An Identification and Expression Analysis of the ABCG Genes Related to Benzaldehyde Transportation among Three Prunus Species
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Collection and Analysis of Volatile Components
2.3. Extraction and Analysis of the Endogenous Compounds in Flowers
2.4. Identification of ABC Gene Family Members
2.5. Physical and Chemical Properties and Subcellular Location of PmABC, PpABC, and PaABC Members
2.6. Collinearity Analysis of ABC Genes and Phylogenetic Tree Construction
2.7. ABCG Genes Expression Patterns Analysis
2.8. Weighted Gene Co-Expression Network Analysis (WGCNA)
2.9. Quantitative Expression Analysis of PmABC Members
3. Results
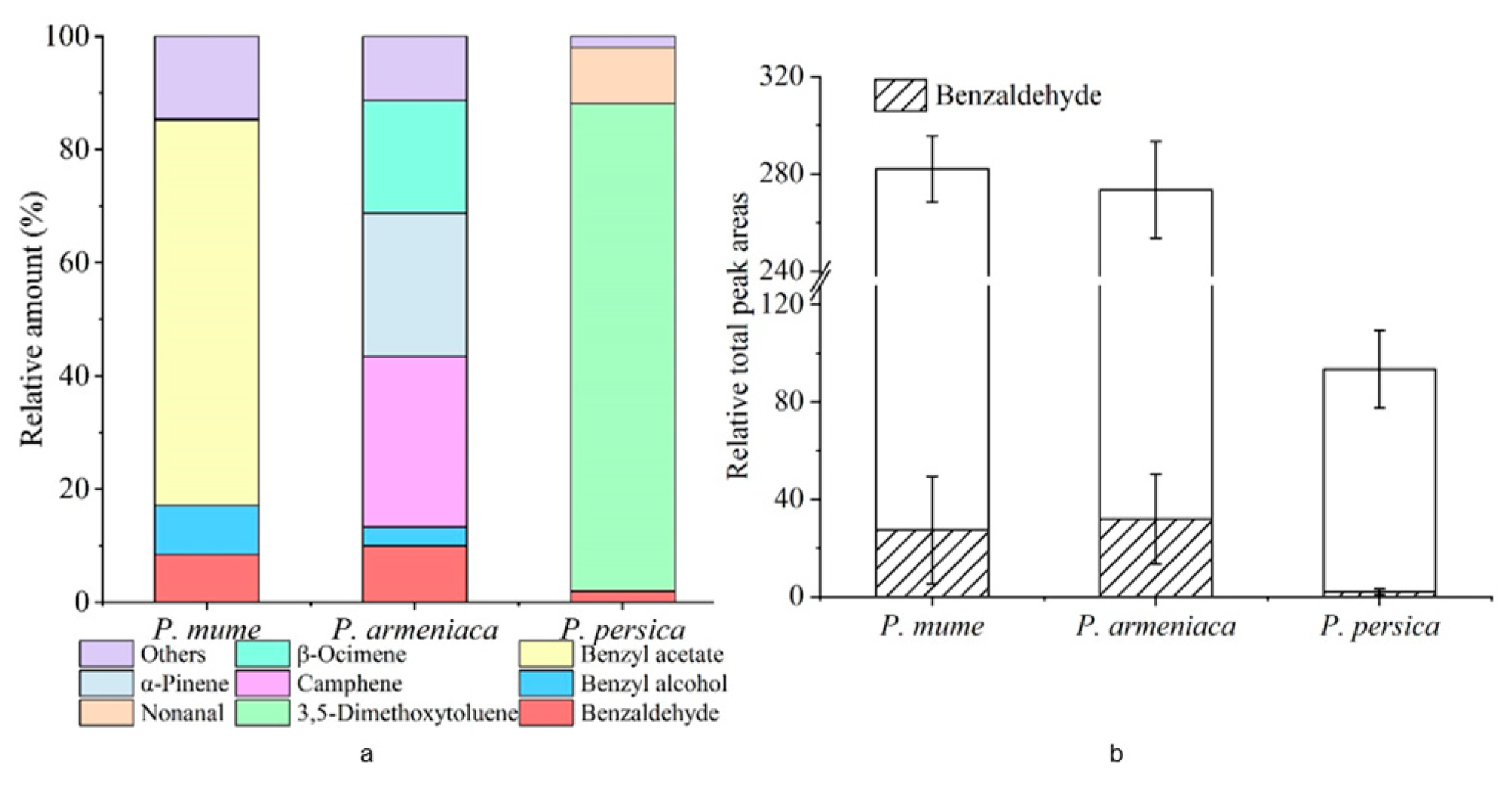
3.1. Floral Scent Components Analysis in P. mume, P. armeniaca, and P. persica
3.2. Identification of ABC Family Members in P. mume, P. armeniaca, and P. persica
3.3. Interspecific Collinearity Analysis of PmABC, PaABC, and PpABC
3.4. Phylogenetic Analysis of the ABCG Genes in P. mume, P. armeniaca, and P. persica
3.5. Transcription Expression Analysis of PmABCG, PaABCG and PpABCG Family Members
3.6. WGCNA Analysis between Benzaldehyde Volatilization and PmABCGs Gene Expression
3.7. qRT-RCR Analysis of the Candidate ABCG Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mark, R.; Duemmel, M.J. Comparison of drought resistance among Prunus species from divergent habitats. Tree Physiol. 1992, 11, 369–380. [Google Scholar] [CrossRef]
- Xue, S.; Shi, T.; Luo, W.; Ni, X.; Iqbal, S.; Ni, Z.; Huang, X.; Yao, D.; Shen, Z.; Gao, Z. Comparative analysis of the complete chloroplast genome among Prunus mume, P. armeniaca, and P. salicina. Hortic. Res. 2019, 6, 89. [Google Scholar] [CrossRef] [Green Version]
- Tan, Q.; Li, S.; Zhang, Y.; Chen, M.; Wen, B.; Jiang, S.; Chen, X.; Fu, X.; Li, D.; Wu, H.; et al. Chromosome-level genome assemblies of five Prunus species and genome-wide association studies for key agronomic traits in peach. Hortic. Res. 2021, 8, 213. [Google Scholar] [CrossRef]
- Dudareva, N.; Klempien, A.; Muhlemann, J.K.; Kaplan, I. Biosynthesis, function and metabolic engineering of plant volatile organic compounds. New Phytol. 2013, 198, 16–32. [Google Scholar] [CrossRef]
- Zhang, T.; Bao, F.; Yang, Y.; Hu, L.; Ding, A.; Ding, A.; Wang, J.; Cheng, T.; Zhang, Q. A comparative analysis of floral scent compounds in intraspecific cultivars of Prunus mume with different corolla colours. Molecules 2020, 25, 145. [Google Scholar] [CrossRef] [Green Version]
