Abstract
The Hosta hybrid cultivar ‘So Sweet’, an important ornamental and widely used horticultural plant, is noted for its rich, fragrant white flowers. The main aroma components of Hosta flowers are terpenoids, mainly monoterpenes. Until now, the terpene synthases responsible for terpene production in Hosta were not described. In this study, two terpene synthase (TPS) genes (HsTPS1 and HsTPS2) were cloned and characterized to further study their function. Furthermore, the volatile terpenes of Hosta ’So Sweet’ in two flower development stages from two in vitro enzyme tests were analyzed by gas chromatography–mass spectrometry (GC–MS). We analyzed the expression levels of two genes at four different developmental stages using quantitative real-time PCR, while localization was analyzed using Nicotina benthamiana leaves. In vitro, the two proteins were identified to mainly produce linalool and nerol. In addition, the active products of the two recombinant proteins were (E,E)-farnesol and (E,E)-farnesal, respectively, using farnesyl pyrophosphate as a substrate. The high expression of HsTPS1 and HsTPS2 was correlated with the release of components of Hosta flowers. To our knowledge, this is the first time that the terpene synthase genes of Hosta species have been isolated and identified, providing an opportunity to study the terpene metabolic pathways in Hosta species.
1. Introduction
All volatile organic compounds (VOCs) are divided into several classes, including terpenoids, phenylpropanoids/benzenoids, fatty acid derivatives, and amino acid derivatives, in addition to a few species-/genus-specific compounds not represented in these major classes [1]. Terpenoids are the most diverse plant secondary metabolites, and they play an important role in the survival and evolution of plants in different ecological environments. The metabolic pathways of VOCs in plants have been extensively studied, especially in flowering plants with high concentrations of terpene compounds [2], including Lilium [3], Rosa chinensis [4], and Orchidaceae [5]. Various publications have estimated that the number of distinct terpenoid compounds (an inclusive term used to describe terpenes and terpene derivatives from different pathways) could be more than 2300 individual structures described in higher plants [6,7]. These products have been widely used in the pharmaceutical, flavor fragrance, and biofuel industries [2]. In general, isoprenoid precursors in plants are produced from interconverted C5 isoprenoyl pyrophosphate (IPP) and its allyl isomer dimethylallyl pyrophosphate (DMAPP), synthesized via the methvaleric acid and methylerythritol phosphate pathways [8]. IPP and DMAPP are synthesized via the MVA and MEP pathways and subsequently condensed to geranyl diphosphate (GPP), farnesyl diphosphate (FPP), and geranylgeranyl diphosphate (GGPP) by prenyl-PP synthases [9]. GPP and GGPP are localized in plastids, while FPP is localized in the cytosol. The key enzymes in the biosynthetic pathways of terpenoids [10] convert the above substrates into various terpenoids with different types and structures [11,12]. In the study of Aharoni et al., the FaNES1 enzyme produced in E. coli was capable of generating both linalool and nerolidol when supplied by in vitro enzyme activity with GPP or FPP [13]. In a recent study, three rose monoterpene synthases were functionally characterized in vitro; one had linalool synthase activity, whereas the other two had dual linalool/nerolidol synthase activity [14]. So far, numerous TPS genes have been isolated from a variety of plants, and their ecological roles have been extensively studied. To our knowledge, previous studies on TPS genes were mainly derived from dicotyledons, with only a few derived from monocotyledons, such as Alstroemeria [15], Hedychium coronarium [16], and Freesia and Lilium cultivars [17,18]. Given the importance of floral fragrances in speciation and evolution [19,20], more TPS genes derived from monocotyledons should be studied.
Plants from the genus Hosta belonging to the Liliaceae family are the most well-known plants and irreplaceable resources for applied gardening and landscaping [21]. They were later classified by botanists as Asparagaceae [22]. H. plantaginea (Lam.) Aschers, the most famous species of the genus, originally from China, is widely cultivated worldwide for its leaves and flowers, along with strong adaptability to different environments, as well as a long blooming period [23,24,25]. H. ‘So Sweet’ originated from the hybridization of H. plantaginea with H. ‘Fragrant Bouquet’ [26,27]. These plants are also important in traditional Chinese medicine [28,29]. The main floral components of Hosta are terpenes, of which monoterpenes account for the majority.
The main objectives of the present study were to isolate and clone the two TPS genes of H. ‘So Sweet’, and to functionally express and characterize the recombinant terpene synthases using in vitro systems. The expression pattern of the TPS involved in the formation and emission of terpenoids in different flower development stages was also elucidated. This research is expected to lay the foundation for the study of Hosta plantaginea terpene synthase genes. We hope that these results can provide new insight into terpene biosynthesis in monocotyledons.
2. Materials and Methods
2.1. Plant Material
Hosta ‘So Sweet’ plants were grown in the horticulture nursery of the Conservation and Exploitation of Wild Resources of Changbai Mountain, Jilin Agricultural University with the temperature set at 25–30 °C in natural light and 18–20 °C in the dark (longitude: 116°13′9.4116″ E, latitude: 39°59′55.8312″ N; 67 m above sea level). The cultivation substrate was garden soil, yellow sand soil, and perlite in a ratio of 3:1:1. The collection period of the flower samples was from mid-August to mid-September. For natural volatile compound analysis, fresh flowers were immediately placed into 100 mL vials, and then quickly transported to the laboratory for scent collection. All samples at four flower developmental stages were immediately frozen in liquid nitrogen and stored at −80 °C until required. The flower developmental stages are presented in Figure S1.
2.2. RNA Extraction and cDNA Synthesis
Total RNA was extracted using a mini BEST Plant RNA extraction kit (TaKaRa, Kyoto, Japan) according to the manufacturer’s instructions. Complementary DNA was synthesized in a final reaction volume of 20 μL using a PrimeScriptTM RT regent kit with gDNA eraser (TaKaRa, Japan).
2.3. Cloning of HsTPS1 and HsTPS2 Genes
Terpene synthase genes in Hosta flowers were screened from a previously reported transcriptome database (NCBI BioSample database: no. PRJNA542483). The sequences were aligned against the nonredundant protein database via the BlastX algorithm (https://www.ncbi.nlm.nih.gov/ accessed on 30 January 2020). HsTPS1 and HsTPS2 were selected and named. The full-length sequence was cloned using pairs of primers designed by Primer Premier 5 (http://www.premierbiosoft.com/primerdesign/ accessed on 10 February 2020). The reaction was carried out in a final volume of 20 μL containing 1 μL of cDNA (0.2 μg), 4–6 pmol of each primer, and 10 μL PrimeSTAR Max Premix from the PrimeSTAR Max DNA Polymerase kit (TaKaRa, Japan) using the following regime: 94 °C for 3 min, 35 cycles at 98 °C for 10 s, 52 °C for 5 s, 72 °C for 20 s, with an extension time of 7 min at 72 °C. The PCR products were cloned into the pEASY-Blunt Simple Cloning Vector (TransGen Biotech, Beijing, China); then, competent E. coli JM109 cells were transformed, and a positive single colony was screened and sequenced. The primers and gene sequence data are shown in Tables S1 and S2.
2.4. Sequence Analysis
For the purpose of phylogenetic tree analysis, amino acid sequences of HsTPS proteins and their homologs in other plant species were aligned using Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo/ accessed on 1 March 2020) by choosing the neighbor-joining method in MEGA 6 (www.megasoftware.net accessed on 1 March 2020) with bootstrap analysis (1000 replicates) [30,31]. Multiple sequence alignment was submitted to ESPript 3.0 (https://espript.ibcp.fr/ESPript/cgi-bin/ESPript.cgi accessed on 1 March 2020) [32]. Conservative motifs such as RRx8W, DDxxD, and DDxxTxxxxE were indicated in gray. The Newick file exported from MEGA 6 was uploaded to ITOL (https://itol.embl.de/ accessed on 1 March 2020) for further annotation [31,33].
2.5. Quantitative Real-Time PCR Analysis
Quantitative RT-PCR was performed to compare the gene expression abundance of HsTPS1 and HsTPS2. Quantitative RT-PCR was performed by an ABI 7500 real-time PCR system using 2× RealStar Green Fast Mixture with ROX II (GenStar, Beijing China). The experimental regime was as follows: 95 °C for 2 min flowed by 40 cycles at 95 °C for 15 s and 55 °C for 30 s. To normalize the expression values, the 2−ΔΔCt method was used to process experimental values [34]. Hosta Actin was used as the reference gene. All the reactions were carried out according to the manufacturer’s protocol in a 20 μL volume using 1 μL of reverse transcribed cDNA as template and 200 nM of each of the primers. All primer and gene sequences are provided in Tables S1 and S2.
