Comprehensive Analysis of N6-Methyladenosine Regulatory Genes from Citrus grandis and Expression Profilings in the Fruits of “Huajuhong” (C. grandis “Tomentosa”) during Various Development Stages
Abstract
:1. Introduction
2. Materials and Methods
2.1. Identification and Chromosomal Localization of the m6A Regulatory Genes from C. grandis
2.2. Phylogenetic Analysis of the m6A Regulatory Genes from C. grandis, S. lycopersicum, and A. thaliana
2.3. Gene Structure Analysis, Cis-Element Analysis, and Heat Map Construction of the m6A Regulatory Genes from C. grandis
2.4. Plant Materials and Treatments
2.5. RNA Isolation and RT-PCR Analysis
3. Results
3.1. Identification of the m6A Regulatory Genes from C. grandis
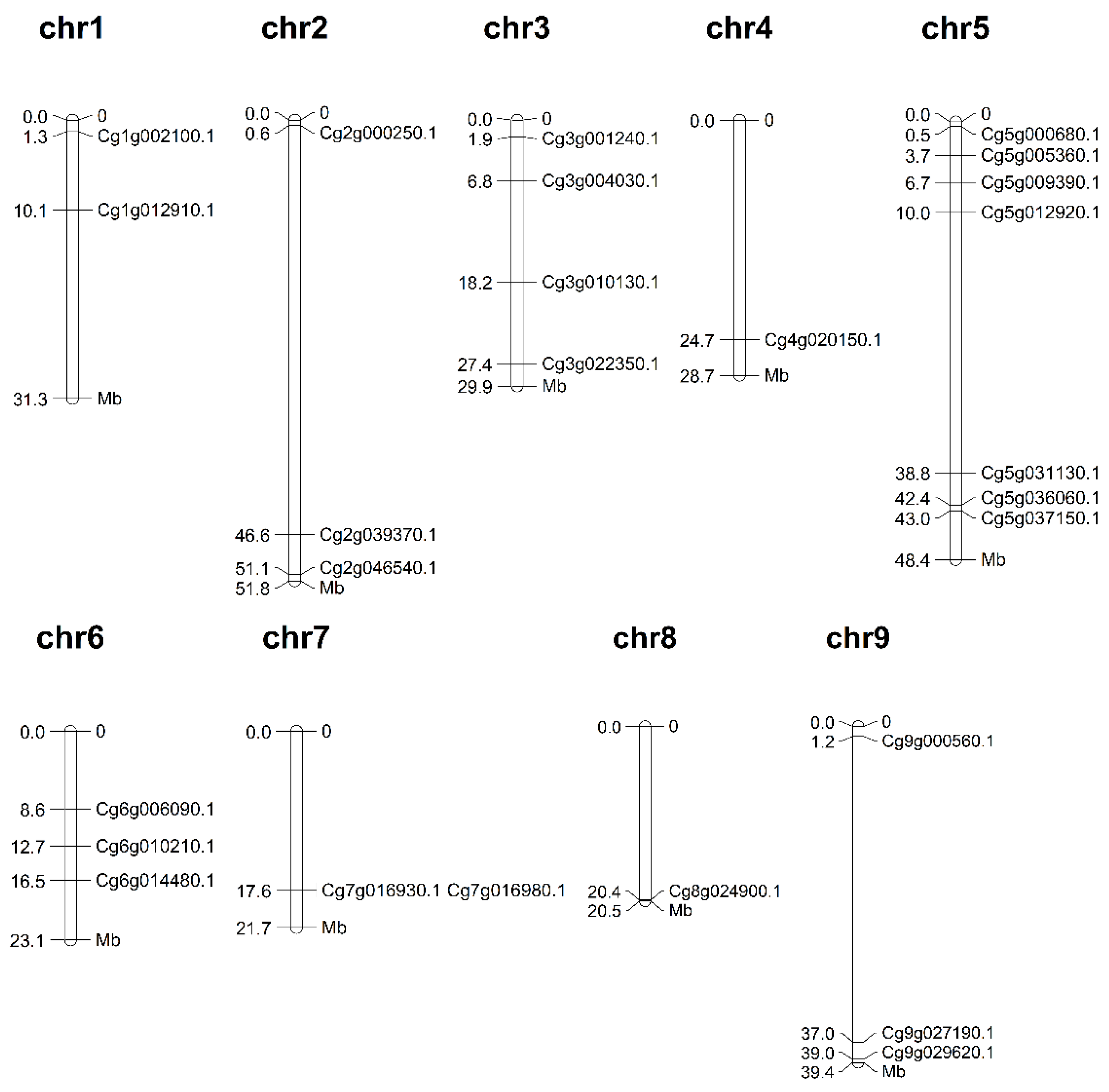
3.2. Chromosomal Localization of the m6A Regulatory Genes from C. grandis
3.3. Phylogenetic Analysis of the m6A Regulatory Genes from C. grandis, S. lycopersicum, and A. thaliana
3.4. Gene Structure Analysis of the m6A Regulatory Genes from C. grandis
3.5. Cis-Element Analysis of the m6A Regulatory Genes from C. grandis
3.6. Expression Patterns in the Fruits of C. grandis “Tomentosa” at Four Growth Points
4. Discussion
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Conflicts of Interest
References
- Matouk, C.C.; Marsden, P.A. Epigenetic regulation of vascular endothelial gene expression. Circ. Res. 2008, 102, 873–887. [Google Scholar] [CrossRef] [PubMed]
- Luger, K.; Richmond, T.J. The histone tails of the nucleosome. Curr. Opin. Genet. Dev. 1998, 8, 140–146. [Google Scholar] [CrossRef]
- Zhang, X. The epigenetic landscape of plants. Science 2008, 320, 489–492. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Dominissini, D.; Rechavi, G.; He, C. Gene expression regulation mediated through reversible m(6)A RNA methylation. Nat. Rev. Genet. 2014, 15, 293–306. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Zhang, S.; Zhou, C.; Xie, S.; Chen, G.; Tian, C.; Xu, K.; Lin, Y.; Lai, Z.; Guo, Y. Genome-Wide Investigation of N6-Methyladenosine Regulatory Genes and Their Roles in Tea (Camellia sinensis) Leaves during Withering Process. Front. Plant Sci. 2021, 12, 702303. [Google Scholar] [CrossRef]
- Yang, Y.; Hsu, P.J.; Chen, Y.S.; Yang, Y.G. Dynamic transcriptomic m(6)A decoration: Writers, erasers, readers and functions in RNA metabolism. Cell Res. 2018, 28, 616–624. [Google Scholar] [CrossRef] [Green Version]
- Saito, K. Phytochemical genomics—A new trend. Curr. Opin. Plant Biol. 2013, 16, 373–380. [Google Scholar] [CrossRef] [Green Version]