- Adebesin, F.; Widhalm, J.R.; Boachon, B.; Lefèvre, F.; Pierman, B.; Lynch, J.H.; Alam, I.; Junqueira, B.; Benke, R.; Ray, S.; et al. Emission of volatile organic compounds from petunia flowers is facilitated by an ABC transporter. Science 2017, 356, 1386–1388. [Google Scholar] [CrossRef] [Green Version]
- Niinemets, Ü.; Arneth, A.; Kuhn, U.; Monson, R.K.; Peñuelas, J.; Staudt, M. The emission factor of volatile isoprenoids: Stress, acclimation, and developmental responses. Biogeosciences 2010, 7, 2203–2223. [Google Scholar] [CrossRef] [Green Version]
- Ward, A.; Reyes, C.L.; Yu, J.; Roth, C.B.; Chang, G. Flexibility in the ABC transporter MsbA: Alternating access with a twist. Proc. Natl. Acad. Sci. USA 2007, 104, 19005–19010. [Google Scholar] [CrossRef] [Green Version]
- Davies, T.G.E.; Coleman, J.O.D. The Arabidopsis thaliana ATP-binding cassette proteins: An emerging superfamily. Plant Cell Environ. 2000, 23, 431–443. [Google Scholar] [CrossRef] [Green Version]
- Rajsz, A.; Warzybok, A.; Migocka, M. Genes encoding cucumber full-size ABCG proteins show different responses to plant growth regulators and sclareolide. Plant Mol. Biol. Rep. 2016, 34, 720–736. [Google Scholar] [CrossRef]
- Verrier, P.J.; Bird, D.; Burla, B.; Dassa, E.; Forestier, C.; Geisler, M.; Klein, M.; Kolukisaoglu, Ü.; Lee, Y.; Martinoia, E.; et al. Plant ABC proteins–a unified nomenclature and updated inventory. Trends Plant Sci. 2008, 13, 151–159. [Google Scholar] [CrossRef] [Green Version]
- Dhara, A.; Raichaudhuri, A. ABCG transporter proteins with beneficial activity on plants. Phytochemistry 2021, 184, 112663. [Google Scholar] [CrossRef]
- Banasiak, J.; Jasiński, M. Defence, Symbiosis and ABCG Transporters. In Plant ABC Transporters; Geisler, M., Ed.; Springer: Cham, Switzerland, 2014; Volume 22, pp. 163–184. [Google Scholar]
- Choi, H.; Jin, J.Y.; Choi, S.; Hwang, J.U.; Kim, Y.Y.; Suh, M.C.; Lee, Y. An ABCG/WBC-type ABC transporter is essential for transport of sporopollenin precursors for exine formation in developing pollen. Plant J. 2011, 65, 181–193. [Google Scholar] [CrossRef]
- Schiestl, F.P. The evolution of floral scent and insect chemical communication. Ecol. Lett. 2010, 13, 643–656. [Google Scholar] [CrossRef]
- Kelley, K.C.; Schilling, A.B. Quantitative variation in chemical defense within and among subgenera of Cicindela. J. Chem. Ecol. 1998, 24, 451–472. [Google Scholar] [CrossRef]
- Hao, R.; Du, D.; Wang, T.; Yang, W.; Zhang, Q. A comparative analysis of characteristic floral scent compounds in Prunus mume and related species. Biosci. Biotechnol. Biochem. 2014, 78, 1640–1647. [Google Scholar] [CrossRef] [Green Version]
- Pasteels, J.M.; Grégoire, J.C.; Rowell-Rahier, M. The chemical ecology of defense in arthropods. Annu. Rev. Entomol. 1983, 28, 263–289. [Google Scholar] [CrossRef]