2.6. Expression of Recombinant HsTPS1, HsTPS2 in E. coli and In Vitro Enzyme Assay
The full-length sequences of HsTPS genes without terminator were amplified using specific primers and then inserted into the pET-28a expression vector with a T7 promoter. The recombinant expression vector was inserted into Rosetta (DE3) competent E. coli cells by heat shock and sequenced to confirm its identity. The E. coli was grown in Luria–Bertani (LB) medium according to the instructions for competent cells. The recombinant protein was induced by 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG), incubated for 16–20 h at 16 °C with shaking at 185 rpm. The soluble protein was purified using a 6× His-Tagged Protein Purification Kit (Cowin Biosciences, Beijing, China) following the manufacturer’s instructions. The reaction mixture used for enzyme activity consisted of 50–100 μg of recombinant HsTPS protein, 1 mM GPP (Sigma) or FPP (Sigma) as the substrate, 25 mM Tris-HCl (pH7.8), 5 mM dithiothreitol as the buffer, and 10 mM MgCl2 as the cofactor in a total volume of 2 mL. Enzyme activity was measured after 2 h incubation in hexane at 30 °C.
2.7. GC–MS Analysis
The GC–MS analyses of the volatile compounds from Hosta flower tissues and enzymatic activities were conducted using an Agilent 6890N GC (gas chromatograph) combined with an Agilent 5975 mass detector. A 30 m × 0.25 mm × 0.10 μm HP-1MS capillary column was equipped with the instrument. Helium gas was used as the carrier gas, at a flow rate of 0.9 mL/min. The temperature program settings were: 3 min at 60 °C after injection, followed by a 10 °C/min temperature ramp from 60 °C to 280 °C, and held for 2 min. Temperatures of the injector and detector were 250 °C. Different aroma components formed their respective chromatographic peaks, and the retention index was determined by searching the NIST08 mass spectral library (http://webbook.nist.gov/chemistry accessed on 1 March 2020). The results were analyzed using C6–C19 alkane standard substances. Multiple comparisons were performed to assist the qualitative retrieval of mass spectrometry data. The relative quantitative analysis was performed through a peak normalization procedure. Peak areas were normalized as percentages and used to determine the relative amounts of the volatile compounds.
2.8. Subcellular Localization of HsTPS Proteins
The subcellular localization of both HsTPS1 and HsTPS2 protein sequences was using a computer-based prediction server (http://www.cbs.dtu.dk/services/TargetP/ accessed on 5 March 2020) [35]. The genes of HsTPS1 and HsTPS2 intact ORF sequences were cloned into the pCambia-1300 expression vector, by replacing the terminator with GFP (green fluorescent protein). Nicotiana benthamiana leaves were infected by Agrobacteria cultures independently expressing the two TPSs [36]. After 48 h of transformation, infiltrated leaves were visualized by laser scanning confocal microscopy. Images were exported and spliced using Photoshop (www.photoshop.com/ accessed on 1 March 2020).
3. Results
3.1. Phylogenetic Analysis of TPS Genes from H. ‘So Sweet’
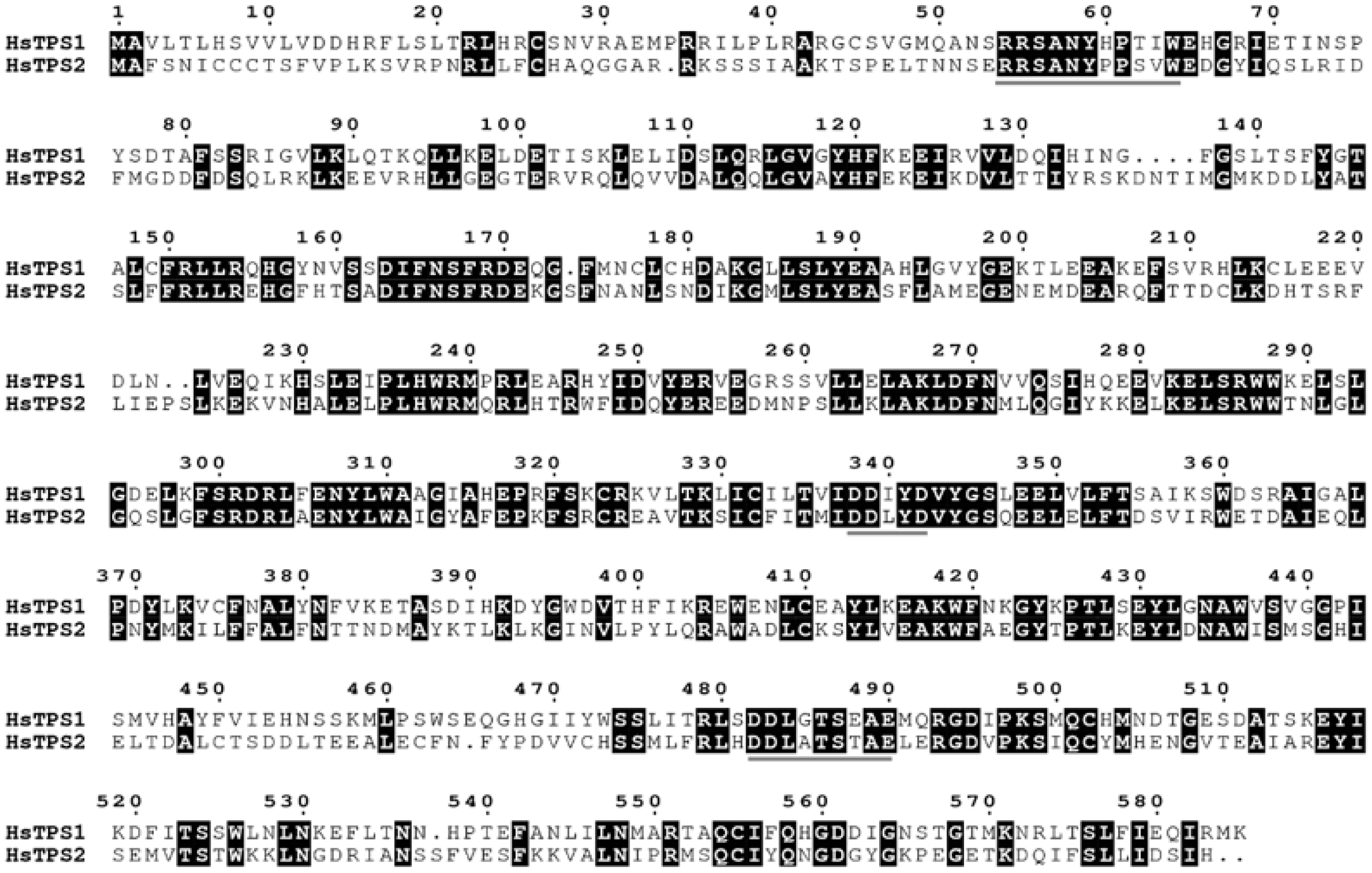
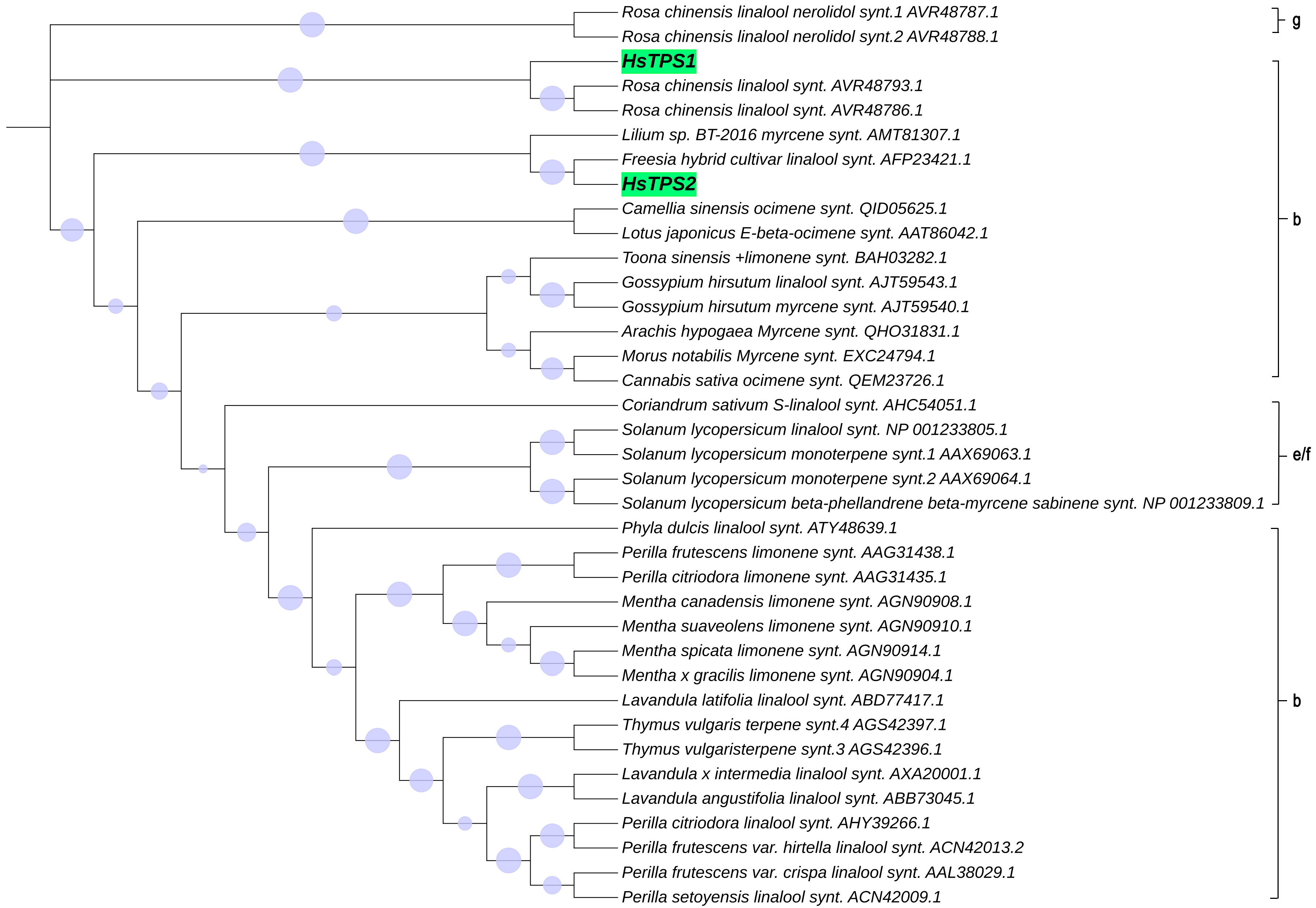