- Yue, H.; Nie, X.; Yan, Z.; Weining, S. N6-methyladenosine regulatory machinery in plants: Composition, function and evolution. Plant Biotechnol. J. 2019, 17, 1194–1208. [Google Scholar] [CrossRef]
- Sledz, P.; Jinek, M. Structural insights into the molecular mechanism of the m(6)A writer complex. eLife 2016, 5, e18434. [Google Scholar] [CrossRef]
- Wang, P.; Doxtader, K.A.; Nam, Y. Structural Basis for Cooperative Function of Mettl3 and Mettl14 Methyltransferases. Mol. Cell 2016, 63, 306–317. [Google Scholar] [CrossRef] [Green Version]
- Liang, Z.; Riaz, A.; Chachar, S.; Ding, Y.; Du, H.; Gu, X. Epigenetic Modifications of mRNA and DNA in Plants. Mol. Plant 2020, 13, 14–30. [Google Scholar] [CrossRef] [PubMed]
- Shen, L.; Liang, Z.; Gu, X.; Chen, Y.; Teo, Z.W.; Hou, X.; Cai, W.M.; Dedon, P.C.; Liu, L.; Yu, H. N(6)-Methyladenosine RNA Modification Regulates Shoot Stem Cell Fate in Arabidopsis. Dev. Cell 2016, 38, 186–200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruzicka, K.; Zhang, M.; Campilho, A.; Bodi, Z.; Kashif, M.; Saleh, M.; Eeckhout, D.; El-Showk, S.; Li, H.; Zhong, S.; et al. Identification of factors required for m(6) A mRNA methylation in Arabidopsis reveals a role for the conserved E3 ubiquitin ligase HAKAI. New Phytol. 2017, 215, 157–172. [Google Scholar] [CrossRef] [Green Version]
- Scarrow, M.; Chen, N.; Sun, G. Insights into the N(6)-methyladenosine mechanism and its functionality: Progress and questions. Crit. Rev. Biotechnol. 2020, 40, 639–652. [Google Scholar] [CrossRef]
- Martinez-Perez, M.; Aparicio, F.; Lopez-Gresa, M.P.; Belles, J.M.; Sanchez-Navarro, J.A.; Pallas, V. Arabidopsis m(6)A demethylase activity modulates viral infection of a plant virus and the m(6)A abundance in its genomic RNAs. Proc. Natl. Acad. Sci. USA 2017, 114, 10755–10760. [Google Scholar] [CrossRef] [Green Version]
- Zhou, L.; Tian, S.; Qin, G. RNA methylomes reveal the m(6)A-mediated regulation of DNA demethylase gene SlDML2 in tomato fruit ripening. Genome Biol. 2019, 20, 156. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Addepalli, B.; Hunt, A.G. A novel endonuclease activity associated with the Arabidopsis ortholog of the 30-kDa subunit of cleavage and polyadenylation specificity factor. Nucleic Acids Res. 2007, 35, 4453–4463. [Google Scholar] [CrossRef] [Green Version]
- Arribas-Hernandez, L.; Bressendorff, S.; Hansen, M.H.; Poulsen, C.; Erdmann, S.; Brodersen, P. An m(6)A-YTH Module Controls Developmental Timing and Morphogenesis in Arabidopsis. Plant Cell 2018, 30, 952–967. [Google Scholar] [CrossRef] [Green Version]
