Transcriptional Regulation in Leaves of Cut Chrysanthemum (Chrysanthemum morifolium) ‘FenDante’ in Response to Post-Harvest Ethylene Treatment
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Ethylene Treatment
2.2. Chlorophyll Content and Relative Water Content Measurements
2.3. RNA Extraction, cDNA Library Construction, and Sequencing
2.4. Sequencing Assembly and Annotation
2.5. qRT-PCR Analysis
2.6. Data Analyses
3. Results
3.1. Phenotypic Characterization of Cut Chrysanthemum ‘FenDante’ with Ethylene and 1-MCP Treatments
3.2. Transcriptome Sequencing and Data Analysis
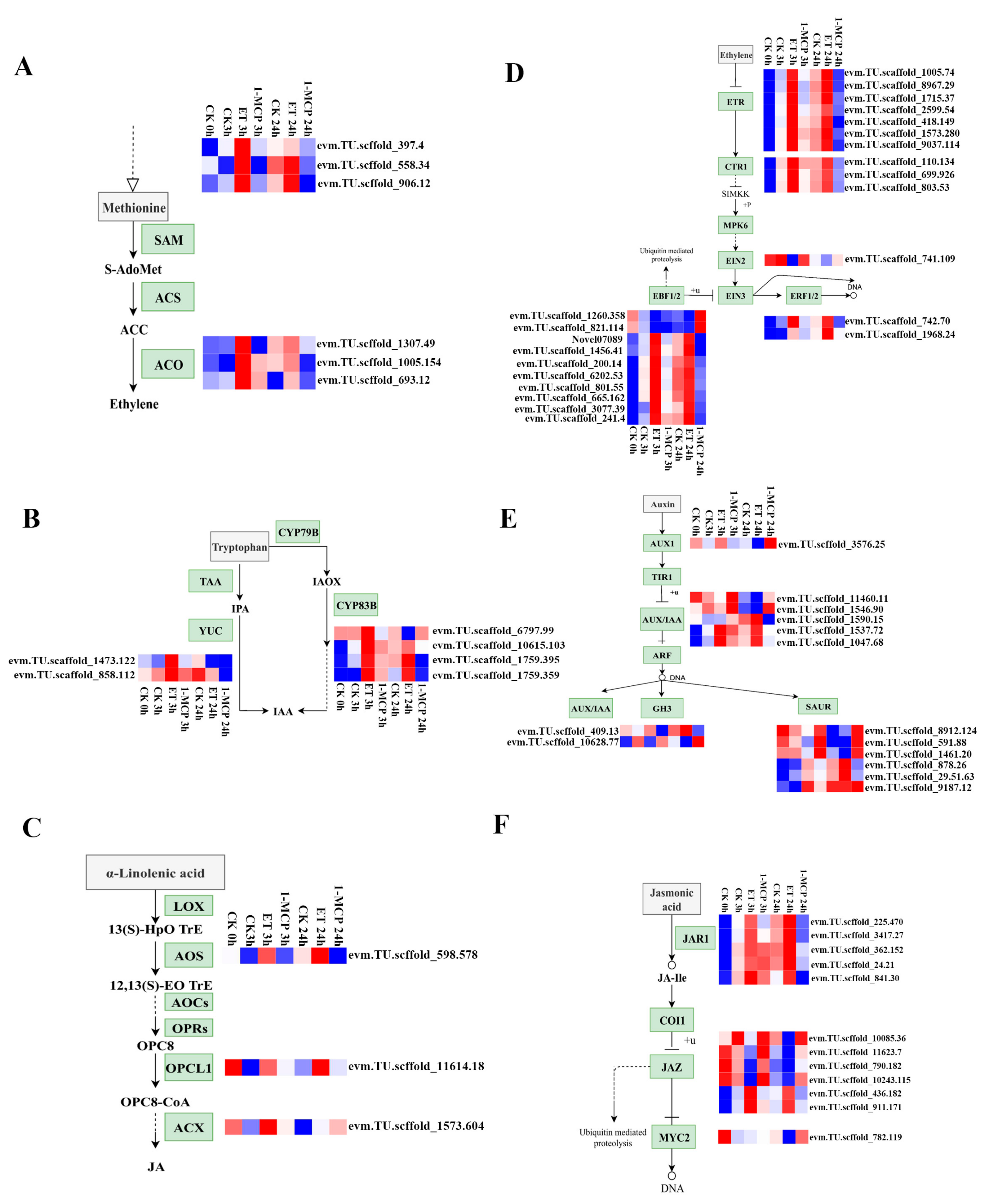
3.3. DEGs Related to Plant Hormone Biosynthesis and Signaling
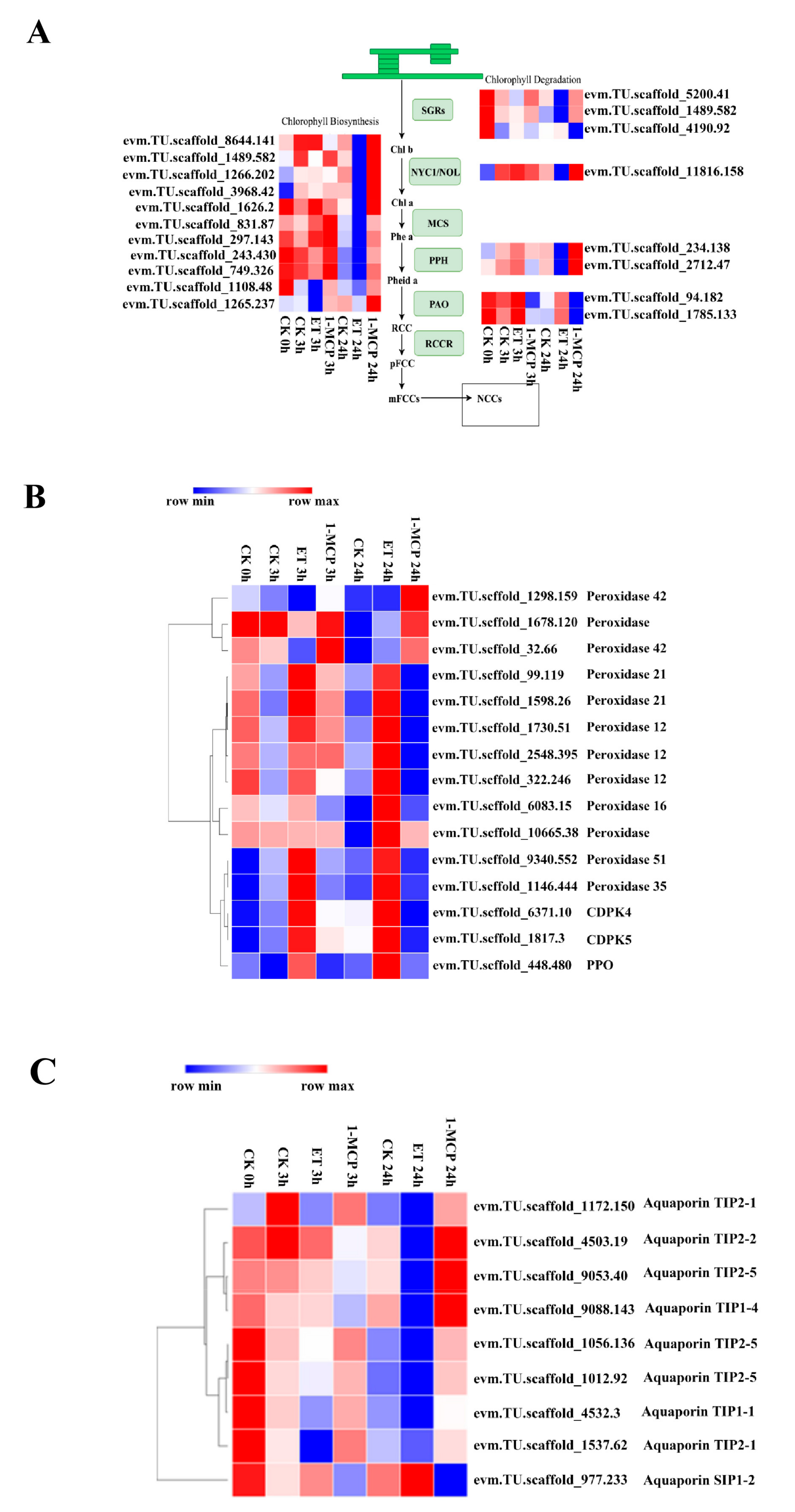
3.4. DEGs Related to Chlorophyll Synthesis and Degradation and the ROS Signaling Pathway
3.5. DEGs Related to Aquaporins
3.6. Identification of Differentially Expressed Transcription Factors (TFs) with Ethylene and 1-MCP Treatment
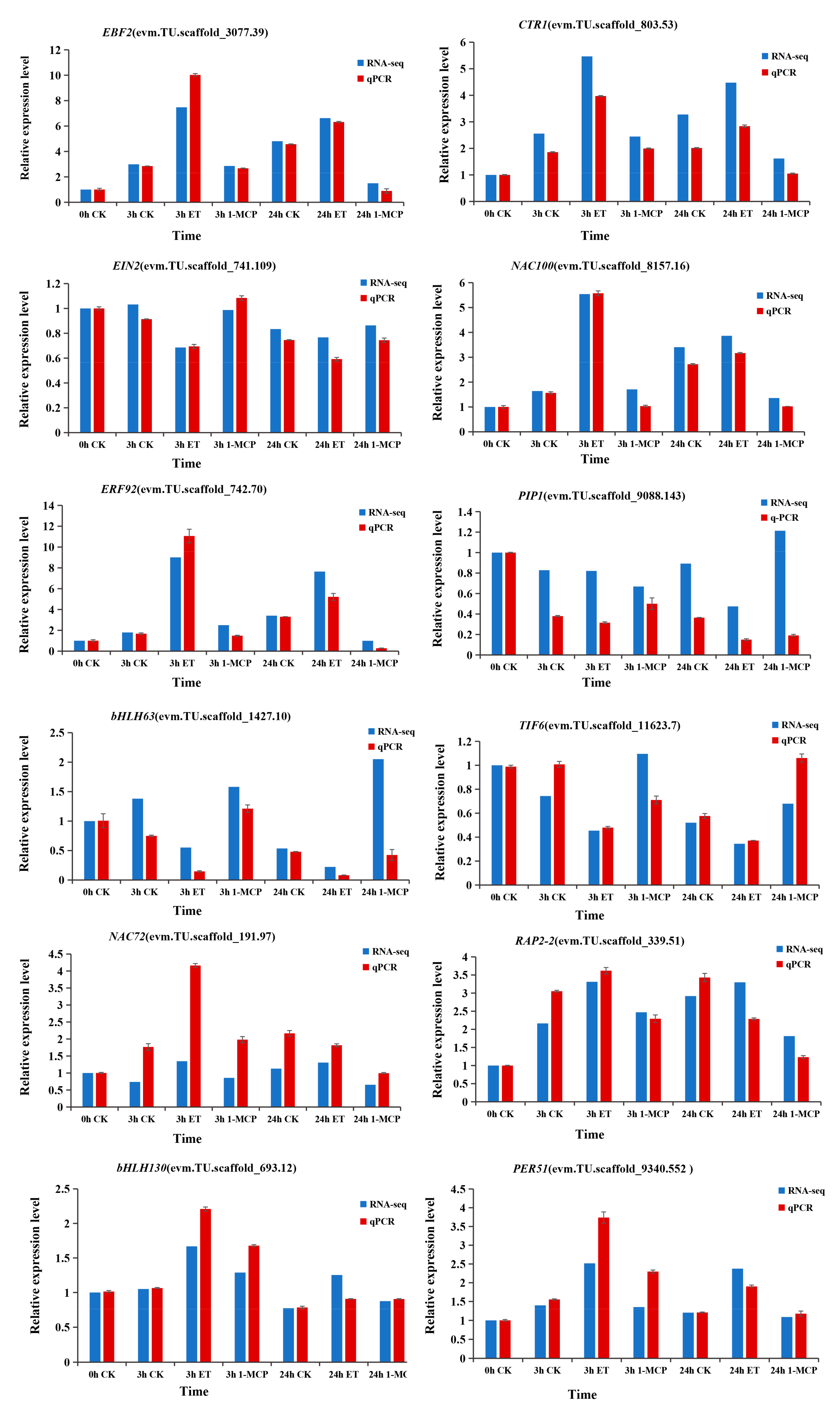
3.7. Validation of the DEGs by Quantitative Reverse Transcription-Polymerase Chain Reaction (qRT-PCR)
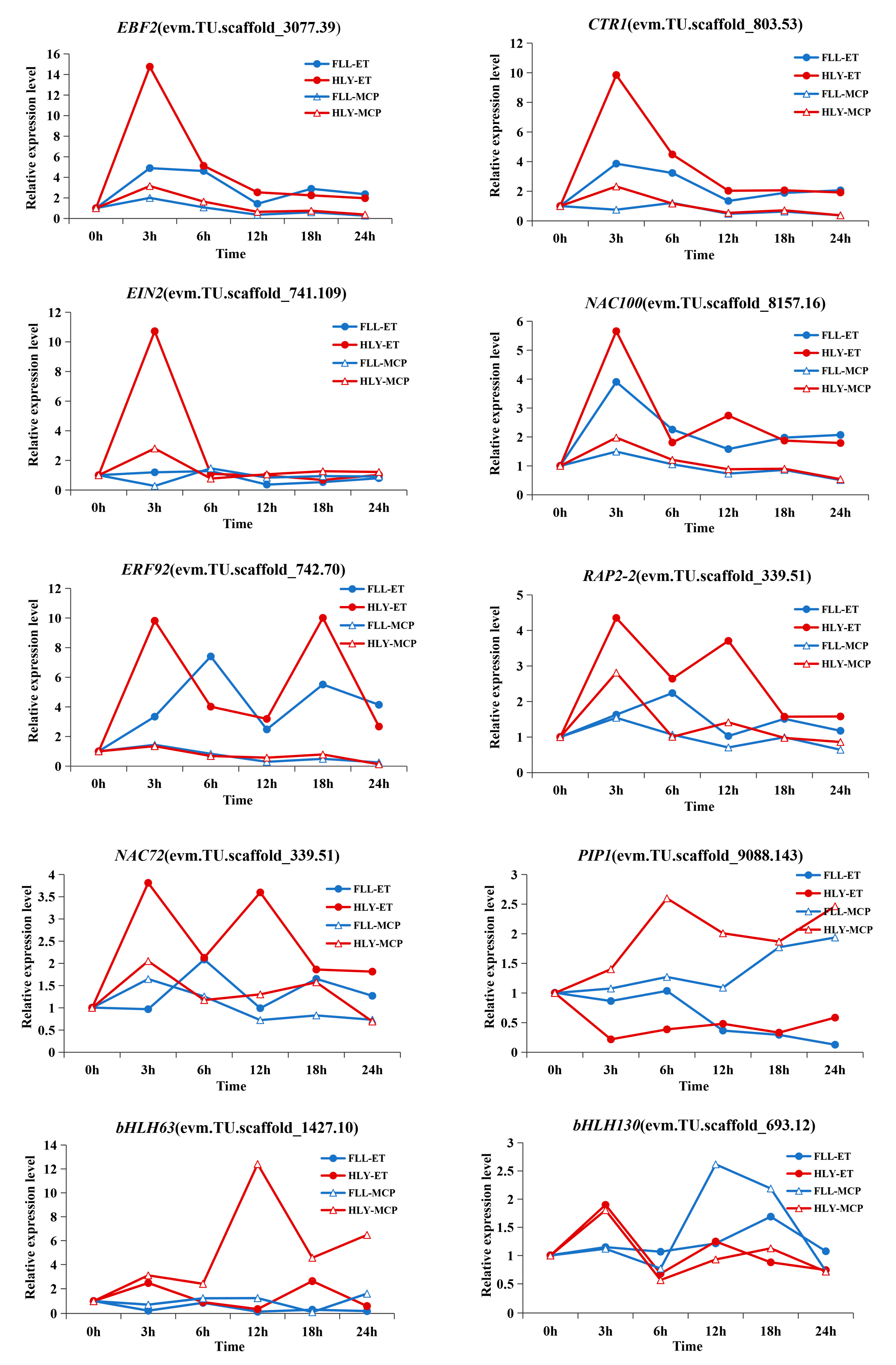
3.8. Expression Patterns of Transcription Factor DEGs in Leaves of Different Ethylene-Sensitive Cultivars
4. Discussion
4.1. DEGs Involved in Chlorophyll Synthesis and Degradation
4.2. DEGs Related to Aquaporins
4.3. DEGs Involved in Plant Hormones ET, JA, and IAA Biosynthesis and Signaling
4.4. DEGs of Transcription Factors
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lin, Z.F.; Zhong, S.; Grierson, D. Recent advances in ethylene research. J. Exp. Bot. 2009, 60, 3311–3336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iqbal, N.; Khan, N.A.; Ferrante, A.; Trivellini, A.; Francini, A.; Khan, M.I.R. Ethylene Role in Plant Growth, Development and Senescence: Interaction with Other Phytohormones. Front. Plant Sci. 2017, 8, 475. