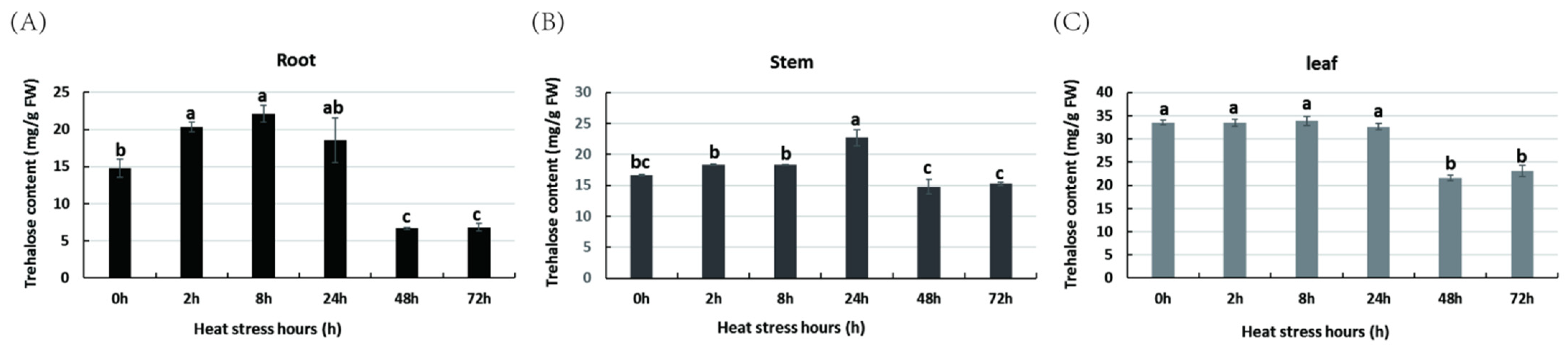
3.1. Dynamics of the Trehalose Levels in the Leaf, Stem, and Root Tissues of R. chinensis under Heat Stress
3.2. Identification and Analysis of the Characteristics of TPS Family Members in R. chinensis
3.3. Gene Structure and cis-Acting Element Analyses of RcTPS Genes
3.4. Protein Domain Analyses and Multiple Sequence Alignment of the TPS Family in R. chinensis
3.5. Evolution Analysis of RcTPS Genes
3.6. Expression Pattern Analysis of RcTPS Genes under Heat Stress
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Li, B.; Gao, K.; Ren, H.; Tang, W. Molecular mechanisms governing plant responses to high temperatures. J. Integr. Plant Biol. 2018, 60, 757–779. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhu, J.; Gong, Z.; Zhu, J. Abiotic stress responses in plants. Nat. Rev. Genet. 2022, 23, 104–119. [Google Scholar] [CrossRef] [PubMed]
- Yatsyshyn, V.Y.; Kvasko, A.Y.; Yemets, A.I. Genetic approaches in research on the role of trehalose in plants. Cytol. Genet. 2017, 51, 371–383. [Google Scholar] [CrossRef]
- Lunn, J.E.; Delorge, I.; Figueroa, C.M.; Van Dijck, P.; Stitt, M. Trehalose metabolism in plants. Plant J. 2014, 79, 544–567. [Google Scholar] [CrossRef] [PubMed]
- Acosta-Perez, P.; Camacho-Zamora, B.D.; Espinoza-Sanchez, E.A.; Gutierrez-Soto, G.; Zavala-Garcia, F.; Jazmin Abraham-Juarez, M.; Ramona Sinagawa-Garcia, S. Characterization of trehalose-6-phosphate synthase and trehalose-6-phosphate phosphatase genes and analysis of its differential expression in maize (Zea mays) seedlings under drought stress. Plants 2020, 9, 315. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, C.; Wang, Z.; Kong, B.; Lin, T. Exogenous trehalose differentially modulate antioxidant defense system in wheat callus during water deficit and subsequent recovery. Plant Growth Regul. 2013, 70, 275–285. [Google Scholar] [CrossRef]
- Fichtner, F.; Lunn, J.E. The role of trehalose 6-phosphate (Tre6P) in plant metabolism and development. Annu. Rev. Plant Biol. 2021, 72, 737–760. [Google Scholar] [CrossRef]
- Vandesteene, L.; Lopez-Galvis, L.; Vanneste, K.; Feil, R.; Maere, S.; Lammens, W.; Rolland, F.; Lunn, J.E.; Avonce, N.; Beeckman, T.; et al. Expansive evolution of the trehalose-6-phosphate phosphatase gene family in Arabidopsis. Plant Physiol. 2012, 160, 884–896. [Google Scholar] [CrossRef] [Green Version]
- Van Houtte, H.; López-Galvis, L.; Vandesteene, L.; Beeckman, T.; Van Dijck, P. Redundant and non-redundant roles of the trehalose-6-phosphate phosphatases in leaf growth, root hair specification and energy-responses in Arabidopsis. Plant Signal. Behav. 2013, 8, e23209. [Google Scholar] [CrossRef] [Green Version]
- Wahl, V.; Ponnu, J.; Schlereth, A.; Arrivault, S.; Langenecker, T.; Franke, A.; Feil, R.; Lunn, J.E.; Stitt, M.; Schmid, M. Regulation of flowering by trehalose-6-phosphate signaling in Arabidopsis thaliana. Science 2013, 339, 704–707. [Google Scholar] [CrossRef]