- Pontier, D.; Picart, C.; El Baidouri, M.; Roudier, F.; Xu, T.; Lahmy, S.; Llauro, C.; Azevedo, J.; Laudie, M.; Attina, A.; et al. The m(6)A pathway protects the transcriptome integrity by restricting RNA chimera formation in plants. Life Sci. Alliance 2019, 2, e201900393. [Google Scholar] [CrossRef] [Green Version]
- Velasco, R.; Licciardello, C. A genealogy of the citrus family. Nat. Biotechnol. 2014, 32, 640–642. [Google Scholar] [CrossRef]
- Tian, Y.; Fan, R.; Zeng, J. Comprehensive Analysis of Jumonji Domain C Family from Citrus grandis and Expression Profilings in the Exocarps of “Huajuhong” (Citrus grandis “Tomentosa”) during Various Development Stages. Horticulturae 2021, 7, 592. [Google Scholar] [CrossRef]
- Fan, R.; Zhu, C.; Qiu, D.; Zeng, J. Comparison of the bioactive chemical components and antioxidant activities in three tissues of six varieties of Citrus grandis ‘Tomentosa’ fruits. Int. J. Food Prop. 2019, 22, 1848–1862. [Google Scholar] [CrossRef] [Green Version]
- Berardini, T.Z.; Reiser, L.; Li, D.; Mezheritsky, Y.; Muller, R.; Strait, E.; Huala, E. The Arabidopsis information resource: Making and mining the “gold standard” annotated reference plant genome. Genesis 2015, 53, 474–485. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bombarely, A.; Menda, N.; Tecle, I.Y.; Buels, R.M.; Strickler, S.; Fischer-York, T.; Pujar, A.; Leto, J.; Gosselin, J.; Mueller, L.A. The Sol Genomics Network (solgenomics.net): Growing tomatoes using Perl. Nucleic Acids Res. 2011, 39, D1149–D1155. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Letunic, I.; Bork, P. 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 2018, 46, D493–D496. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; Hurwitz, D.I.; Marchler, G.H.; Song, J.S.; et al. CDD/SPARCLE: The conserved domain database in 2020. Nucleic Acids Res. 2020, 48, D265–D268. [Google Scholar] [CrossRef] [Green Version]
- Artimo, P.; Jonnalagedda, M.; Arnold, K.; Baratin, D.; Csardi, G.; de Castro, E.; Duvaud, S.; Flegel, V.; Fortier, A.; Gasteiger, E.; et al. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 2012, 40, W597–W603. [Google Scholar] [CrossRef]
- Zuo, C.; Liu, H.; Lv, Q.; Chen, Z.; Tian, Y.; Mao, J.; Chu, M.; Ma, Z.; An, Z.; Chen, B. Genome-Wide Analysis of the Apple (Malus domestica) Cysteine-Rich Receptor-Like Kinase (CRK) Family: Annotation, Genomic Organization, and Expression Profiles in Response to Fungal Infection. Plant Mol. Biol. Report. 2019, 38, 14–24. [Google Scholar] [CrossRef]