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hunter, D.; Yoo, S.D.; Butcher, S.M.; McManus, M.T. Expression of 1-Aminocyclopropane-1-Carboxylate Oxidase during Leaf Ontogeny in White Clover1. Plant Physiol. 1999, 120, 131–142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grbić, V.; Bleecker, A.B. Ethylene regulates the timing of leaf senescence in Arabidopsis. Plant J. 1995, 8, 595–602. [Google Scholar] [CrossRef]
- Wang, H.; Stier, G.; Lin, J.; Liu, G.; Zhang, Z.; Chang, Y.; Reid, M.S.; Jiang, C.-Z. Transcriptome changes associated with delayed flower senescence on transgenic petunia by inducing expression of etr1-1, a mutant ethylene receptor. PLoS ONE. 2013, 8, e65800. [Google Scholar] [CrossRef] [PubMed]
- Qiu, K.; Li, Z.; Yang, Z.; Chen, J.; Wu, S.; Zhu, X.; Gao, S.; Gao, J.; Ren, G.; Kuai, B.; et al. EIN3 and ORE1 Accelerate Degreening during Ethylene-Mediated Leaf Senescence by Directly Activating Chlorophyll Catabolic Genes in Arabidopsis. PLoS Genet. 2015, 11, e1005399. [Google Scholar] [CrossRef]
- Lim, P.O.; Kim, H.J.; Nam, H.G. Leaf senescence. Annu. Rev. Plant Biol. 2007, 58, 115–136. [Google Scholar] [CrossRef] [Green Version]
- Guo, Y. Towards systems biological understanding of leaf senescence. Plant Mol. Biol. 2013, 82, 519–528. [Google Scholar] [CrossRef]
- Watanabe, M.; Balazadeh, S.; Tohge, T.; Erban, A.; Giavalisco, P.; Kopka, J.; Mueller-Roeber, B.; Fernie, A.R.; Hoefgen, R. Comprehensive Dissection of Spatiotemporal Metabolic Shifts in Primary, Secondary, and Lipid Metabolism during Developmental Senescence in Arabidopsis. Plant Physiol. 2013, 162, 1290–1310. [Google Scholar] [CrossRef] [Green Version]