- Hu, Z.; Shen, Y.; Luo, Y.; Shen, F.; Gao, H.; Gao, R. Aldehyde volatiles emitted in succession from mechanically damaged leaves of poplar cuttings. J. Plant Biol. 2008, 51, 269–275. [Google Scholar] [CrossRef]
- Li, S.; Chen, L.; Xu, Y.; Wang, L.; Wang, L. Identification of floral fragrances in tree peony cultivars by gas chromatography–mass spectrometry. Sci. Hortic. 2012, 142, 158–165. [Google Scholar] [CrossRef]
- Hao, R.; Yang, S.; Zhang, Z.; Zhang, Y.; Chang, J.; Qiu, C. Identification and specific expression patterns in flower organs of ABCG genes related to floral scent from Prunus mume. Sci. Hortic. 2021, 288, 110218. [Google Scholar] [CrossRef]
- Kondo, M.; Oyama-Okubo, N.; Ando, T.; Marchesi, E.; Nakayama, M. Floral scent diversity is differently expressed in emitted and endogenous components in Petunia axillaris lines. Ann. Bot. 2006, 98, 1253–1259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Q.; Chen, W.; Sun, L.; Zhao, F.; Huang, B.; Yang, W.; Tao, Y.; Wang, J.; Yuan, Z.; Fan, G.; et al. The genome of Prunus mume. Nat. Commun. 2012, 3, 1318. [Google Scholar] [CrossRef] [PubMed]
- Verde, I.; Jenkins, J.; Dondini, L.; Micali, S.; Pagliarani, G.; Vendramin, E.; Paris, R.; Aramini, V.; Gazza, L.; Rossini, L.; et al. The Peach v2. 0 release: High-resolution linkage mapping and deep resequencing improve chromosome-scale assembly and contiguity. BMC Genom. 2017, 18, 225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, F.; Zhang, J.; Wang, S.; Yang, L.; Luo, Y.; Gao, S.; Zhang, M.; Wu, S.; Hu, S.; Sun, H.; et al. The apricot (Prunus armeniaca L.) genome elucidates Rosaceae evolution and beta-carotenoid synthesis. Hortic. Res. 2019, 6, 128. [Google Scholar] [CrossRef] [Green Version]
- Lane, T.S.; Rempe, C.S.; Davitt, J.; Staton, M.E.; Peng, Y.; Soltis, D.E.; Melkonian, M.; Deyholos, M.; Leebens-Mack, J.H.; Chase, M.; et al. Diversity of ABC transporter genes across the plant kingdom and their potential utility in biotechnology. BMC Biotechnol. 2016, 16, 47. [Google Scholar] [CrossRef] [Green Version]
- Sun, T.; Jia, D.; Huang, L.; Shao, Y.; Ma, F. Comprehensive genomic identification and expression analysis of the nucleobase-ascorbate transporter (NAT) gene family in apple. Sci. Hortic. 2016, 198, 473–481. [Google Scholar] [CrossRef]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.; Marra, M. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.P.; Tang, H.B.; Debarry, J.D.; Tan, X.; Li, J.P.; Wang, X.Y.; Lee, T.; Jin, H.Z.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Ding, Y.; Wang, X.; Chen, H.; Cao, H.; Niu, L.; Pan, L.; Lu, Z.; Cui, G.; Zeng, W.; et al. Analysis of PpGLV gene family suggests that PpGLV4 peptide coordinates auxin and ethylene signaling in peach. Sci. Hortic. 2019, 246, 12–20. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef] [Green Version]