- Xie, T.; Chen, C.; Li, C.; Liu, J.; Liu, C.; He, Y. Genome-wide investigation of WRKY gene family in pineapple: Evolution and expression profiles during development and stress. BMC Genom. 2018, 19, 490. [Google Scholar] [CrossRef] [Green Version]
- Hu, R.; Qi, G.; Kong, Y.; Kong, D.; Gao, Q.; Zhou, G. Comprehensive analysis of NAC domain transcription factor gene family in Populus trichocarpa. BMC Plant Biol. 2010, 10, 145. [Google Scholar] [CrossRef] [Green Version]
- Ren, J.; Wen, L.; Gao, X.; Jin, C.; Xue, Y.; Yao, X. DOG 1.0: Illustrator of protein domain structures. Cell Res. 2009, 19, 271–273. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Lescot, M.; Dehais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouze, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Fan, R.; Yang, Q.; Hu, C.; Sheng, O.; Deng, G.; Dong, T.; Li, C.; Peng, X.; Bi, F.; et al. Genome-Wide Identification and Characterization of the NAC Transcription Factor Family in Musa Acuminata and Expression Analysis during Fruit Ripening. Int. J. Mol. Sci. 2020, 21, 634. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takeuchi, T.; Yamazaki, Y.; Katoh-Fukui, Y.; Tsuchiya, R.; Kondo, S.; Motoyama, J.; Higashinakagawa, T. Gene trap capture of a novel mouse gene, jumonji, required for neural tube formation. Genes Dev. 1995, 9, 1211–1222. [Google Scholar] [CrossRef] [Green Version]
- Akimoto, K.; Katakami, H.; Kim, H.J.; Ogawa, E.; Sano, C.M.; Wada, Y.; Sano, H. Epigenetic inheritance in rice plants. Ann. Bot. 2007, 100, 205–217. [Google Scholar] [CrossRef] [Green Version]
- Shen, Y.; Conde, E.S.N.; Audonnet, L.; Servet, C.; Wei, W.; Zhou, D.X. Over-expression of histone H3K4 demethylase gene JMJ15 enhances salt tolerance in Arabidopsis. Front. Plant Sci. 2014, 5, 290. [Google Scholar] [CrossRef] [Green Version]
- Sharma, A.; Singh, K.; Almasan, A. Histone H2AX phosphorylation: A marker for DNA damage. Methods Mol. Biol. 2012, 920, 613–626. [Google Scholar] [CrossRef]
- Scutenaire, J.; Deragon, J.M.; Jean, V.; Benhamed, M.; Raynaud, C.; Favory, J.J.; Merret, R.; Bousquet-Antonelli, C. The YTH Domain Protein ECT2 Is an m(6)A Reader Required for Normal Trichome Branching in Arabidopsis. Plant Cell 2018, 30, 986–1005. [Google Scholar] [CrossRef] [Green Version]
- Miao, Z.; Zhang, T.; Qi, Y.; Song, J.; Han, Z.; Ma, C. Evolution of the RNA N (6)-Methyladenosine Methylome Mediated by Genomic Duplication. Plant Physiol. 2020, 182, 345–360. [Google Scholar] [CrossRef] [Green Version]