- Martins, M.C.M.; Hejazi, M.; Fettke, J.; Steup, M.; Feil, R.; Krause, U.; Arrivault, S.; Vosloh, D.; Figueroa, C.M.; Ivakov, A. Feedback inhibition of starch degradation in Arabidopsis leaves mediated by trehalose 6-phosphate. Plant Physiol. 2013, 163, 1142–1163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lunn, J.E. Gene families and evolution of trehalose metabolism in plants. Funct. Plant Biol. 2007, 34, 550–563. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Zhang, Z.; Liu, J.; Chen, M.; Pan, R.; Hu, W.; Guan, Y.; Hu, J. Seed priming with spermidine and trehalose enhances chilling tolerance of rice via different mechanisms. J. Plant Growth Regul. 2020, 39, 669–679. [Google Scholar] [CrossRef]
- Xie, D.W.; Wang, X.N.; Fu, L.S.; Sun, J.; Zheng, W.; Li, Z.F. Identification of the trehalose-6-phosphate synthase gene family in winter wheat and expression analysis under conditions of freezing stress. J. Genet. 2015, 94, 55–65. [Google Scholar] [CrossRef]
- Xu, Y.; Wang, Y.; Mattson, N.; Yang, L.; Jin, Q. Genome-wide analysis of the Solanum tuberosum (potato) trehalose-6-phosphate synthase (TPS) gene family: Evolution and differential expression during development and stress. BMC Genom. 2017, 18, 926. [Google Scholar] [CrossRef]
- Yang, Y.; Ma, K.; Zhang, T.; Li, L.; Zhang, Q. Characteristics and expression analyses of trehalose-6-phosphate synthase family in Prunus mume reveal genes involved in trehalose biosynthesis and drought response. Biomolecules 2020, 10, 1358. [Google Scholar] [CrossRef]
- Vandesteene, L.; Ramon, M.; Roy, K.L.; Dijck, P.V.; Rolland, F. A single active trehalose-6-P synthase (TPS) and a family of putative regulatory TPS-like proteins in Arabidopsis. Mol. Plant 2010, 3, 406–419. [Google Scholar] [CrossRef]
- Delorge, I.; Figueroa, C.M.; Feil, R.; Lunn, J.E.; Van Dijck, P. Trehalose-6-phosphate synthase 1 is not the only active TPS in Arabidopsis thaliana. Biochem. J. 2015, 466, 283–290. [Google Scholar] [CrossRef]
- Almeida, A.M.; Villalobos, E.; Araújo, S.S.; Leyman, B.; Van Dijck, P.; Alfaro-Cardoso, L.; Fevereiro, P.S.; Torné, J.M.; Santos, D.M. Transformation of tobacco with an Arabidopsis thaliana gene involved in trehalose biosynthesis increases tolerance to several abiotic stresses. Euphytica 2005, 146, 165–176. [Google Scholar] [CrossRef]
- Liu, X.; Fu, L.; Qin, P.; Sun, Y.; Liu, J.; Wang, X. Overexpression of the wheat trehalose 6-phosphate synthase 11 gene enhances cold tolerance in Arabidopsis thaliana. Gene 2019, 710, 210–217. [Google Scholar] [CrossRef]
- Wang, C.L.; Zhang, S.C.; Qi, S.D.; Zheng, C.C.; Wu, C.A. Delayed germination of Arabidopsis seeds under chilling stress by overexpressing an abiotic stress inducible GhTPS11. Gene 2016, 575, 206–212. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Li, W.M.; Wang, W. Trehalose: Protector of antioxidant enzymes or reactive oxygen species scavenger under heat stress? Environ. Exp. Bot. 2008, 63, 378–384. [Google Scholar] [CrossRef]
- Wu, X.; Hou, Z.; Huang, C.; Chen, Q.; Gao, W.; Zhang, J. Cloning, purification and characterization of trehalose-6-phosphate synthase from Pleurotus tuoliensis. PeerJ 2018, 6, e5230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Z.; Xing, W.; Luo, P.; Zhang, F.; Jin, X.; Zhang, M. Comparative transcriptome analysis of Rosa chinensis ‘Slaters’ crimson China’ provides insights into the crucial factors and signaling pathways in heat stress response. Plant Physiol. Bioch. 2019, 142, 312–331. [Google Scholar] [CrossRef]