The RNA of Hosta flower was reverse-transcribed into cDNA and analyzed by PCR. HsTPS1 (1758 bp) and HsTPS2 (1770 bp) encode 586 and 590 amino acids, respectively, with molecular weights of 67.48 and 68.1 kDa. The HsTPS1 full-length sequence shared 56.79% amino acid identity with the (R)-linalool synthase from Magnolia champaca (ART66980.1). In addition, HsTPS1 had 56.69% and 56.50% homology with trans-ocimene synthase from Litsea cubeba (AEJ91554.1) and Cinamomum micranthum f. kanehirae (RWR 88332.1). The homology of HsTPS1 with monoterpene synthase from a Lilium hybrid cultivar (QBP79170.1) was 52.90%. The HsTPS2 full-length sequence shared 63.06% and 61.75% identity with the linalool synthase from a Freesia hybrid cultivar (AQM50913.1) and 54.02% identity with the myrcene synthase from Lilium sp. BT-2016 (AMT81307.1). HsTPS1 and HsTPS2 shared 44.46% identity with each other (Figure 1). Terpenoid synthases are usually categorized into seven branches, named TPS-a, TPS-b, TPS-c, TPS-d, TPS-e/f, and TPS-g [6]. The following motifs have already been reported to be implicated in catalysis by TPS: an anarginine-enriched region, often appearing as an RRx8W motif; a specific linalool synthase region, often appearing as (M/L)L(S/Q/N) L(F/Y)EAS; asparticacid-enriched and glutamic acid-enriched regions, appearing as DDXXD and NSE/DTE motifs [37,38]. The RRx8W motif is characteristic of the N-terminal domain of monoterpene synthases, as well as the TPS-b subfamily in angiosperms and the TPS-d subfamily in gymnosperms [39,40,41]. It has also been demonstrated to be isomerized in the first step from geranyl diphosphate to linalalyl diphosphate [42,43]. These motifs may be involved in the linalool synthase activity of HsTPS1 and 2. The LSLYEA(S/A) motif of HsTPS was located in a highly similar region to that seen in other mTPSs, considered to be an active site peptide. The highly conserved DDXXD motif is generally recognized as a binding site for the metal ion-chelated diphosphate ester substrate [44]. In contrast, NSE/DTE motifs are less conserved, and they are known to be combined with divalent metal ion cofactors in enzyme activity [37,45]; they were present as the DDL(G/A)TS(E/T)AE motif in the HsTPS amino-acid sequences. In general, monoterpenes tend to cluster by genus rather than function [37]. Thirty-five different mTPSs from twenty-seven different species and two HsTPSs were aligned in this study, and a phylogenetic tree was constructed (Figure 2). It is worth mentioning that most of the reported linalool/nerolidol synthases belong to the TPS-g group. This is a subfamily closely related to the TPS-b group, which comprises angiosperm acyclic terpene synthases. Both TPSs were clustered into the TPS-b group, close to the TPS-g group.
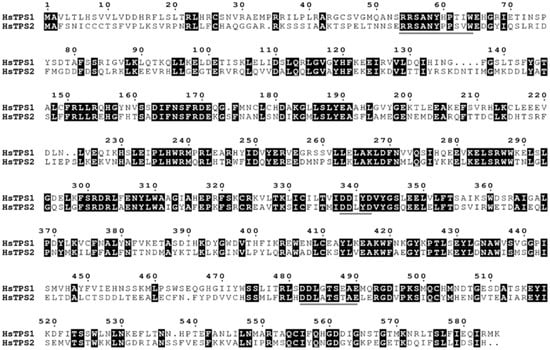
Figure 1.
Protein sequence alignment of the two HsTPS was constructed using ClustalW and was edited using Escript 3.0. The three conserved motifs, RRx8W, DDxxD, and DDxxTxxxE, characteristic of terpene synthases, were overlined in gray.
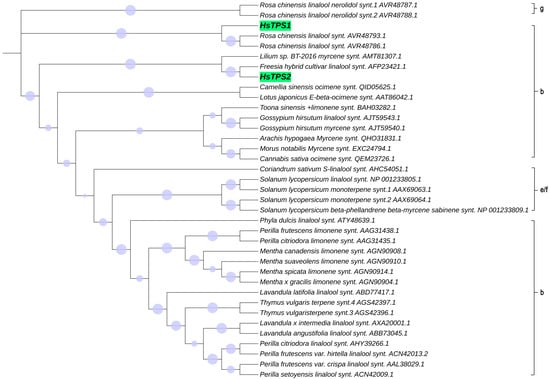
Figure 2.
Results of phylogenetic analysis of terpene synthase proteins from H. ‘So Sweet’ (HsTPS1,HsTPS2) and other plants, done using 1000 replications for bootstrapping by MEGA 6 program. Circle at the branches of the tree shows bootstrap values.