- Breeze, E.; Harrison, E.; McHattie, S.; Hughes, L.; Hickman, R.; Hill, C.; Kiddle, S.; Kim, Y.-S.; Penfold, C.A.; Jenkins, D.; et al. High-resolution temporal profling of transcripts during Arabidopsis leaf senescence reveals a distinct chronology of processes and regulation. Plant Cell 2011, 23, 873–894. [Google Scholar] [CrossRef] [Green Version]
- Roberts, I.N.; Caputo, C.; Criado, M.V.; Funk, C. Senescence-associated proteases in plants. Physiol. Plant. 2012, 145, 130–139. [Google Scholar] [CrossRef] [PubMed]
- Van Breusegem, F.; Dat, J.F. Reactive oxygen species in plant cell death. Plant Physiol. 2006, 141, 384–390. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Srivalli, S.; Khanna-Chopra, R. Delayed wheat flag leaf senescence due to removal of spikelets is associated with increased activities of leaf antioxidant enzymes, reduced glutathione/oxidized glutathione ratio and oxidative damage to mitochondrial proteins. Plant Physiol. Biochem. 2009, 47, 663–670. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Gan, S. AtNAP, a NAC family transcription factor, has an important role in leaf senescence. Plant J. 2006, 46, 601–612. [Google Scholar] [CrossRef] [PubMed]
- Christiansen, M.W.; Gregersen, P.L. Members of the barley NAC transcription factor gene family show differential co-regulation with senescence-associated genes during senescence of flag leaves. J. Exp. Bot. 2014, 65, 4009–4022. [Google Scholar] [CrossRef] [Green Version]
- Zhang, K.; Xia, X.; Zhang, Y.; Gan, S.-S. An ABA-regulated and Golgi-localized protein phosphatase controls water loss during leaf senescence in Arabidopsis. Plant J. 2012, 69, 667–678. [Google Scholar] [CrossRef]