- Jaakola, L.; Pirttilä, A.; Halonen, M.; Hohtola, A. Isolation of high quality RNA from bilberry (Vaccinium myrtillus L.) fruit. Mol. Biotechnol. 2001, 19, 201–203. [Google Scholar] [CrossRef]
- Wang, T.; Lu, J.; Xu, Z.; Yang, W.; Wang, J.; Cheng, T.; Zhang, Q. Selection of suitable reference genes for miRNA expression normalization by qRT-PCR during flower development and different genotypes of Prunus mume. Sci. Hortic. 2014, 169, 130–137. [Google Scholar] [CrossRef]
- Adnan, M.; Morton, G.; Hadi, S. Analysis of rpoS and bolA gene expression under various stress-induced environments in planktonic and biofilm phase using 2−ΔΔCT method. Mol. Cell Biochem. 2011, 357, 275–282. [Google Scholar] [CrossRef] [PubMed]
- Knudsen, J.T. Floral scent differentiation among coflowering, sympatric species of Geonoma (Arecaceae). Plant Speci. Biol. 1999, 14, 137–142. [Google Scholar] [CrossRef]
- Suinyuy, T.N.; Donaldson, J.S.; Johnson, S.D. Geographical variation in cone volatile composition among populations of the African cycad Encephalartos villosus. Biol. J. Linn. Soc. 2012, 106, 514–527. [Google Scholar] [CrossRef]
- Gong, W.C.; Chen, G.; Liu, C.Q.; Dunn, B.L.; Sun, W.B. Comparison of floral scent between and within Buddleja fallowiana and Buddleja officinalis (Scrophulariaceae). Biochem. Syst. Ecol. 2014, 55, 322–328. [Google Scholar] [CrossRef]
- Jantzen, F.; Lynch, J.H.; Kappel, C.; Höfflin, J.; Skaliter, O.; Wozniak, N.; Sicard, A.; Sas, C.; Adebesin, F.; Ravid, J. Retracing the molecular basis and evolutionary history of the loss of benzaldehyde emission in the genus Capsella. New Phytol. 2019, 224, 1349–1360. [Google Scholar] [CrossRef] [Green Version]
- El-Sayed, A.M.; Sporle, A.; Colhoun, K.; Furlong, J.; White, R.; Suckling, D.M. Scents in orchards: Floral volatiles of four stone fruit crops and their attractiveness to pollinators. Chemoecology 2018, 28, 39–49. [Google Scholar] [CrossRef]
- Townsend, G.F. Benzaldehyde: A new repellent for driving bees. Bee. World 1963, 44, 146–149. [Google Scholar] [CrossRef]
- Hoballah, M.E.; Stuurman, J.; Turlings, T.C.J.; Guerin, P.M.; Connetable, S.; Kuhlemeier, C. The composition and timing of flower odour emission by wild Petunia axillaris coincide with the antennal perception and nocturnal activity of the pollinator Manduca sexta. Planta 2005, 222, 141–150. [Google Scholar] [CrossRef] [Green Version]