3.2. Functional Characterization of Recombinant HsTPS1 and HsTPS2
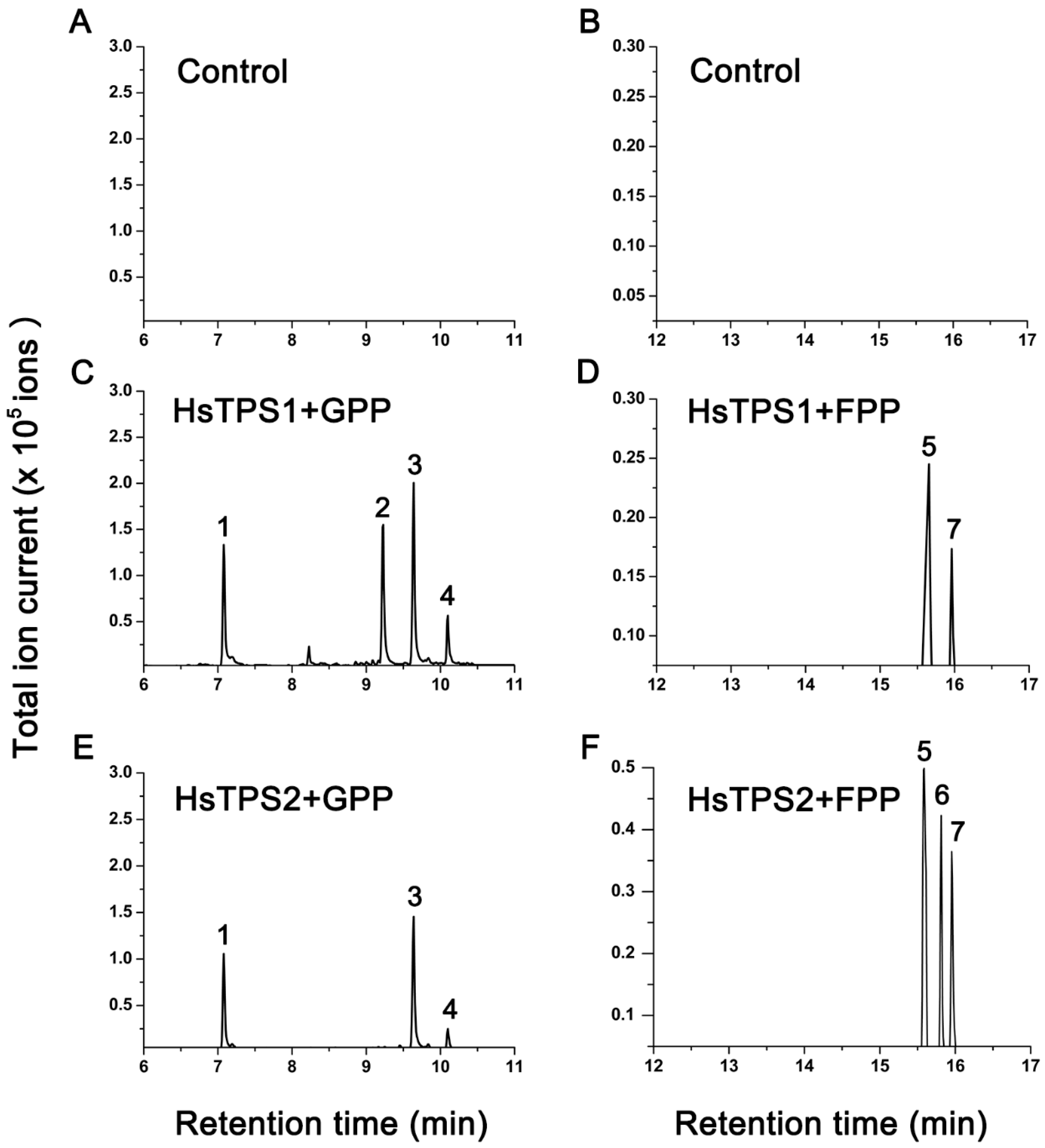
The recombinant proteins were obtained from the E. coli Rosetta (DE3) strain using a pET-28a expression vector. The soluble portion of the protein induced by low temperature was purified and incubated with GPP and FPP substrates, and the products were analyzed by GC–MS (Figure 3). The two recombinant proteins were active when incubated with GPP substrates and produced monoterpenes. Incubation of recombinant HsTPS1 with GPP mainly produced linalool, citronellene, and nerol. In addition, minor amounts of geranial were detected. Incubation of recombinant HsTPS2 with GPP produced linalool, nerol, and geranial. After incubation with FPP, both recombinant proteins showed activity and produced sesquiterpenes in lower amounts. Recombinant HsTPS1 used FPP to produce (E,E)-farnesol and (E,E)-farnesal. In addition to the above two sesquiterpenes, recombinant HsTPS2 protein also produced a small amount of nerolidol.
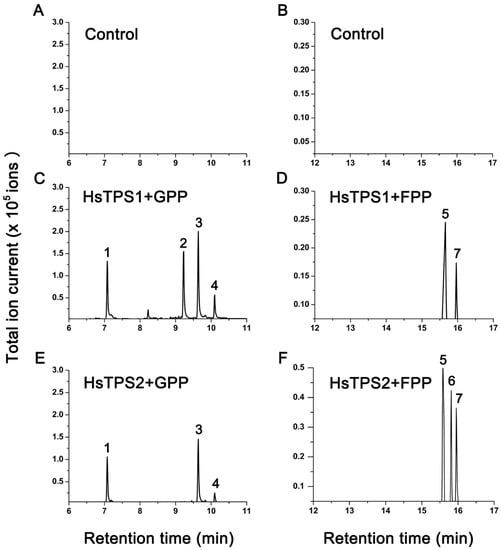
Figure 3.
Total ion chromatogram of the products formed by incubating extracts of empty vector (control) with GPP and FPP. GC–MS analysis of products formed by recombinant HsTPS1 and HsTPS2 in the substrate GPP and FPP. Only sample peaks that were significantly higher than the negative control were marked with letters. Corresponding compounds are: 1 = linalool (retention time = 7.087 min); 2 = citronellene (retention time = 9.229 min); 3 = nerol (retention time = 9.639 min); 4 = geranial (retention time = 10.100 min); 5 = (E,E)-farnesal (retention time = 15.587 min); 6 = nerolidol (retention time = 15.796 min); 7 = (E,E)-farnesol (retention time = 15.961 min).
3.3. Terpenoids Production in Hosta and Expressional Analysis of HsTPS1 and HsTPS2 Genes
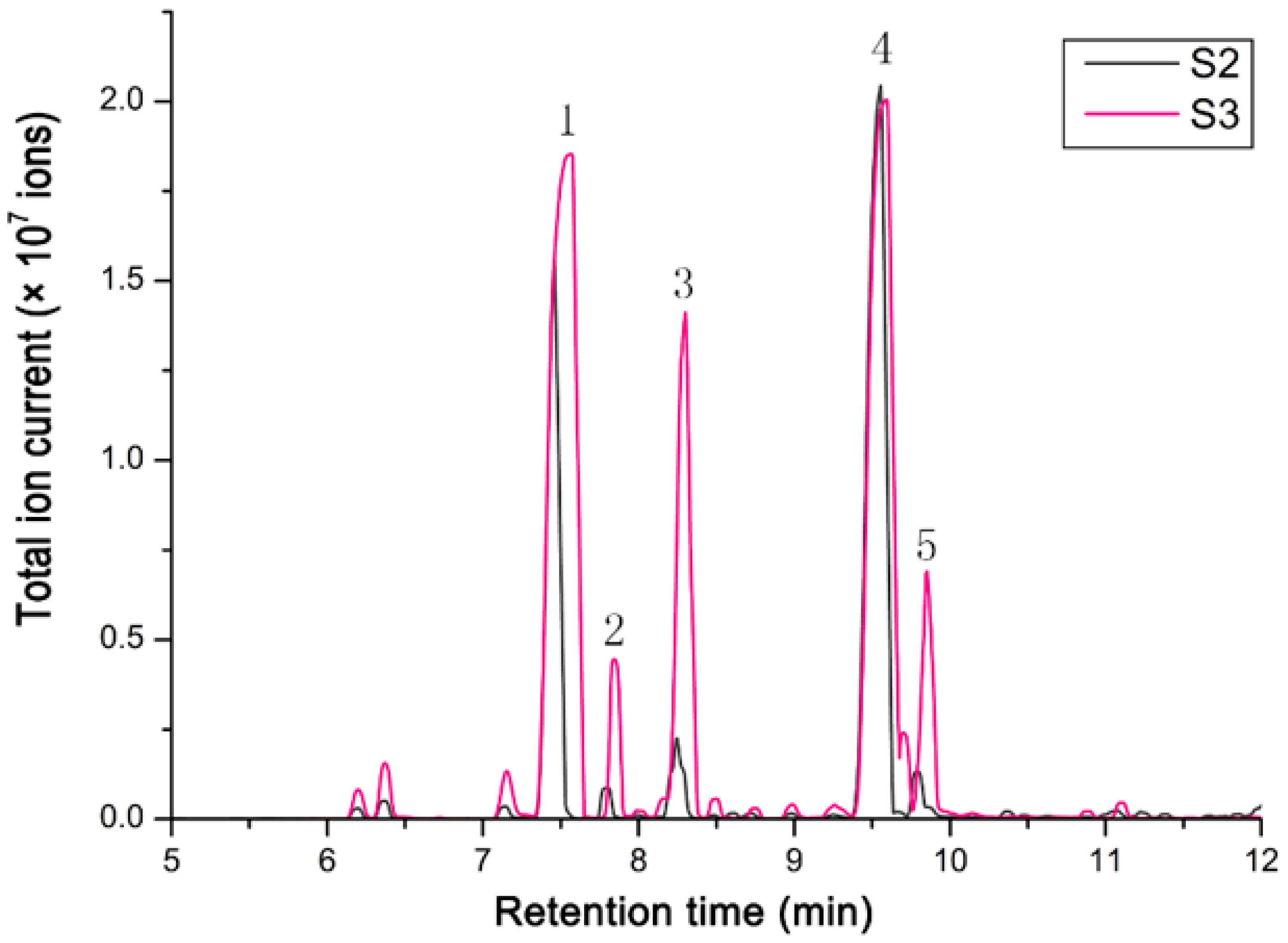
Terpenoids are the dominant scent components in Hosta flowers, mainly including linalool, myrcene, limonene, and (E)-β-ocimene [21]. The volatile compounds in flowers of H. ‘So Sweet’ were analyzed by hexane extraction and GC–MS. The test results showed that the relative content of terpenoids in H. ‘So Sweet’ was 91.78–93% of the total volatile compounds, with a small amount of phenylpropionic acid compounds and fatty family compounds. Fifteen terpenoids were detected at S2 (the stage of flower buds 1 day before full bloom), while twenty-two were detected at S3 (fully bloom flowers for 1 day). The data are shown in Table 1. The total ion chromatograms of scent components emitted from the flowers at two different developmental stages are shown in Figure 4. Linalool and β-pinene were the main compounds, accounting for 52.23% and 28.79% at S2. At S3, the contents of linalool and β-pinene accounted for 40.76% and 37.13% of the total compounds. In addition, there were three major components, d-limonene, α-phellandrene, and β-phellandrene, which varied greatly between two stages.