- Kim, J.H.; Woo, H.R.; Kim, J.; Lim, P.O.; Lee, I.C.; Choi, S.H.; Hwang, D.; Nam, H.G. Trifurcate feed-forward regulation of agedependent cell death involving miR164 in Arabidopsis. Science 2009, 323, 1053–1057. [Google Scholar] [CrossRef] [Green Version]
- Kim, H.J.; Hong, S.H.; Kim, Y.W.; Lee, I.H.; Jun, J.H.; Phee, B.-K.; Rupak, T.; Jeong, H.; Lee, Y.; Hong, B.S.; et al. Gene regulatory cascade of senescence-associated NAC transcription factors activated by ETHYLENE-INSENSITIVE2-mediated leaf senescence signalling in Arabidopsis. J. Exp. Bot. 2014, 65, 4023–4036. [Google Scholar] [CrossRef] [Green Version]
- Rushton, D.L.; Tripathi, P.; Rabara, R.C.; Lin, J.; Ringler, P.; Boken, A.K.; Langum, T.J.; Smidt, L.; Boomsma, D.D.; Emme, N.J.; et al. WRKY transcription factors: Key components in abscisic acid signalling. Plant Biotechnol. J. 2012, 10, 2–11. [Google Scholar] [CrossRef]
- Miao, Y.; Laun, T.; Zimmermann, P.; Zentgraf, U. Targets of the WRKY53 transcription factor and its role during leaf senescence in Arabidopsis. Plant Mol. Biol. 2014, 55, 853–867. [Google Scholar] [CrossRef]
- da Silva, J.A.T.; Shinoyama, H.; Aida, R.; Matsushita, Y.; Raj, S.K.; Chen, F. Chrysanthemum Biotechnology: Quo vadis? Crit. Rev. Plant Sci. 2013, 32, 21–52. [Google Scholar] [CrossRef]
- Reid, M.S.; Wu, M.-J. Ethylene and flower senescence. Plant Growth Regul. 1992, 11, 37–43. [Google Scholar] [CrossRef]
- Reyes-Arribas, T.; Barrett, J.E.; Huber, D.J.; Nell, T.A.; Clark, D.G. Leaf senescence in a non-yellowing cultivar of chrysanthemum (Dendranthema grandiflora). Physiol. Plant. 2001, 111, 540–544. [Google Scholar] [CrossRef] [PubMed]
- Ferrante, A.; Tognoni, F.; Mensuali-Sodi, A.; Serra, G. Treatment with thidiazuron for preventing leaf yellowing in cut tulips and chrysanthemum. Acta Hortic. 2003, 357–363. [Google Scholar] [CrossRef]
