Comparative Analysis of Complete Chloroplast Genome and Phenotypic Characteristics of Japanese Apricot Accessions
Abstract
:1. Introduction
2. Materials and Methods
2.1. Genetic Materials and Agro-Morphological Characterization
2.2. Sample Preparations, DNA Extraction and Sequencing
2.3. Assembly, Annotation and Analysis of the Chloroplast Genome Sequences
2.4. Analysis of Repeat Sequences and Single-Sequence Repeats
2.5. Genome Comparison and Sequence Divergence of Chloroplast Genomes
2.6. Evolutionary Analysis
3. Results
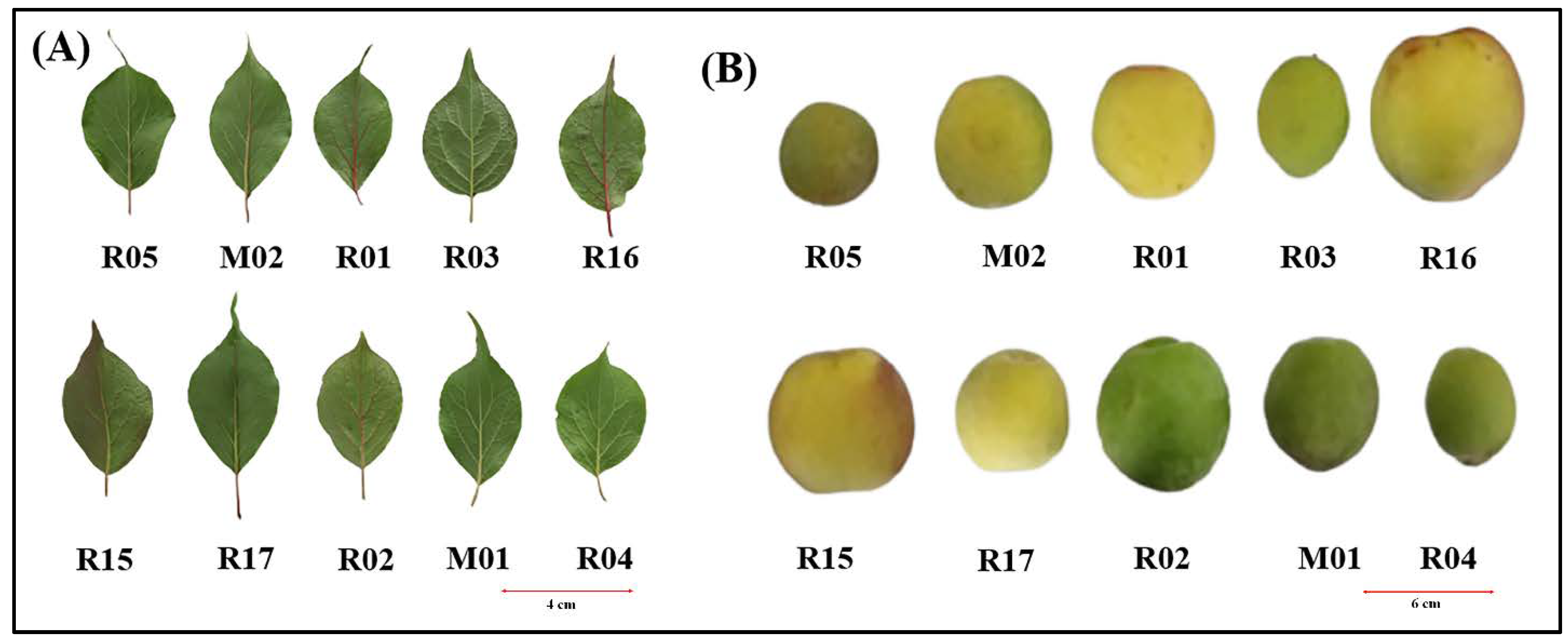
3.1. Agro-Morphological and Fruit Quality Characteristics among the Ten Prunus mume Accessions
3.2. Structural Features and Gene Content of the Chloroplast Genome in Prunus mume Accessions
3.3. Protein-Coding Gene Capacity and Codon Usage Analysis
3.4. Simple Sequence Repeats and Repetitive Sequence Analysis
3.5. Analysis of Nucleotide Diversity in P. mume Accessions
3.6. Inverted Repeat Expansion and Contraction
3.7. Sequence Analysis of ndhF-ycf1 Genes from Ten Prunus mume Accessions
3.8. Selective Pressure Analyses
3.9. Phylogenetic Analysis among Prunus mume Accessions
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| cp | chloroplast |
| bp | base pairs |
| SSC | Small single-copy |
| IR | Inverted repeat |
| LSC | Large single copy |
| SSR | Single-sequence Repeat |
| RSCU | Relative synonymous codon usage |
| TSS | Total soluble solids |
| TAC | Titratable acid content |
References
- Bock, R.; Knoop, V. Genomics of Chloroplasts and Mitochondria; Springer Science & Business Media: New York, NY, USA, 2012; Volume 35. [Google Scholar]
- Ris, H.; Plaut, W. Ultrastructure of DNA-containing areas in the chloroplast of Chlamydomonas. J. Cell Biol. 1962, 13, 383–391. [Google Scholar] [CrossRef] [PubMed]
- Timmis, J.N.; Ayliffe, M.A.; Huang, C.Y.; Martin, W. Endosymbiotic gene transfer: Organelle genomes forge eukaryotic chromosomes. Nat. Rev. Genet. 2004, 5, 123–135. [Google Scholar] [CrossRef] [PubMed]
- Dyall, S.D.; Brown, M.T.; Johnson, P.J. Ancient invasions: From endosymbionts to organelles. Science 2004, 304, 253–257. [Google Scholar] [CrossRef]
- Price, D.C. Genome Elucidates Origin of Photosynthesis Cyanophora paradoxa. Science 2012, 1213561, 335. [Google Scholar] [CrossRef] [PubMed]
- Mustárdy, L.; Buttle, K.; Steinbach, G.; Garab, G.Z. The three-dimensional network of the thylakoid membranes in plants: Quasihelical model of the granum-stroma assembly. Plant Cell 2008, 20, 2552–2557. [Google Scholar] [CrossRef]