Table 1.
Terpenoids identified from Hosta flower volatiles of two different developmental floral stages using GC–MS analysis. All compounds were identified using the NIST database and quantified by comparing the peak area with the internal standard ethyl decylate. Data are presented as means (±SE, n = 3).
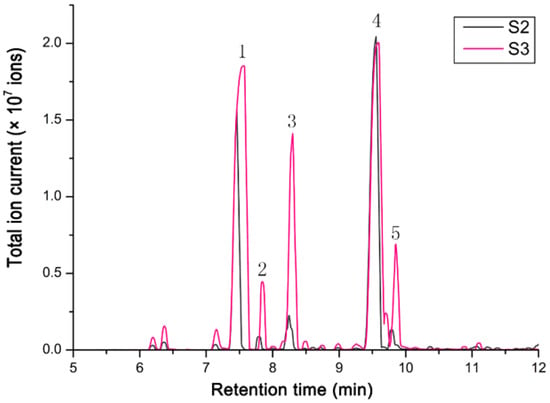
Figure 4.
The total ion chromatogram of scent components emitted from the flowers of two different developmental floral stages. The stage of flower buds one day before full bloom (S2) and full-bloom flowers for one day (S3). 1 = β-Pinene; 2 = α-Phellandrene; 3 = D-Limonene; 4 = Linalool; 5 = β-Phellandrene.
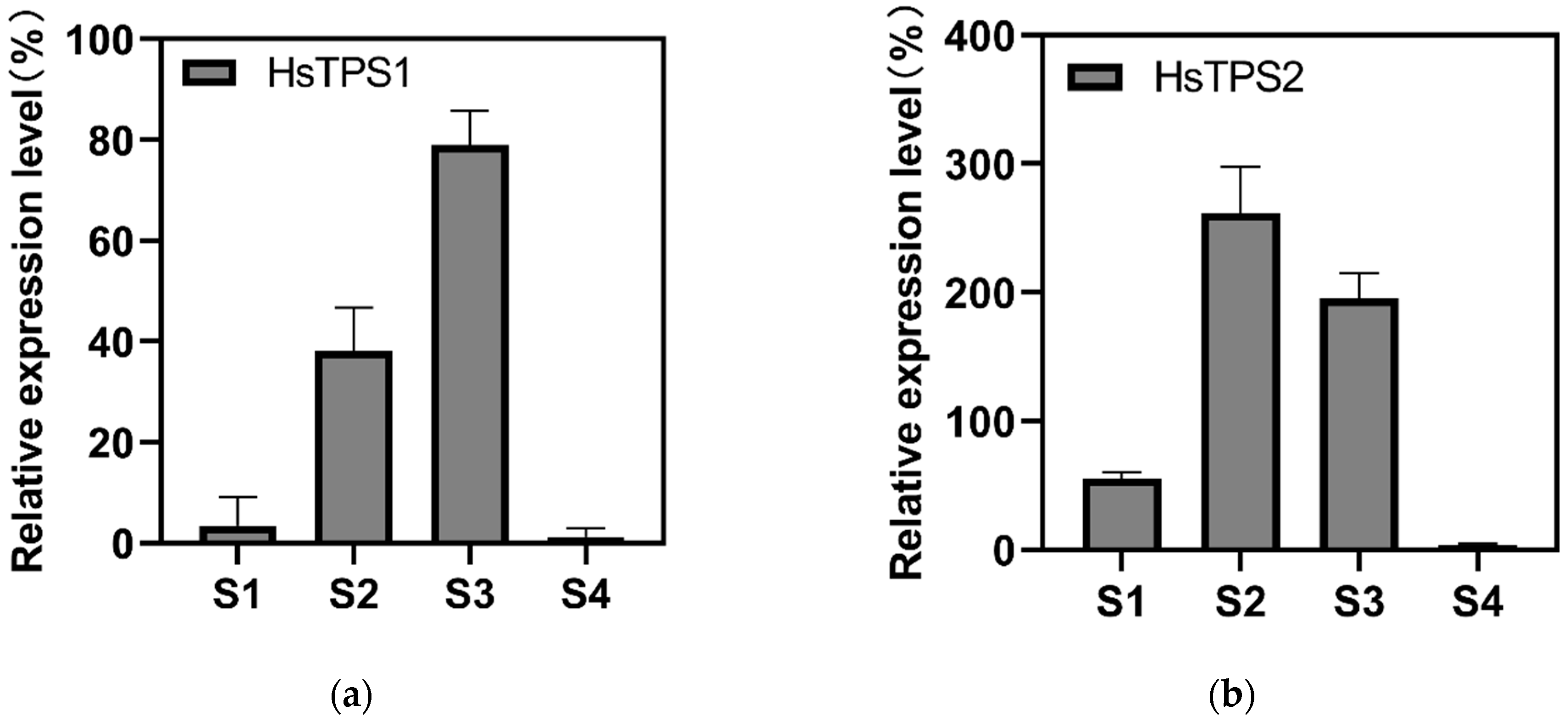
The expression of HsTPS1 and HsTPS2 genes was consequently evaluated during petal development using quantitative RT-PCR to compare their transcription. The expression level of HsTPS2 was much higher than that of HsTPS1. The expression patterns of the two HsTPSs were higher at S2 and S3 compared with S1 and S4. HsTPS1 expression was highest at S3, while HsTPS2 expression was highest at S2. The expression levels of HsTPS1 and HsTPS2 were significantly decreased at S4 (Figure 5). Linalool was released and accumulated before flowering, with peak values reaching the maximum after flowering (Figure 4).
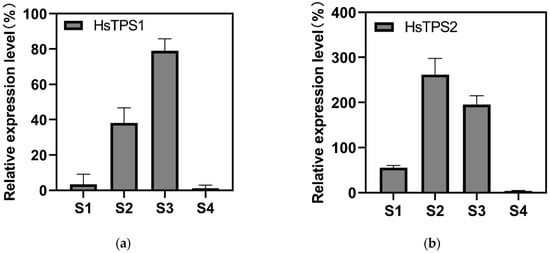
Figure 5.
Relative expression analysis was performed by qRT-PCR using Actin as reference genes. (a) Relative expression levels of HsTPS1; (b) Relative expression levels of HsTPS2. The relative transcription level in tissue with the highest expression quantity was set to 1 (100%). Each bar represents the mean value ±SE of three biological and three technical replicates (p-value < 0.05). The stage of flower buds two or three days before full bloom (S1); flower buds one day before full bloom (S2); full-bloom flowers for one day (S3); and flowers one day after full bloom (S4).