- Hu, J.; Manduzio, S.; Kang, H. Epitranscriptomic RNA Methylation in Plant Development and Abiotic Stress Responses. Front. Plant Sci. 2019, 10, 500. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, J.; Wan, J.; Gao, X.; Zhang, X.; Jaffrey, S.R.; Qian, S.B. Dynamic m(6)A mRNA methylation directs translational control of heat shock response. Nature 2015, 526, 591–594. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, J.; Wan, J.; Shu, X.E.; Mao, Y.; Liu, X.M.; Yuan, X.; Zhang, X.; Hess, M.E.; Bruning, J.C.; Qian, S.B. N(6)-Methyladenosine Guides mRNA Alternative Translation during Integrated Stress Response. Mol. Cell 2018, 69, 636–647.e637. [Google Scholar] [CrossRef] [Green Version]
- Duan, H.C.; Wei, L.H.; Zhang, C.; Wang, Y.; Chen, L.; Lu, Z.; Chen, P.R.; He, C.; Jia, G. ALKBH10B Is an RNA N(6)-Methyladenosine Demethylase Affecting Arabidopsis Floral Transition. Plant Cell 2017, 29, 2995–3011. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kramer, M.C.; Janssen, K.A.; Palos, K.; Nelson, A.D.L.; Vandivier, L.E.; Garcia, B.A.; Lyons, E.; Beilstein, M.A.; Gregory, B.D. N(6)-methyladenosine and RNA secondary structure affect transcript stability and protein abundance during systemic salt stress in Arabidopsis. Plant Direct. 2020, 4, e00239. [Google Scholar] [CrossRef] [PubMed]
- Xia, E.; Tong, W.; Hou, Y.; An, Y.; Chen, L.; Wu, Q.; Liu, Y.; Yu, J.; Li, F.; Li, R.; et al. The Reference Genome of Tea Plant and Resequencing of 81 Diverse Accessions Provide Insights into Its Genome Evolution and Adaptation. Mol. Plant 2020, 13, 1013–1026. [Google Scholar] [CrossRef]
- Zaccara, S.; Ries, R.J.; Jaffrey, S.R. Reading, writing and erasing mRNA methylation. Nat. Rev. Mol. Cell Biol. 2019, 20, 608–624. [Google Scholar] [CrossRef]
- Tian, R.; Yang, Y.; Chen, M. Genome-wide survey of the amino acid transporter gene family in wheat (Triticum aestivum L.): Identification, expression analysis and response to abiotic stress. Int. J. Biol. Macromol. 2020, 162, 1372–1387. [Google Scholar] [CrossRef]
- Pellagatti, A.; Boultwood, J. Splicing factor gene mutations in the myelodysplastic syndromes: Impact on disease phenotype and therapeutic applications. Adv. Biol. Regul. 2017, 63, 59–70. [Google Scholar] [CrossRef]
- Sun, Z.; Wang, X.; Qiao, K.; Fan, S.; Ma, Q. Genome-wide analysis of JMJ-C histone demethylase family involved in salt-tolerance in Gossypium hirsutum L. Plant Physiol. Biochem. 2021, 158, 420–433. [Google Scholar] [CrossRef]