- Neuhaus, H.; Emes, M. Nonphotosynthetic metabolism in plastids. Annu. Rev. Plant Biol. 2000, 51, 111–140. [Google Scholar] [CrossRef]
- Li, X.; Tan, W.; Sun, J.; Du, J.; Zheng, C.; Tian, X.; Zheng, M.; Xiang, B.; Wang, Y. Comparison of Four Complete Chloroplast Genomes of Medicinal and Ornamental Meconopsis Species: Genome Organization and Species Discrimination. Sci. Rep. 2019, 9, 10567. [Google Scholar] [CrossRef]
- Xue, S.; Shi, T.; Luo, W.; Ni, X.; Iqbal, S.; Ni, Z.; Huang, X.; Yao, D.; Shen, Z.; Gao, Z. Comparative analysis of the complete chloroplast genome among Prunus mume, P. armeniaca, and P. salicina. Hortic. Res. 2019, 6, 89. [Google Scholar] [CrossRef]
- Peredo, E.L.; King, U.M.; Les, D.H. The plastid genome of Najas flexilis: Adaptation to submersed environments is accompanied by the complete loss of the NDH complex in an aquatic angiosperm. PLoS ONE 2013, 8, e68591. [Google Scholar]
- Köhler, M.; Reginato, M.; Souza-Chies, T.T.; Majure, L.C. Insights into chloroplast genome evolution across opuntioideae (Cactaceae) reveals robust yet sometimes conflicting phylogenetic topologies. Front. Plant Sci. 2020, 11, 729. [Google Scholar] [CrossRef]
- Hodel, R.G.; Zimmer, E.; Wen, J.J. A phylogenomic approach resolves the backbone of Prunus (Rosaceae) and identifies signals of hybridization and allopolyploidy. Mol. Phylogenetics Evol. 2021, 160, 107118. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Shi, C.; Gao, L.-Z. Plastid genome sequence of a wild woody oil species, Prinsepia utilis, provides insights into evolutionary and mutational patterns of Rosaceae chloroplast genomes. PLoS ONE 2013, 8, e73946. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Dong, W.; Liu, B.; Xu, C.; Yao, X.; Gao, J.; Corlett, R.T. Comparative analysis of complete chloroplast genome sequences of two tropical trees Machilus yunnanensis and Machilus balansae in the family Lauraceae. Front. Plant Sci. 2015, 6, 662. [Google Scholar] [CrossRef] [PubMed]
- Amar, M.H. Biotechnology. ycf 1-ndh F genes, the most promising plastid genomic barcode, sheds light on phylogeny at low taxonomic levels in Prunus persica. J. Genet. Eng. 2020, 18, 42. [Google Scholar]
- Numaguchi, K.; Akagi, T.; Kitamura, Y.; Ishikawa, R.; Ishii, T. Interspecific introgression and natural selection in the evolution of Japanese apricot (Prunus mume). Plant J. 2020, 104, 1551–1567. [Google Scholar] [CrossRef]