- Jiang, C.; Xu, J.; Zhang, H.; Zhang, X.; Shi, J.; Li, M.; Ming, F. A cytosolic class I small heat shock protein, RcHSP17.8, of Rosa chinensis confers resistance to a variety of stresses to Escherichia coli, yeast and Arabidopsis thaliana. Plant Cell Environ. 2009, 32, 1046–1059. [Google Scholar] [CrossRef]
- Suzuki, N.; Bajad, S.; Shuman, J.; Shulaev, V.; Mittler, R. The transcriptional co-activator MBF1c is a key regulator of thermotolerance in Arabidopsis thaliana. J. Biol. Chem. 2008, 283, 9269–9275. [Google Scholar] [CrossRef] [Green Version]
- Finn, R.D.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Mistry, J.; Mitchell, A.L.; Potter, S.C.; Punta, M.; Qureshi, M.; Sangrador-Vegas, A.; et al. The Pfam protein families database: Towards a more sustainable future. Nucleic. Acids. Res. 2016, 44, D279–D285. [Google Scholar] [CrossRef]
- Hu, B.; Jin, J.; Guo, A.-Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools, a Toolkit for Biologists integrating various HTS-data handling tools with a user-friendly interface. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Guo, X.; Yu, C.; Luo, L.; Wan, H.; Zhen, N.; Xu, T.; Tan, J.; Pan, H.; Zhang, Q. Transcriptome of the floral transition in Rosa chinensis ‘Old Blush’. BMC Genom. 2017, 18, 199. [Google Scholar] [CrossRef] [Green Version]
- Paul, M.J.; Primavesi, L.F.; Jhurreea, D.; Zhang, Y. Trehalose metabolism and signaling. Annu. Rev. Plant Biol. 2008, 59, 417–441. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, B.; Fu, L.; Zhang, D.; He, X.; Chen, Q.; Peng, M.; Zhang, J. Interspecies and intraspecies analysis of trehalose contents and the biosynthesis pathway gene family reveals crucial roles of trehalose in osmotic-stress tolerance in cassava. Int. J. Mol. Sci. 2016, 17, 1077. [Google Scholar] [CrossRef] [PubMed]
- Sanghera, G.S.; Wani, S.H.; Hussain, W.; Singh, N.B. Engineering cold stress tolerance in crop plants. Curr. Genom. 2011, 12, 30–43. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Usadel, B.; Blaesing, O.E.; Gibon, Y.; Retzlaff, K.; Hoehne, M.; Guenther, M.; Stitt, M. Global transcript levels respond to small changes of the carbon status during progressive exhaustion of carbohydrates in Arabidopsis rosettes. Plant Physiol. 2008, 146, 1834–1861. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kong, W.-W.; Huang, C.-Y.; Chen, Q.; Zou, Y.-J.; Zhao, M.-R.; Zhang, J.-X. Nitric oxide is involved in the regulation of trehalose accumulation under heat stress in Pleurotus eryngii var. tuoliensis. Biotechnol. Lett. 2012, 34, 1915–1919. [Google Scholar] [CrossRef]
- Miranda, J.A.; Avonce, N.; Suarez, R.; Thevelein, J.M.; Van Dijck, P.; Iturriaga, G. A bifunctional TPS-TPP enzyme from yeast confers tolerance to multiple and extreme abiotic-stress conditions in transgenic Arabidopsis. Planta 2007, 226, 1411–1421. [Google Scholar] [CrossRef] [Green Version]
- Lyu, J.I.; Park, J.H.; Kim, J.K.; Bae, C.H.; Jeong, W.J.; Min, S.R.; Liu, J.R. Enhanced tolerance to heat stress in transgenic tomato seeds and seedlings overexpressing a trehalose-6-phosphate synthase/phosphatase fusion gene. Plant Biotechnol. Rep. 2018, 12, 399–408. [Google Scholar] [CrossRef]