3.4. In Vivo Localization of the Two TPSs
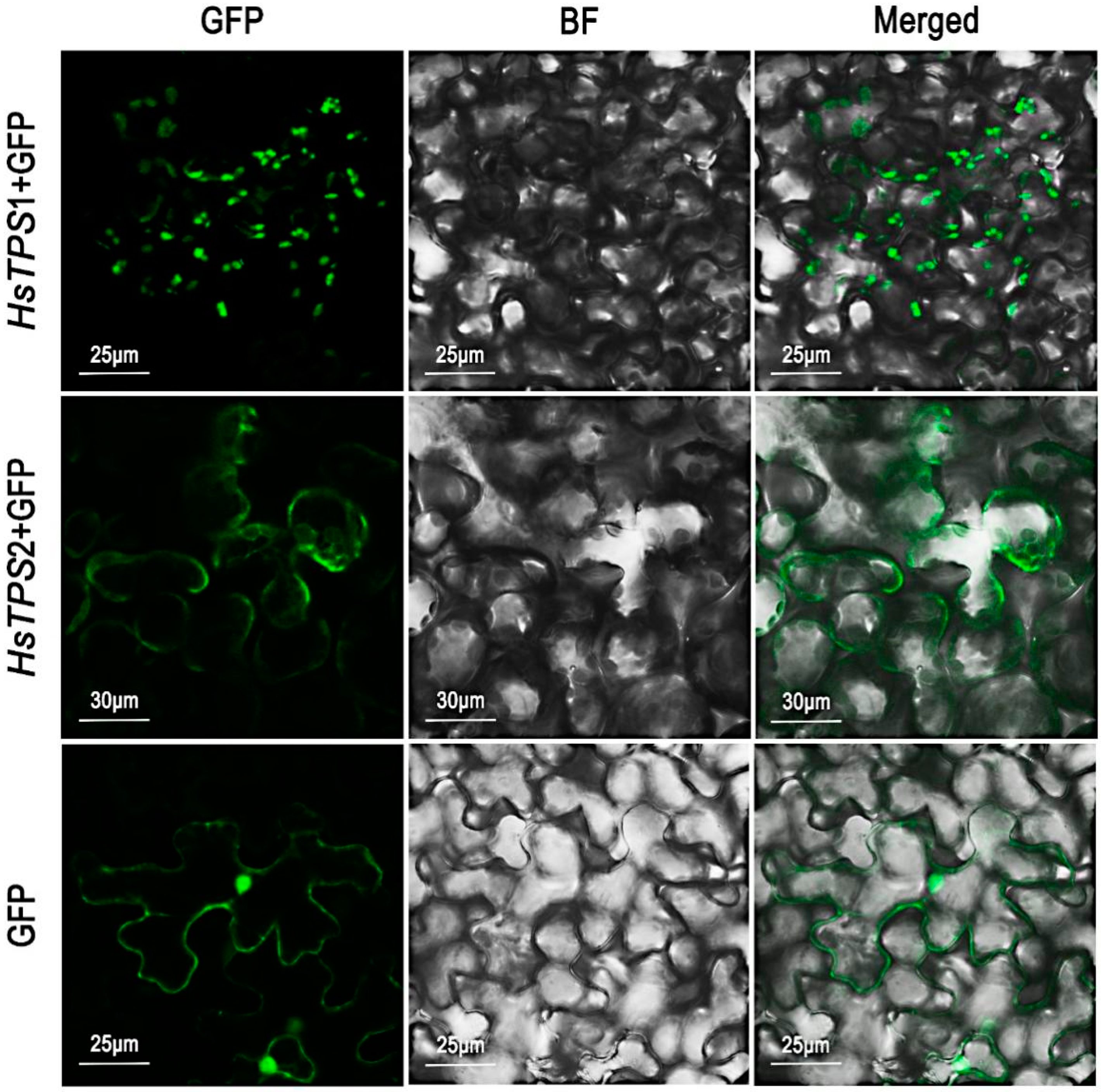
Most monoterpene synthases are thought to be localized in plastids, while sesquiterpene synthases are generally thought to be localized in the cytosol. It is typically believed that monoterpene synthases and sesquiterpene synthases use GPP and FPP as their substrates, respectively [1,6]. Bioinformatic analyses of the two TPSs were performed using the online software TargetP 2.0, and the two TPS genes were predicted with high probability to be chloroplast transport peptides, indicating the possibility of chloroplast localization. The reliability value of the prediction result was lower than the general value (>4), indicating low reliability. To test the above prediction, Nicotiana benthamiana leaves were infected by Agrobacteria cultures independently expressing the two TPSs [36]. The results of scanning laser confocal microscopy showed that HsTPS1 was probably localized in plastids, whereas HsTPS2 was localized in the cytosol (Figure 6).
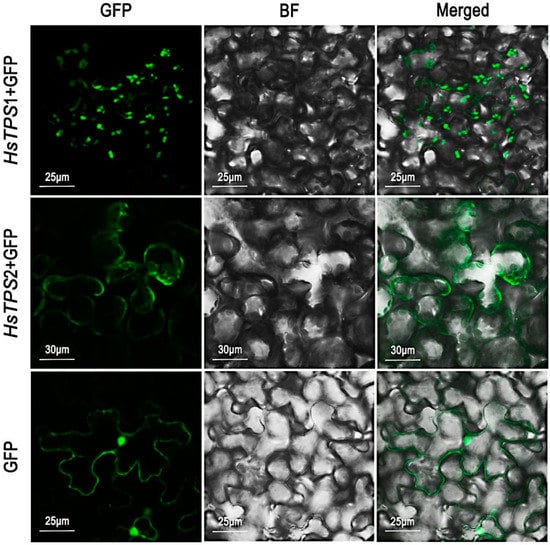
Figure 6.
Subcellular localization of HsTPS1 and HsTPS2 by transient transformation of N. benthamiana with Agrobacterium carrying a pCambia 1300 expression vector, where the HsTPS1 and HsTPS2 coding sequence was fused with the GFP coding sequence, respectively. GFP, GFP fluorescence detected in the green channel; BF, brightfield image; Merged, merged green and BF channel images.
4. Discussion
In the present work, we isolated and functionally analyzed two terpene synthase genes from the Hosta flower for the first time, and we used qRT-PCR to investigate the expression of these genes in flowers at four different developmental stages. There was a significant correlation between the release of linalool in Hosta and the expression of HsTPS genes. To date, several terpene synthases have been found to be bifunctional in vitro but monofunctional in planta [46]. Some terpene synthases synthesize linalool and nerolol in vitro but exhibit monoterpene or sesquiterpene biosynthesis activity in planta [47]. These studies suggest that some sesquiterpene synthases may have evolved from a subset of monoterpene synthases adapted to use farnesyl diphosphate as a substrate [48]. In a recent study, PamTPS1 (Plectranthus amboinicus) was identified as a linalool/nerolidol synthase with the ability to exclusively produce linalool and nerolidol. Thus, some enzymes possess both monoterpene synthase and sesquiterpene synthase activities [49]. Recombinant SaNES/LIS (Santalum album L.) is a bifunctional enzyme that showed the biosynthesis of (E)-nerolidol from farnesyl diphosphate and of linalool from geranyl diphosphate [50]. In a study of Clematis florida, recombinant proteins CfTPS1 and CfTPS2 were demonstrated to catalyze the conversion of geranyl diphosphate to linalool. In addition, CfTPS1 and CfTPS2 produced nerolidol from farnesyl diphosphate [51]. Although recombinant proteins of HsTPS1 and HsTPS2 showed enzyme activity when provided with GPP and FPP as substrates in vitro, respectively, their role in plants should be inferred in conjunction with subcellular localization. Different localization results were obtained in the subcellular localization analysis of tobacco. The exact role of HsTPS in Hosta was not clarified in this study; hence, the possibility of HsTPS acting as both a monoterpene synthase and a sesquiterpene synthase in plants cannot be ruled out. Further experiments are needed to provide evidence for more reliable conclusions.