- Wu, C.-S.; Wang, Y.-N.; Hsu, C.-Y.; Lin, C.-P.; Chaw, S.-M. Loss of different inverted repeat copies from the chloroplast genomes of Pinaceae and cupressophytes and influence of heterotachy on the evaluation of gymnosperm phylogeny. Genome Biol. Evol. 2011, 3, 1284–1295. [Google Scholar] [CrossRef]
- My, C. China Fruit Records-Mei; China Forestry Press: Beijing, China, 1999; pp. 97–188. [Google Scholar]
- Shimada, T.; Haji, T.; Yamaguchi, M.; Takeda, T.; Nomura, K.; Yoshida, M. Classification of mume (Prunus mume Sieb. et Zucc.) by RAPD assay. J. Jpn. Soc. Hortic. 1994, 63, 543–551. [Google Scholar] [CrossRef]
- Reyes, E.; Nadot, S.; Von Balthazar, M.; Schoenenberger, J.; Sauquet, H. Testing the impact of morphological rate heterogeneity on ancestral state reconstruction of five floral traits in angiosperms. Sci. Rep. 2018, 8, 9473. [Google Scholar] [CrossRef]
- Stevens, P. Angiosperm Phylogeny Website. Available online: http://www.mobot.org/MOBOT/research/APweb (accessed on 12 January 2022).
- Sauquet, H.; Von Balthazar, M.; Magallón, S.; Doyle, J.A.; Endress, P.K.; Bailes, E.J.; de Morais, E.B.; Bull-Hereñu, K.; Carrive, L.; Chartier, M. The ancestral flower of angiosperms and its early diversification. J. Nat. Commun. 2017, 8, 16047. [Google Scholar] [CrossRef]
- Byrne, M.; Macdonald, B.; Brand, J. Phylogeography and divergence in the chloroplast genome of Western Australian Sandalwood (Santalum spicatum). Heredity 2003, 91, 389–395. [Google Scholar] [CrossRef]
- Heritage, S. MBASR: Workflow-simplified ancestral state reconstruction of discrete traits with MrBayes in the R environment. bioRxiv 2021. [Google Scholar]
- Gao, L.; Su, Y.J.; Wang, T. Plastid genome sequencing, comparative genomics, and phylogenomics: Current status and prospects. J. Syst. Evol. 2010, 48, 77–93. [Google Scholar] [CrossRef]
- Mayer, N.A.; Pereira, F.M.; Môro, F.V.J.R.B.d.F. Caracterização morfológica de três genótipos de umezeiro selecionados como porta-enxertos para pessegueiro. Rev. Bras. De Frutic. 2008, 30, 716–722. [Google Scholar] [CrossRef]
- Cao, J.K.J.W.B.; Zhao, Y.M. Study on Physiology and Biochemistry of Fruits and Vegetables after Harvest; China Light Industry Press: Beijing, China, 2007; pp. 2054–2125. [Google Scholar]
- Simko, T.W.a.V. R Package “Corrplot”: Visualization of a Correlation Matrix (Version 0.84). Available online: https://github.com/taiyun/corrplot (accessed on 15 June 2017).