- Satoh, S.; Watanabe, M.; Chisaka, K.; Narumi, T. Suppressed leaf senescence in Chrysanthemum transformed with a mutated ethylene receptor gene mDG-ERS1 (etr1-4). J. Plant Biol. 2008, 51, 424–427. [Google Scholar] [CrossRef]
- Ren, L.; Sun, J.; Chen, S.; Gao, J.; Dong, B.; Liu, Y.; Xia, X.; Wang, Y.; Liao, Y.; Teng, N.; et al. A transcriptomic analysis of Chrysanthemum nankingense provides insights into the basis of low temperature tolerance. BMC Genom. 2014, 15, 844. [Google Scholar] [CrossRef] [Green Version]
- Gonen, T.; Walz, T. The structure of aquaporins. Q. Rev. Biophys. 2006, 39, 361–396. [Google Scholar] [CrossRef]
- Petridou, M.; Voyiatzi, C.; Voyiatzis, D. Methanol, ethanol and other compunds retard leaf senenscence and improve the vase life and quality of cut chrysanthemum flowers. Postharvest Biol. Technol. 2001, 23, 79–83. [Google Scholar] [CrossRef]
- Sakuraba, Y.; Park, S.-Y.; Kim, Y.-S.; Wang, S.-H.; Yoo, S.-C.; Hörtensteiner, S.; Paek, N.-C. Arabidopsis STAY-GREEN2 is a negative regulator of chlorophyll degradation during leaf senescence. Mol. Plant 2014, 7, 1288–1302. [Google Scholar] [CrossRef] [Green Version]
- Sakuraba, Y.; Kim, D.; Kim, Y.-S.; Hörtensteiner, S.; Paek, N.-C. Arabidopsis STAYGREEN-LIKE (SGRL) promotes abiotic stress-induced leaf yellowing during vegetative growth. FEBS Lett. 2014, 588, 3830–3837. [Google Scholar] [CrossRef] [Green Version]
- Maurel, C.; Verdoucq, L.; Luu, D.-T.; Santoni, V. Plant Aquaporins: Membrane Channels with Multiple Integrated Functions. Annu. Rev. Plant Biol. 2008, 59, 595–624. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hachez, C.; Heinen, R.B.; Draye, X.; Chaumont, F. The expression pattern of plasma membrane aquaporins in maize leaf highlights their role in hydraulic regulation. Plant Mol. Biol. 2008, 68, 337–353. [Google Scholar] [CrossRef] [PubMed]