- Moerkercke, A.V. The Floral Volatile Phenylpropanoid/Benzenoid Pathway in Petunia. Ph.D. Thesis, University of Amsterdam, Amsterdam, The Netherlands, 2011. [Google Scholar]
- Kretzschmar, T.; Burla, B.; Lee, Y.; Martinoia, E.; Nagy, R. Functions of ABC transporters in plants. Essays Biochem. 2011, 50, 145–160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moitra, K.; Dean, M. Evolution of ABC transporters by gene duplication and their role in human disease. Biol. Chem. 2011, 392, 29–37. [Google Scholar] [CrossRef] [PubMed]
- Xiong, J.; Feng, J.; Yuan, D.; Zhou, J.; Miao, W. Tracing the structural evolution of eukaryotic ATP binding cassette transporter superfamily. Sci. Rep. 2015, 5, 16724. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Collauto, A.; Mishra, S.; Litvinov, A.; Mchaourab, H.S.; Goldfarb, D. Direct Spectroscopic Detection of ATP Turnover Reveals Mechanistic Divergence of ABC Exporters. Structure 2017, 25, 1264–1274.e3. [Google Scholar] [CrossRef] [PubMed]
- Foster, A.J.; Gervais, P.; Philippe, T.; Seguin, A. Transcriptome analysis of poplar during leaf spot infection with Sphaerulina spp. PLoS ONE 2015, 10, e0138162. [Google Scholar] [CrossRef] [Green Version]
- Srikant, S. Evolutionary history of ATP-binding cassette proteins. FEBS Lett. 2020, 594, 3882–3897. [Google Scholar] [CrossRef]
- Srikant, S.; Gaudet, R.; Murray, A.W. Selecting for altered substrate specificity reveals the evolutionary flexibility of ATP-binding cassette transporters. Curr. Biol. 2020, 30, 1689–1702.e6. [Google Scholar] [CrossRef] [Green Version]







Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hao, R.; Chang, J.; Qiu, C.; Yang, S. An Identification and Expression Analysis of the ABCG Genes Related to Benzaldehyde Transportation among Three Prunus Species. Horticulturae 2022, 8, 475. https://doi.org/10.3390/horticulturae8060475
Hao R, Chang J, Qiu C, Yang S. An Identification and Expression Analysis of the ABCG Genes Related to Benzaldehyde Transportation among Three Prunus Species. Horticulturae. 2022; 8(6):475. https://doi.org/10.3390/horticulturae8060475
Chicago/Turabian StyleHao, Ruijie, Jun Chang, Chen Qiu, and Shuting Yang. 2022. "An Identification and Expression Analysis of the ABCG Genes Related to Benzaldehyde Transportation among Three Prunus Species" Horticulturae 8, no. 6: 475. https://doi.org/10.3390/horticulturae8060475
APA StyleHao, R., Chang, J., Qiu, C., & Yang, S. (2022). An Identification and Expression Analysis of the ABCG Genes Related to Benzaldehyde Transportation among Three Prunus Species. Horticulturae, 8(6), 475. https://doi.org/10.3390/horticulturae8060475