Overall, the results showed that there was a certain correlation between the high expression of HsTPS genes and the accumulation of the flower terpene content at S2 and S3. The release of linalool and its rich monoterpene compounds in Hosta plants was directly related to the level of TPS. It is speculated that HsTPS1 and HsTPS2 are involved in the mass production of linalool in Hosta, while there may also be many multifunctional enzymes similar to HsTPS1 and HsTPS2 in Hosta.
These studies lay a foundation for the study of terpene release in Hosta flowers, fill the gap of floral scent research in Liliaceae plants, and provide ideas for further study of the Liliaceae gene and cell engineering. The production of large amounts of linalool in Hosta flowers may be facilitated by various enzymes such as HsTPS1 and HsTPS2, a hypothesis that needs to be further verified. In the future, the identification and expression of key TPS genes can provide information for the study of the correlation between the TPS family and terpene synthesis in Hosta.
5. Conclusions
In this study, we identified and functionally characterized, for the first time, two TPS genes of a Hosta species. These two key enzymes were placed in the evolutionary perspective of TPS families in various plants, and further related studies have been carried out. This study lays the foundation for further understanding the production process of terpenoids. Studies of terpene synthase are of great significance in terms of medicinal value and the genetic breeding of Hosta and other horticultural plants. More functional characterization and detailed chemical analysis of TPS will contribute to a better understanding of the evolution of TPS in Hosta and, more broadly, how fragrant Hosta evolved. We hope that more attention will be paid to Hosta plants with rich terpenoids. This is potentially a step toward improving the fragrance and, hence, the ornamental and economic value of Hosta plants. We believe that Hosta flowers should also be further evaluated, especially hybrids such as ‘So Sweet’, which may be a good source for studying terpene synthase activity and genetic engineering. Hosta flowers can also be used as raw materials in the spice industry.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/horticulturae8050447/s1.
Author Contributions
Conception and design of the study: B.C., S.Q., H.L. and S.L. (Shuying Liu); Realization of the experiments: X.Z., B.C., Y.L., Z.H., Q.Y., L.H. and S.L. (Sijia Liu); Analysis of the data: B.C., G.C. and S.Q.; Writing of the manuscript: B.C., X.Z., Y.L. and Q.Y.; Revising the content of the manuscript: S.L. (Shuying Liu), H.L. and B.C. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Project of Science and Technology Department of Jilin Province (20190301046NY) and the Strategic Priority Research Program of the Chinese Academy of Sciences (grant no. XDA28080400). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The raw/processed data required to reproduce these findings cannot be shared at this time, as the data also forms part of an ongoing study.
Acknowledgments
We would like to thank Zhou Zhengwen for his assistance with the plant growth, associate Liu shuying for provision of material and useful discussions, and Liu baofeng for invaluable technical assistance with the GC–MS. We are very grateful to Liu hongzhang for his advice and support.
Conflicts of Interest
The authors declare no conflict of interest or personal relationships that could have appeared to influence the work reported in this paper.
References
- Dudareva, N.; Klempien, A.; Muhlemann, J.K.; Kaplan, I. Biosynthesis, function and metabolic engineering of plant volatile organic compounds. New Phytol. 2013, 198, 16–32. [Google Scholar] [CrossRef] [PubMed]
- Pichersky, E.; Raguso, R.A. Why do plants produce so many terpenoid compounds? New Phytol. 2018, 220, 692–702. [Google Scholar] [CrossRef] [PubMed]
- Du, F.; Wang, T.; Fan, J.-M.; Liu, Z.-Z.; Zong, J.-X.; Fan, W.-X.; Han, Y.-H.; Grierson, D. Volatile composition and classification of Lilium flower aroma types and identification, polymorphisms, and alternative splicing of their monoterpene synthase genes. Hortic. Res. 2019, 6, 110. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Li, M.; Zhang, X.; Kong, W.; Bendahmane, M.; Bao, M.; Fu, X. Tissue-Specific Expression of the Terpene Synthase Family Genes in Rosa chinensis and Effect of Abiotic Stress Conditions. Genes 2022, 13, 547. [Google Scholar] [CrossRef]
- Huang, L.-M.; Huang, H.; Chuang, Y.-C.; Chen, W.-H.; Wang, C.-N.; Chen, H.-H. Evolution of Terpene Synthases in Orchidaceae. Int. J. Mol. Sci. 2021, 22, 6947. [Google Scholar] [CrossRef]
- Chen, F.; Tholl, D.; Bohlmann, J.; Pichersky, E. The family of terpene synthases in plants: A mid-size family of genes for specialized metabolism that is highly diversified throughout the kingdom. Plant J. Cell Mol. Biol. 2011, 66, 212–229. [Google Scholar] [CrossRef]
- Nomani, M.; Noori, S.A.S.; Tohidfar, M.; Ramshini, H. Overexpression of TPS2 gene to increase thymol content using Agrobacterium tumefaciens-mediated transformation in Trachyspermum ammi (Qom ecotype)—Sciencedirect. Ind. Crop. Prod. 2018, 130, 63–70. [Google Scholar] [CrossRef]
- Muhlemann, J.K.; Klempien, A.; Dudareva, N. Floral volatiles: From biosynthesis to function. Plant Cell Environ. 2014, 37, 1936–1949. [Google Scholar] [CrossRef]
- Vranová, E.; Coman, D.; Gruissem, W. Network analysis of the MVA and MEP pathways for isoprenoid synthesis. Annu. Rev. Plant Biol. 2013, 64, 665–700. [Google Scholar] [CrossRef]
- Tholl, D. Terpene synthases and the regulation, diversity and biological roles of terpene metabolism. Curr. Opin. Plant Biol. 2006, 9, 297–304. [Google Scholar] [CrossRef]
- Knudsen, J.T.; Eriksson, R.; Gershenzon, J.; Stahl, B. Diversity and distribution of floral scent. Bot. Rev. 2006, 72, 1–120. [Google Scholar] [CrossRef]
- Abbas, F.; Ke, Y.; Yu, R.; Yue, Y.; Amanullah, S.; Jahangir, M.M.; Fan, Y. Volatile terpenoids: Multiple functions, biosynthesis, modulation and manipulation by genetic engineering. Planta 2017, 246, 803–816. [Google Scholar] [CrossRef] [PubMed]
- Aharoni, A.; Giri, A.P.; Verstappen, F.W.; Bertea, C.M.; Sevenier, R.; Sun, Z.; Jongsma, M.A.; Schwab, W.; Bouwmeester, H.J. Gain and loss of fruit flavor compounds produced by wild and cultivated strawberry species. Plant Cell 2004, 16, 3110–3131. [Google Scholar] [CrossRef] [PubMed]
- Magnard, J.L.; Bony, A.R.; Bettini, F.; Campanaro, A.; Blerot, B.; Baudino, S.; Jullien, F. Linalool and linalool nerolidol synthases in roses, several genes for little scent. Plant Physiol. Biochem. 2018, 127, 74–87. [Google Scholar] [CrossRef] [PubMed]
- Aros, D.; Gonzalez, V.; Allemann, R.K.; Müller, C.T.; Rosati, C.; Rogers, H.J. Volatile emissions of scented Alstroemeria genotypes are dominated by terpenes, and a myrcene synthase gene is highly expressed in scented Alstroemeria flowers. J. Exp. Bot. 2012, 63, 2739–2752. [Google Scholar] [CrossRef]
- Yue, Y.; Yu, R.; Fan, Y. Characterization of two monoterpene synthases involved in floral scent formation in Hedychium coronarium. Planta 2014, 240, 745–762. [Google Scholar] [CrossRef]