- Kamaluddin, M.; Zwiazek, J.J. Ethylene Enhances Water Transport in Hypoxic Aspen. Plant Physiol. 2002, 128, 962–969. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, N.; Xue, J.; Li, Y.; Liu, X.; Dai, F.; Jia, W.; Luo, Y.; Gao, J. Rh-PIP2;1, a Rose Aquaporin Gene, Is Involved in Ethylene-Regulated Petal Expansion. Plant Physiol. 2008, 148, 894–907. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, W.; Yin, X.; Wang, L.; Tian, J.; Yang, R.; Liu, D.; Yu, Z.; Ma, N.; Gao, J. Involvement of rose aquaporin RhPIP1;1 in ethylene-regulated petal expansion through interaction with RhPIP2;1. Plant Mol. Biol. 2013, 83, 219–233. [Google Scholar] [CrossRef]
- Qiao, H.; Chang, K.N.; Yazaki, J.; Ecker, J.R. Interplay between ethylene, ETP1/ETP2 F-box proteins, and degradation of EIN2 triggers ethylene responses in Arabidopsis. Genes Dev. 2009, 23, 512–521. [Google Scholar] [CrossRef] [Green Version]
- Törnroth-Horsefield, S.; Wang, Y.; Hedfalk, K.; Johanson, U.; Karlsson, M.; Tajkhorshid, E.; Neutze, R.; Kjellbom, P. Structural mechanism of plant aquaporin gating. Nature 2006, 439, 688–694. [Google Scholar] [CrossRef]
- Qing, D.; Yang, Z.; Li, M.; Wong, W.S.; Guo, G.; Liu, S.; Guo, H.; Li, N. Quantitative and Functional Phosphoproteomic Analysis Reveals that Ethylene Regulates Water Transport via the C-Terminal Phosphorylation of Aquaporin PIP2;1 in Arabidopsis. Mol. Plant 2015, 9, 158–174. [Google Scholar] [CrossRef] [Green Version]
- Katsir, L.; Schilmiller, A.L.; Staswick, P.E.; He, S.Y.; Howe, G.A. COI1 is a critical component of a receptor for jasmonate and the bacterial virulence factor coronatine. Proc. Natl. Acad. Sci. USA 2008, 105, 7100–7105. [Google Scholar] [CrossRef] [Green Version]