- Doyle, J.J.; Doyle, J.L. A Rapid DNA Isolation Procedure for Small Quantities of Fresh Leaf Tissue. Phytochem. Bull. 1987, 19, 11–15. [Google Scholar]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Jansen, R.K.; Saski, C.; Lee, S.-B.; Hansen, A.K.; Daniell, H. Complete plastid genome sequences of three rosids (Castanea, Prunus, Theobroma): Evidence for at least two independent transfers of rpl22 to the nucleus. Mol. Biol. Evol. 2011, 28, 835–847. [Google Scholar] [CrossRef] [PubMed]
- Capella-Gutiérrez, S.; Silla-Martínez, J.M.; Gabaldón, T. trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Darriba, D.; Posada, D.; Kozlov, A.M.; Stamatakis, A.; Morel, B.; Flouri, T. ModelTest-NG: A new and scalable tool for the selection of DNA and protein evolutionary models. Mol. Biol. Evol. Bioinform. 2020, 37, 291–294. [Google Scholar] [CrossRef]
- Gao, F.; Chen, C.; Arab, D.A.; Du, Z.; He, Y.; Ho, S.Y. EasyCodeML: A visual tool for analysis of selection using CodeML. Ecol. Evol. Bioinform. 2019, 9, 3891–3898. [Google Scholar] [CrossRef]
- Lewis, P.O. A likelihood approach to estimating phylogeny from discrete morphological character data. Syst. Biol. 2001, 50, 913–925. [Google Scholar] [CrossRef] [PubMed]
- Pagel, M. The maximum likelihood approach to reconstructing ancestral character states of discrete characters on phylogenies. Syst. Biol. 1999, 48, 612–622. [Google Scholar] [CrossRef]
- Jeon, J.-H.; Kim, S.-C. Comparative analysis of the complete chloroplast genome sequences of three closely related East-Asian wild roses (Rosa sect. Synstylae; Rosaceae). Genes 2019, 10, 23. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Zhou, T.; Duan, D.; Yang, J.; Feng, L.; Zhao, G. Comparative analysis of the complete chloroplast genomes of five Quercus species. Front. Plant Sci. 2016, 7, 959. [Google Scholar] [CrossRef] [Green Version]
- Du, Z.; Lu, K.; Zhang, K.; He, Y.; Wang, H.; Chai, G.; Shi, J.; Duan, Y. The chloroplast genome of Amygdalus L. (Rosaceae) reveals the phylogenetic relationship and divergence time. BMC Genom. 2021, 22, 645. [Google Scholar] [CrossRef]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef]
- Michu, E. A short guide to phylogeny reconstruction. Plant Soil Environ. 2007, 53, 442. [Google Scholar] [CrossRef]
- Tzonev, R.; Haji, T.; Yamaguchi, M. A contribution to the apricot taxonomy: Investigation on the flowers, leaves and shoots of some European cultivars and Asian Prunus species. In Proceedings of the XI International Symposium on Apricot Culture 488, Veria-Makedonia, Greece, 25 May 1997; pp. 243–246. [Google Scholar]
- Chartier, M.; Jabbour, F.; Gerber, S.; Mitteroecker, P.; Sauquet, H.; von Balthazar, M.; Staedler, Y.; Crane, P.R.; Schönenberger, J. The floral morphospace–a modern comparative approach to study angiosperm evolution. New Phytol. 2014, 204, 841–853. [Google Scholar] [CrossRef]
- Quast, E.; Vieira, I.; Nogueira, A.; Schmidt, F.L. Chemical and physical characterization of mume fruit collected from different locations and at different maturity stages in São Paulo State. Food Sci. Technol. 2013, 33, 441–445. [Google Scholar] [CrossRef]
- Mratinić, E.; Rakonjac, V.; Milatović, D. Genetic parameters of yield and morphological fruit and stone properties in apricot. Genetika 2007, 39, 315–324. [Google Scholar] [CrossRef]