- Jiang, W.; Fu, F.L.; Zhang, S.Z.; Wu, L.; Li, W.C. Cloning and characterization of functional trehalose-6-phosphate synthase gene in maize. J. Plant Biol. 2010, 53, 134–141. [Google Scholar] [CrossRef]
- Jin, Q.; Hu, X.; Li, X.; Wang, B.; Wang, Y.; Jiang, H.; Mattson, N.; Xu, Y. Genome-wide identification and evolution analysis of trehalose-6-phosphate synthase gene family in Nelumbo nucifera. Front. Plant Sci. 2016, 7, 1445. [Google Scholar] [CrossRef]
- Hu, X.; Wu, Z.D.; Luo, Z.Y.; Burner, D.M.; Wu, C.W. Genome-wide analysis of the trehalose-6-phosphate synthase (TPS) gene family and expression profiling of ScTPS genes in sugarcane. Agronomy 2020, 10, 969. [Google Scholar] [CrossRef]
- Dan, Y.; Niu, Y.; Wang, C.; Yan, M.; Liao, W. Genome-wide identification and expression analysis of the trehalose-6-phosphate synthase (TPS) gene family in cucumber (Cucumis sativus L.). PeerJ 2021, 9, e11398. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.-L.; Liu, Y.-J.; Wang, C.-L.; Zeng, Q.-Y. Molecular evolution of trehalose-6-phosphate synthase (TPS) gene family in Populus, Arabidopsis and rice. PLoS ONE 2012, 7, e42438. [Google Scholar] [CrossRef] [PubMed]
- Du, L.; Qi, S.; Ma, J.; Xing, L.; Fan, S.; Zhang, S.; Li, Y.; Shen, Y.; Zhang, D.; Han, M. Identification of TPS family members in apple (Malus x domestica Borkh.) and the effect of sucrose sprays on TPS expression and floral induction. Plant Physiol. Biochem. 2017, 120, 10–23. [Google Scholar] [CrossRef] [PubMed]
- Mu, M.; Lu, X.K.; Wang, J.J.; Wang, D.L.; Yin, Z.J.; Wang, S.; Fan, W.L.; Ye, W.W. Genome-wide identification and analysis of the stress-resistance function of the TPS (trehalose-6-phosphate synthase) gene family in cotton. BMC Genom. 2016, 17, 54. [Google Scholar] [CrossRef]
- Ramon, M.; De Smet, I.; Vandesteene, L. Extensive expression regulation and lack of heterologous enzymatic activity of the class II trehalose metabolism proteins from Arabidopsis thaliana. Plant Cell Environ. 2009, 32, 1015–1032. [Google Scholar] [CrossRef]
- Chary, S.N.; Hicks, G.R.; Choi, Y.G. Trehalose-6-phosphate synthase/phosphatase regulates cell shape and plant architecture in Arabidopsis. Plant Physiol. 2008, 146, 97–107. [Google Scholar] [CrossRef] [Green Version]
- Lin, M.; Jia, R.; Li, J.; Zhang, M.; Chen, H.; Zhang, D.; Chen, X. Evolution and expression patterns of the trehalose-6-phosphate synthase gene family in drumstick tree (Moringa oleifera Lam.). Planta 2018, 248, 999–1015. [Google Scholar] [CrossRef]
- Kim, H.; Seomun, S.; Yoon, Y.; Jang, G. Jasmonic acid in plant abiotic stress tolerance and interaction with abscisic acid. Agronomy 2021, 11, 1886. [Google Scholar] [CrossRef]
- Jones, A.M. A new look at stress: Abscisic acid patterns and dynamics at high-resolution. New Phytol. 2016, 210, 38–44. [Google Scholar] [CrossRef]
- Ramon, M.; Rolland, F. Plant development: Introducing trehalose metabolism. Trends. Plant Sci. 2007, 12, 185–188. [Google Scholar] [CrossRef]