- Gao, F.; Liu, B.; Li, M.; Gao, X.; Fang, Q.; Liu, C.; Ding, H.; Wang, L.; Gao, X. Identification and characterization of terpene synthase genes accounting for volatile terpene emissions in flowers of Freesia x hybrida. J. Exp. Bot. 2018, 69, 4249–4265. [Google Scholar] [CrossRef]
- Abbas, F.; Ke, Y.; Yu, R.; Fan, Y. Functional characterization and expression analysis of two terpene synthases involved in floral scent formation in Lilium ‘Siberia’. Planta 2019, 249, 71–93. [Google Scholar] [CrossRef]
- Parachnowitsch, A.L.; Burdon, R.C.; Raguso, R.A.; Kessler, A. Natural selection on floral volatile production in Penstemon digitalis: Highlighting the role of linalool. Plant Signal. Behav. 2013, 8, e22704. [Google Scholar] [CrossRef]
- Adler, L.S.; Irwin, R.E. What you smell is more important than what you see? Natural selection on floral scent. New Phytol. 2012, 195, 510–511. [Google Scholar] [CrossRef]
- Liu, J.X.; Zhao, C.H.; Liu, X.R.; Xi, Y.Z.; Zhang, Y.L. Pollen morphology of Hosta Tratt. in China and its taxonomic signifificance. Plant Syst. Evol. 2011, 294, 99–107. [Google Scholar] [CrossRef]
- Ryu, S.H.; Walck, J.L.; Hidayati, S.N.; Rhie, Y.H.; Yang, J.C.; Lee, S.Y. Embryo growth and dormancy in seeds of six Hosta species native to Korea. Plant Species Biol. 2019, 34, 166–173. [Google Scholar] [CrossRef]
- Jo, H.; Kim, M. A new variety of Hosta (Liliaceae): Hosta clausa var. Geumgangensis. Korean J. Plant Taxon. 2016, 46, 306–313. [Google Scholar] [CrossRef][Green Version]
- He, J.W.; Yang, L.; Zhong, G.Y. Research progress in chemical constituents, pharmacological activities, clinical practices and quality control of folk medicine Hosta plantaginea. Chin. Tradit. Herb. Drugs 2016, 47, 4295–4300. [Google Scholar] [CrossRef]
- Yang, L.; He, J.W. Hosta plantaginea (Lam.) Aschers (Yuzan): An overview on its botany, traditional use, phytochemistry, quality control and pharmacology. R. Soc. Chem. Adv. 2019, 9, 35050. [Google Scholar] [CrossRef] [PubMed]
- Grenfell, D.; Shadrack, M. The Color Encyclopedia of Hostas; Timber Press: London, UK, 2004; p. 85. [Google Scholar]
- Schmid, W.G. The Genus Hosta; Timber Press: London, UK, 2009; p. 278. [Google Scholar]
- Yang, L.; Jiang, S.T.; Zhou, Q.G.; Zhong, G.Y.; He, J.W. Chemical Constituents from the Flower of Hosta plantaginea with Cyclooxygenases Inhibition and Antioxidant Activities and Their Chemotaxonomic Significance. Molecules 2017, 22, 1825. [Google Scholar] [CrossRef]
- Yang, L.; He, J. Traditional uses, phytochemistry, pharmacology and toxicological aspects of the genus Hosta (Liliaceae): A comprehensive review. J. Ethnopharmacol. 2021, 265, 113323. [Google Scholar] [CrossRef]
- Sievers, F.; Higgins, D.G. Clustal Omega for making accurate alignments of many protein sequences. Protein Sci. A Publ. Protein Soc. 2018, 27, 135–145. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Robert, X.; Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014, 42, 320–324. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, 256–259. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Armenteros, J.J.A.; Salvatore, M.; Emanuelsson, O.; Winther, O.; von Heijne, G.; Elofsson, A.; Nielsen, H. Detecting sequence signals in targeting peptides using deep learning. Life Sci. Alliance 2019, 2, e201900429. [Google Scholar] [CrossRef]
- Sindarovska, Y.; Kuchuk, M. Long-Term Potato Virus X (PVX)-Based Transient Expression of Recombinant GFP Protein in Nicotiana benthamiana Culture In Vitro. Plants 2021, 10, 2187. [Google Scholar] [CrossRef] [PubMed]
- Bergman, M.E.; Davis, B.; Phillips, M.A. Medically Useful Plant Terpenoids: Biosynthesis, Occurrence, and Mechanism of Action. Molecules 2019, 24, 3961. [Google Scholar] [CrossRef] [PubMed]
- Martin, D.M.; Aubourg, S.; Schouwey, M.B.; Daviet, L.; Schalk, M.; Toub, O.; Lund, S.T.; Bohlmann, J. Functional annotation, genome organization and phylogeny of the grapevine (Vitis vinifera) terpene synthase gene family based on genome assembly, FLcDNA cloning, and enzyme assays. BMC Plant Biol. 2010, 10, 226. [Google Scholar] [CrossRef] [PubMed]
- Zhou, F.; Pichersky, E. More is better: The diversity of terpene metabolism in plants. Curr. Opin. Plant Biol. 2020, 55, 1–10. [Google Scholar] [CrossRef]
- Keilwagen, J.; Lehnert, H.; Berner, T.; Budahn, H.; Nothnagel, T.; Ulrich, D.; Dunemann, F. The Terpene Synthase Gene Family of Carrot (Daucus carota L.): Identification of QTLs and Candidate Genes Associated with Terpenoid Volatile Compounds. Front. Plant Sci. 2017, 8, 1930. [Google Scholar] [CrossRef]
- Chen, Z.; Vining, K.J.; Qi, X.; Yu, X.; Zheng, Y.; Liu, Z.; Fang, H.; Li, L.; Bai, Y.; Liang, C.; et al. Genome-Wide Analysis of Terpene Synthase Gene Family in Mentha longifolia and Catalytic Activity Analysis of a Single Terpene Synthase. Genes 2021, 12, 518. [Google Scholar] [CrossRef]
- Farré-Armengol, G.; Filella, I.; Llusià, J.; Peñuelas, J. β-Ocimene, a Key Floral and Foliar Volatile Involved in Multiple Interactions between Plants and Other Organisms. Molecules 2017, 22, 1148. [Google Scholar] [CrossRef]
- Leferink, N.G.H.; Ranaghan, K.E.; Battye, J.; Johannissen, L.O.; Hay, S.; van der Kamp, M.; Mulholland, A.J.; Scrutton, N.S. Taming the Reactivity of Monoterpene Synthases To Guide Regioselective Product Hydroxylation. ChemBioChem 2019, 21, 985–990. [Google Scholar] [CrossRef] [PubMed]
- Desai, J.; Liu, Y.-L.; Wei, H.; Liu, W.; Ko, T.-P.; Guo, R.-T.; Oldfield, E. Structure, Function, and Inhibition ofStaphylococcus aureusHeptaprenyl Diphosphate Synthase. ChemMedChem 2016, 11, 1915–1923. [Google Scholar] [CrossRef] [PubMed]
- Christianson, D.W. Structural and Chemical Biology of Terpenoid Cyclases. Chem. Rev. 2017, 117, 11570–11648. [Google Scholar] [CrossRef] [PubMed]
- Mostafa, S.; Wang, Y.; Zeng, W.; Jin, B. Floral Scents and Fruit Aromas: Functions, Compositions, Biosynthesis, and Regulation. Front. Plant Sci. 2022, 13, 860157. [Google Scholar] [CrossRef]
- Green, S.A.; Chen, X.; Nieuwenhuizen, N.J.; Matich, A.J.; Wang, M.Y.; Bunn, B.J.; Yauk, Y.K.; Atkinson, R.G. Identification, functional characterization, and regulation of the enzyme responsible for floral (E)-nerolidol biosynthesis in kiwifruit (Actinidia chinensis). J. Exp. Bot. 2012, 63, 1951–1967. [Google Scholar] [CrossRef] [PubMed]
- Pazouki, L.; Niinemets, Ü. Multi-Substrate Terpene Synthases: Their Occurrence and Physiological Significance. Front. Plant Sci. 2016, 12, 1019. [Google Scholar] [CrossRef]
- Ashaari, N.S.; Ab Rahim, M.H.; Sabri, S.; Lai, K.S.; Song, A.A.; Abdul Rahim, R.; Wan Abdullah, W.M.A.N.; Ong Abdullah, J. Functional characterization of a new terpene synthase from Plectranthus amboinicus. PLoS ONE. 2020, 15, e0235416. [Google Scholar] [CrossRef]
- Zhang, X.; da Silva, J.A.T.; Niu, M.; Zhang, T.; Liu, H.; Zheng, F.; Yuan, Y.; Li, Y.; Fang, L.; Zeng, S.; et al. Functional characterization of an Indian sandalwood (Santalum album L.) dual-localized bifunctional nerolidol/linalool synthase gene involved in stress response. Phytochemistry 2021, 183, 112610. [Google Scholar] [CrossRef]
- Jiang, Y.; Qian, R.; Zhang, W.; Wei, G.; Ma, X.; Zheng, J.; Köllner, T.G.; Chen, F. Composition and Biosynthesis of Scent Compounds from Sterile Flowers of an Ornamental Plant Clematis florida cv. ‘Kaiser’. Molecules 2020, 25, 1711. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).