- Shi, T.; Luo, W.; Li, H.; Huang, X.; Ni, Z.; Gao, H.; Iqbal, S.; Gao, Z. Association between blooming time and climatic adaptation in Prunus mume. Ecol. Evol. 2020, 10, 292–306. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Yan, M.; Ding, Y.; Huo, Y.; Yuan, Z. Characterization and comparative analysis of the complete chloroplast genome sequence from Prunus avium ‘Summit’. PeerJ 2019, 7, e8210. [Google Scholar] [CrossRef]
- Sobreiro, M.B.; Vieira, L.D.; Nunes, R.; Novaes, E.; Coissac, E.; Silva-Junior, O.B.; Grattapaglia, D.; Collevatti, R.G. Chloroplast genome assembly of Handroanthus impetiginosus: Comparative analysis and molecular evolution in Bignoniaceae. Planta 2020, 252, 91. [Google Scholar] [CrossRef] [PubMed]
- Ni, L.; Zhao, Z.; Dorje, G.; Ma, M. The complete chloroplast genome of Ye-Xing-Ba (Scrophularia dentata; Scrophulariaceae), an alpine Tibetan herb. PLoS ONE 2016, 11, e0158488. [Google Scholar] [CrossRef] [Green Version]
- Li, C.; Zheng, Y.; Huang, P. Molecular markers from the chloroplast genome of rose provide a complementary tool for variety discrimination and profiling. Sci. Rep. 2020, 10, 12188. [Google Scholar] [PubMed]
- Lu, R.-S.; Li, P.; Qiu, Y.-X. The complete chloroplast genomes of three Cardiocrinum (Liliaceae) species: Comparative genomic and phylogenetic analyses. Front. Plant Sci. 2017, 7, 2054. [Google Scholar] [CrossRef] [PubMed]
- Ren, T.; Yang, Y.; Zhou, T.; Liu, Z.-L. Comparative plastid genomes of Primula species: Sequence divergence and phylogenetic relationships. Int. J. Mol. Sci. 2018, 19, 1050. [Google Scholar] [CrossRef]
- Kaur, S.; Panesar, P.S.; Bera, M.B.; Kaur, V. Simple sequence repeat markers in genetic divergence and marker-assisted selection of rice cultivars: A review. Crit. Rev. Food Sci. Nutr. 2015, 55, 41–49. [Google Scholar] [CrossRef]
- Shen, X.; Wu, M.; Liao, B.; Liu, Z.; Bai, R.; Xiao, S.; Li, X.; Zhang, B.; Xu, J.; Chen, S. Complete chloroplast genome sequence and phylogenetic analysis of the medicinal plant Artemisia annua. Molecules 2017, 22, 1330. [Google Scholar] [CrossRef]
- Xie, D.-F.; Yu, Y.; Deng, Y.-Q.; Li, J.; Liu, H.-Y.; Zhou, S.-D.; He, X.-J. Comparative analysis of the chloroplast genomes of the Chinese endemic genus Urophysa and their contribution to chloroplast phylogeny and adaptive evolution. Int. J. Mol. Sci. 2018, 19, 1847. [Google Scholar] [CrossRef]
- Li, Y.; Xu, W.; Zou, W.; Jiang, D.; Liu, X. Complete chloroplast genome sequences of two endangered Phoebe (Lauraceae) species. Bot. Stud. 2017, 58, 37. [Google Scholar] [CrossRef] [PubMed]
- Ivanova, Z.; Sablok, G.; Daskalova, E.; Zahmanova, G.; Apostolova, E.; Yahubyan, G.; Baev, V. Chloroplast genome analysis of resurrection tertiary relict Haberlea rhodopensis highlights genes important for desiccation stress response. Front. Plant Sci. 2017, 8, 204. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Yang, M.; Mo, C.; Xie, W.; Liu, C.; Wu, B.; Ma, X. Complete chloroplast genomes of two Siraitia Merrill species: Comparative analysis, positive selection and novel molecular marker development. PLoS ONE 2019, 14, e0226865. [Google Scholar] [CrossRef] [PubMed]
- Xu, F.; He, L.; Gao, S.; Su, Y.; Li, F.; Xu, L. Comparative Analysis of two Sugarcane Ancestors Saccharum officinarum and S. spontaneum based on Complete Chloroplast Genome Sequences and Photosynthetic Ability in Cold Stress. Int. J. Mol. Sci. 2019, 20, 3828. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Li, J.Z.L.; Gao, L.; Xu, J.; Yang, M. Structural and Comparative Analysis of the Complete Chloroplast Genome of Pyrus hopeiensis “Wild Plants with a Tiny Population”and Three Other Pyrus Species. Int. J. Mol. Sci. 2018, 19, 3262. [Google Scholar] [CrossRef]