- Avonce, N.; Leyman, B.; José, O.M.-G.; Dijck, P.V.; Iturriaga, T.G. The arabidopsis trehalose-6-p synthase AtTPS1 gene is a regulator of glucose, abscisic acid, and stress signaling. Plant Physiol. 2004, 136, 3649–3659. [Google Scholar] [CrossRef] [PubMed] [Green Version]



| Gene | Scaffold ID | Predicted Position | CDS (bp) | Deduced Polypeptide | Predicted Subcellular Localization | |||
|---|---|---|---|---|---|---|---|---|
| Length (aa) | MW (kDa) | pI | GRAVY | |||||
| RcTPS1a | Scaffold_5 | 8460464–8470382 | 2934 | 977 | 109.54 | 6.03 | −0.290 | Nuclear |
| RcTPS1b | Scaffold_5 | 23007334–23020393 | 2793 | 930 | 105.29 | 6.23 | −0.391 | Nuclear |
| RcTPS5 | Scaffold_7 | 27956967–27962352 | 2586 | 861 | 97.30 | 5.71 | −0.164 | Cytoplasmic |
| RcTPS6 | Scaffold_4 | 64110270–64114733 | 2565 | 854 | 96.75 | 5.84 | −0.184 | Nuclear |
| RcTPS7a | Scaffold_2 | 47116733–47121620 | 2664 | 887 | 101.08 | 6.18 | −0.264 | Cytoplasmic |
| RcTPS7b | Scaffold_5 | 7851849–7856449 | 2565 | 854 | 96.32 | 5.87 | −0.266 | Cytoplasmic |
| RcTPS9 | Scaffold_7 | 12207610–12212604 | 2592 | 863 | 98.38 | 6.11 | −0.224 | Cytoplasmic |
| RcTPS10 | Scaffold_4 | 64989897–64994372 | 2601 | 866 | 97.48 | 5.85 | −0.205 | Cytoplasmic |
| RcTPS11 | Scaffold_4 | 24462064–24465676 | 2637 | 878 | 99.56 | 5.89 | −0.203 | Cytoplasmic |
| Group | N a | dN/dS(ω) under M0 | 2Δ/M3 vs. M0 b | 2Δ/M8 vs. M7 | M8 Estimates | Selective Position |
|---|---|---|---|---|---|---|
| I | 17 | 0.09691 | 295.922 ** | 44.089 | p1 = 0.02814 ω = 2.49539 (p = 0.59741 q = 5.21453) | 137L, 255S, 388D, 401I, 435P, 503E, 562D *, 632N, 637S, 639Q, 679S |
| II1 | 18 | 0.05352 | 475.124 ** | 17.556 | p1 = 0.01043 ω = 7.57672 (p = 0.26744 q = 3.43511) | 300K, 426A |
| II2 | 16 | 0.08810 | 339.810 ** | 25.457 | p1 = 0.03229 ω = 3.83594 (p = 0.36190 q = 3.30447) | 3L, 54S, 166S, 228R, 409L, 476A, 482R, 506A, 552Q, 682L, 685I, 691S, 716N |
| II3 | 19 | 0.11694 | 712.521 ** | 34.111 | p1 = 0.02958 ω = 9.36648 (p = 0.44335 q = 2.88651) | 25A, 44Y, 622T, 771I, 772M * |
| II4 | 8 | 0.09783 | 318.212 ** | 28.595 | p1 = 0.05407 ω = 2.40780 (p = 0.44632 q = 4.10918) | 20N, 39V, 40P, 46S, 65A, 88K, 113S, 128M, 307E, 315Q, 458D, 459R, 504A, 542R, 630L, 656T, 752Q, 780S, 781S, 786V, 825S, 826T, 828P, 829K, 830L |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wei, X.-R.; Ling, W.; Ma, Y.-W.; Du, J.-L.; Cao, F.-X.; Chen, H.-X.; Chen, J.-R.; Li, Y.-F. Genome-Wide Analysis of the Trehalose-6-Phosphate Synthase Gene Family in Rose (Rosa chinensis) and Differential Expression under Heat Stress. Horticulturae 2022, 8, 429. https://doi.org/10.3390/horticulturae8050429
Wei X-R, Ling W, Ma Y-W, Du J-L, Cao F-X, Chen H-X, Chen J-R, Li Y-F. Genome-Wide Analysis of the Trehalose-6-Phosphate Synthase Gene Family in Rose (Rosa chinensis) and Differential Expression under Heat Stress. Horticulturae. 2022; 8(5):429. https://doi.org/10.3390/horticulturae8050429
Chicago/Turabian StyleWei, Xiao-Ru, Wu Ling, Yu-Wan Ma, Jiao-Lin Du, Fu-Xiang Cao, Hai-Xia Chen, Ji-Ren Chen, and Yu-Fan Li. 2022. "Genome-Wide Analysis of the Trehalose-6-Phosphate Synthase Gene Family in Rose (Rosa chinensis) and Differential Expression under Heat Stress" Horticulturae 8, no. 5: 429. https://doi.org/10.3390/horticulturae8050429