- Li, P.L.; Liu, M.H.; Hu, J.H.; Su, W.W. Systematic chemical profiling of Citrus grandis ‘Tomentosa’ by ultra-fast liquid chromatography/diode-array detector/quadrupole time-of-flight tandem mass spectrometry. J. Pharm. Biomed. Anal. 2014, 90, 167–179. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Liu, Q.; Xie, Z.; Lam, S.; Xu, X. Chromatographic Fingerprint Analysis of Exocarpium Citri Grandis by High-Performance Liquid Chromatography Coupled with Diode-Array Detector. Food Anal. Methods 2014, 8, 1868–1875. [Google Scholar] [CrossRef]





| Genes Names | Forward Primers | Reverse Primers |
|---|---|---|
| Actin | ATCTGCTGGAAGGTGCTGAG | CCAAGCAGCATGAAGATCAA |
| CgMTA | GCGTCCTGAATATTGTTCTGAAGTA | AACTCCAAATTGTCCTAAAATGTCC |
| CgMTB | CAACTCGAAAGCCTGAAGATATGTA | GCAAAGTTTTTAATGTATGCCTCCT |
| CgMTC | AACTTACTCATTCAGAGGGAGCACT | CATCTAGCTTCACCTTCAACCAGTA |
| CgVIR | TGATTTTCTTGCTAGTTTGTTGGAG | CACCAGCTAGTCAAAGTAGAACCAA |
| CgHAKAI | TAGCTAGGGAAGAAGGTATGGTGTC | AGTGTCTTCACAGGCTCATACAATC |
| CgFIP37 | ATGAAATGGTGCTTATGTTACGAGA | AACTGAATCATTCTTGTCCTCTTCC |
| CgALKBH1 | TTGAAAGAGAAAACGTATTCAGCAC | GATCACTTTAAACAGATGAGGCAGA |
| CgALKBH2 | ATTGGAATTCAGATTGGAGAAACTC | TCTTTAGTATCAGCTATGGCAGGTG |
| CgALKBH3 | TGGAAATTCTTTGATTACCTCAACA | ATTGTGTCACTCCTTCACTAGCAAC |
| CgALKBH4 | TCATTCTAAATGGTAATGGAGCTGA | ACTTGTTTGAGGGAGAATTAACCAC |
| CgALKBH5 | TCATTTCCAAACCTTGATGATTCTA | AGGCTGAAAATTAATCCTTCAAATG |
| CgALKBH6 | CATCAATGAATACCAACCTAACCAA | AAATGCATCACCATTAGAACTTTCA |
| CgALKBH7 | CTAGAACCAATTCCAGCTCTTCTTC | ATCAAAGAAGTTAATGATGCAACCA |
| CgALKBH8 | AGGGAAGAATAGATAATCCACATGC | ATTTATCTCGTGCTTCCAAAGGTAG |
| CgALKBH9 | CGAAAGCAGATACAGTCTCAAGAAG | CCCCACCAAATATTAGTACATCTCC |
| CgALKBH10 | CCCATAGATGACTTATTCAATGCTG | AAAATTTCCACCTATGAGCATCTTC |
| CgECT1 | GTTAATACAAGTGGGCAGTTTGTTG | AAGTCATTCCAGTTTGTCCTAGCTG |
| CgECT2 | TAACATGTACAAGCTGGGTTTTTGT | AAATGTTTATTAGGGTTGCCATGAT |
| CgECT3 | ACAGTCTATGAAACCAGTGAGCAAG | AAGCCTTGTTTTTGGTTAGAAAATG |
| CgECT4 | GCTAAACAACAAGTTAGCCTGACAA | TTGAGAGAGCACCAATAATACTTGC |
| CgECT5 | GGAAGCCTTTCATAACTCTGGTAAA | AATCGAAGCCATTTGACTTTAAAAC |
| CgECT6 | GAAGATTTTCCCGAGAGTTACTCAG | ATATGCTGCATCAAGCTTCTTATTG |
| CgECT7 | TTTCTGTTGTTTTCGGTAAATGCTA | CATCCTTAATTATGTGCCACTTGAC |
| CgECT8 | TTCTTTATGTGGTTGCAATGGTACT | TCAAGAATCTTAACCACAGACTTGC |
| CgECT9 | GGAAATGTATAATGCGTCTGACAAC | AATAAACTCAGAGGCTCCTTCGTTA |
| CgECT10 | ACCTTTAGAGGAAATGATGACGAAC | ATCCATAATGTGCTCCAGTCTGATA |
| Gene Names | Annotation Number | AAs | Mw (kDa) | pI | Subcellular Localization |
|---|---|---|---|---|---|
| CgALKBH1 | Cg1g002100.1 | 515 | 57.94 | 6.59 | cyto |
| CgALKBH2 | Cg1g012910.1 | 624 | 68.53 | 7.58 | nucl |
| CgECT1 | Cg2g000250.1 | 711 | 77.99 | 7.57 | nucl |