- Pauwels, L.; Barbero, G.F.; Geerinck, J.; Tilleman, S.; Grunewald, W.; Pérez, A.C.; Chico, J.M.; Bossche, R.V.; Sewell, J.; Gil, E.; et al. NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature 2010, 464, 788–791. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.I.; Murphy, A.S.; Baek, D.; Lee, S.-W.; Yun, D.-J.; Bressan, R.A.; Narasimhan, M.L. YUCCA6 over-expression demonstrates auxin function in delaying leaf senescence in Arabidopsis thaliana. J. Exp. Bot. 2011, 62, 3981–3992. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Cheng, X.; Wang, X.; Li, G.; Wang, B.; Wang, W.; Zhang, N.; Han, Y.; Jiao, B.; Wang, Y.; et al. Glyoxalase I-4 functions downstream of NAC72 to modulate downy mildew resistance in grapevine. Plant J. 2021, 108, 394–410. [Google Scholar] [CrossRef] [PubMed]
- Hu, G.; Lei, Y.; Liu, J.; Hao, M.; Zhang, Z.; Tang, Y.; Chen, A.; Wu, J. The ghr-miR164 and GhNAC100 modulate cotton plant resistance against Verticillium dahlia. Plant Sci. 2020, 293, 110438. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Chahal, A.; Prasad, H.; Walia, A.; Kumar, R.; Dobhal, S. Identification, phylogeny and transcript profiling of ERF family genes during temperature stress treatment in Pea (Pisum sativum L.). Mol. Genet. Genomics 2010, 284, 455–475. [Google Scholar] [CrossRef] [PubMed]
- Hinz, M.; Wilson, I.W.; Yang, J.; Buerstenbinder, K.; Llewellyn, D.; Dennis, E.S.; Sauter, M.; Dolferus, R. Arabidopsis RAP2.2: An ethylene response transcription factor that is important for hypoxia survival. Plant Physiol. 2010, 153, 757–772. [Google Scholar] [CrossRef] [Green Version]