| CgECT2 | Cg2g039370.1 | 701 | 76.31 | 6.26 | nucl |
| CgALKBH3 | Cg2g046540.1 | 245 | 28.73 | 9.50 | nucl |
| CgECT3 | Cg3g001240.1 | 572 | 63.06 | 6.34 | nucl |
| CgALKBH4 | Cg3g004030.1 | 464 | 51.92 | 8.52 | nucl |
| CgALKBH5 | Cg3g010130.1 | 874 | 97.61 | 6.09 | nucl |
| CgMTC | Cg3g022350.1 | 408 | 46.90 | 6.47 | cyto |
| CgECT4 | Cg4g020150.1 | 616 | 68.11 | 4.94 | nucl |
| CgMTA | Cg5g000680.1 | 710 | 78.81 | 5.94 | cyto |
| CgFIP37 | Cg5g005360.1 | 342 | 38.76 | 5.21 | nucl |
| CgECT5 | Cg5g009390.1 | 398 | 45.00 | 6.32 | nucl |
| CgALKBH6 | Cg5g012920.1 | 195 | 22.14 | 6.05 | cyto |
| CgECT6 | Cg5g031130.1 | 611 | 66.88 | 6.51 | nucl |
| CgALKBH7 | Cg5g036060.1 | 547 | 59.98 | 5.56 | cyto |
| CgALKBH8 | Cg5g037150.1 | 260 | 29.84 | 4.50 | nucl |
| CgHAKAI | Cg6g006090.1 | 779 | 83.60 | 8.23 | cyto |
| CgALKBH9 | Cg6g010210.1 | 458 | 51.51 | 9.06 | cyto |
| CgVIR | Cg6g014480.1 | 2199 | 241.16 | 5.34 | nucl |
| CgECT7 | Cg7g016930.1 | 631 | 69.52 | 6.45 | nucl |
| CgALKBH10 | Cg7g016980.1 | 361 | 40.75 | 6.04 | nucl |
| CgECT8 | Cg8g024900.1 | 656 | 71.85 | 5.41 | nucl |
| CgECT9 | Cg9g000560.1 | 696 | 76.43 | 5.88 | nucl |
| CgMTB | Cg9g027190.1 | 1189 | 133.49 | 8.19 | nucl |
| CgECT10 | Cg9g029620.1 | 552 | 61.01 | 6.08 | nucl |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tian, Y.; Zeng, J.; Fan, R. Comprehensive Analysis of N6-Methyladenosine Regulatory Genes from Citrus grandis and Expression Profilings in the Fruits of “Huajuhong” (C. grandis “Tomentosa”) during Various Development Stages. Horticulturae 2022, 8, 462. https://doi.org/10.3390/horticulturae8050462
Tian Y, Zeng J, Fan R. Comprehensive Analysis of N6-Methyladenosine Regulatory Genes from Citrus grandis and Expression Profilings in the Fruits of “Huajuhong” (C. grandis “Tomentosa”) during Various Development Stages. Horticulturae. 2022; 8(5):462. https://doi.org/10.3390/horticulturae8050462
Chicago/Turabian StyleTian, Yuzhen, Jiwu Zeng, and Ruiyi Fan. 2022. "Comprehensive Analysis of N6-Methyladenosine Regulatory Genes from Citrus grandis and Expression Profilings in the Fruits of “Huajuhong” (C. grandis “Tomentosa”) during Various Development Stages" Horticulturae 8, no. 5: 462. https://doi.org/10.3390/horticulturae8050462
APA StyleTian, Y., Zeng, J., & Fan, R. (2022). Comprehensive Analysis of N6-Methyladenosine Regulatory Genes from Citrus grandis and Expression Profilings in the Fruits of “Huajuhong” (C. grandis “Tomentosa”) during Various Development Stages. Horticulturae, 8(5), 462. https://doi.org/10.3390/horticulturae8050462