- Tang, H.; Bi, H.; Liu, B.; Lou, S.; Song, Y.; Tong, S.; Chen, N.; Jiang, Y.; Liu, J.; Liu, H. WRKY33 interacts with WRKY12 protein to up-regulate RAP2. 2 during submergence induced hypoxia response in Arabidopsis thaliana. New Phytol. 2021, 229, 106–125. [Google Scholar] [CrossRef]




| DEG Set | All DEG | Up-Regulated | Down-Regulated |
|---|---|---|---|
| 3 h ET vs. 3 h CK | 738 | 498 | 240 |
| 3 h 1-MCP vs. 3 h CK | 1 | 1 | 0 |
| 3 h ET vs. 3 h 1-MCP | 39 | 19 | 20 |
| 24 h ET vs. 24 h CK | 196 | 127 | 69 |
| 24 h 1-MCP vs. 24 h CK | 202 | 111 | 91 |
| 24 h ET vs. 24 h 1-MCP | 2723 | 992 | 1731 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, R.; Zuo, X.; Chen, Y.; Qian, Z.; Xu, C.; Wang, L.; Chen, S. Transcriptional Regulation in Leaves of Cut Chrysanthemum (Chrysanthemum morifolium) ‘FenDante’ in Response to Post-Harvest Ethylene Treatment. Horticulturae 2022, 8, 573. https://doi.org/10.3390/horticulturae8070573
Liu R, Zuo X, Chen Y, Qian Z, Xu C, Wang L, Chen S. Transcriptional Regulation in Leaves of Cut Chrysanthemum (Chrysanthemum morifolium) ‘FenDante’ in Response to Post-Harvest Ethylene Treatment. Horticulturae. 2022; 8(7):573. https://doi.org/10.3390/horticulturae8070573
Chicago/Turabian StyleLiu, Rui, Xuele Zuo, Yu Chen, Ziyan Qian, Can Xu, Likai Wang, and Sumei Chen. 2022. "Transcriptional Regulation in Leaves of Cut Chrysanthemum (Chrysanthemum morifolium) ‘FenDante’ in Response to Post-Harvest Ethylene Treatment" Horticulturae 8, no. 7: 573. https://doi.org/10.3390/horticulturae8070573
APA StyleLiu, R., Zuo, X., Chen, Y., Qian, Z., Xu, C., Wang, L., & Chen, S. (2022). Transcriptional Regulation in Leaves of Cut Chrysanthemum (Chrysanthemum morifolium) ‘FenDante’ in Response to Post-Harvest Ethylene Treatment. Horticulturae, 8(7), 573. https://doi.org/10.3390/horticulturae8070573