- Kyalo, C.M.; Li, Z.-Z.; Mkala, E.M.; Malombe, I.; Hu, G.-W.; Wang, Q.-F. The First Glimpse of Streptocarpus ionanthus (Gesneriaceae) Phylogenomics: Analysis of Five Subspecies’ Chloroplast Genomes.Plants. Plants 2020, 18, 456. [Google Scholar] [CrossRef]
- Liu, H.; Xia, M.; Xiao, Q.; Fang, J.; Wang, A.; Chen, S.; Zhang, D. Characterization of the complete chloroplast genome of Linnaea borealis, a rare, clonal self-incompatible plant. Mitochondrial DNA Part B 2020, 5, 200–201. [Google Scholar] [CrossRef]
- Thode, V.A.; Lohmann, L.G. Comparative chloroplast genomics at low taxonomic levels: A case study using Amphilophium (Bignonieae, Bignoniaceae). Front. Plant Sci. 2019, 10, 796. [Google Scholar] [CrossRef]
- Tang, H.; Tang, L.; Shao, S.; Peng, Y.; Li, L.; Luo, Y. Chloroplast genomic diversity in Bulbophyllum section Macrocaulia (Bl.) Aver.(Orchidaceae, Epidendroideae, Malaxideae): Insights into species divergence and adaptive evolution. Plant Divers. 2021, 43, 350–361. [Google Scholar] [CrossRef]
- Davis, J.I.; Soreng, R.J. Migration of endpoints of two genes relative to boundaries between regions of the plastid genome in the grass family (Poaceae). Am. J. Bot. 2010, 97, 874–892. [Google Scholar] [CrossRef]
- Huang, H.; Shi, C.; Liu, Y.; Mao, S.-Y.; Gao, L.-Z. Thirteen Camellia chloroplast genome sequences determined by high-throughput sequencing: Genome structure and phylogenetic relationships. BMC Evol. Biol. 2014, 14, 151. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.-J.; Lee, H.-L. Complete chloroplast genome sequences from Korean ginseng (Panax schinseng Nees) and comparative analysis of sequence evolution among 17 vascular plants. DNA Res. 2004, 11, 247–261. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Ma, P.-F.; Li, H.-T.; Hu, G.-X.; Li, D.-Z. Comparative plastomic analysis and insights into the phylogeny of Salvia (Lamiaceae). Plant Divers. 2020, 43, 15–26. [Google Scholar] [CrossRef] [PubMed]







| Prunus mume Accessions Name | Accession Number | City of Origin | Province of Origin | Designation |
|---|---|---|---|---|
| Nanhongmei | MW755873 | Nanjing | Jiangsu | R01 |
| Hongguangmei | MW755879 | Suzhou | Jiangsu | R02 |
| Sichuanqingmei | MW755875 | Dayi | Sichuan | R03 |
| Sichuanbaimei | MW755874 | Dayi | Sichuan | R04 |
| Sichuanhuangmei | MW755877 | Dayi | Sichuan | R05 |
| Ruantiaohongmei | MW755885 | Chaoshan | Zhejiang | R15 |
| Xiaoyezhugan | MW755886 | Chaoshan | Zhejiang | R16 |
| Qingjia No.2 | MW755887 | Chaoshan | Zhejiang | R17 |
| Zaohong | MW759299 | Fenghua | Zhejiang | M01 |
| Changnong No.17 | MW759300 | Changxing | Zhejiang | M02 |
| SSR Type | Repeat Unit | R01 | R04 | R03 | R05 | R02 | R15 | R16 | R17 | M01 | M02 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Mono | A | 63 | 64 | 64 | 64 | 63 | 64 | 64 | 64 | 64 | 64 |
| T | 86 | 85 | 85 | 85 | 88 | 86 | 86 | 86 | 86 | 86 | |
| C | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | |
| G | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | |
| Di | AT | 7 | 7 | 7 | 7 | 7 | 7 | 7 | 7 | 7 | 7 |
| TA | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | |
| TC | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |
| Tri | AAC/AAG/AGA/ GAA/TTG | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 |
| AAT | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | |
| ACC/ACT/AGC/ATC/ ATG/CAA/CAG/CCA | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |
| ATA | 7 | 7 | 7 | 7 | 7 | 7 | 7 | 7 | 7 | 7 | |
| CTT/TCT | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | |
| GAT/GCA/GCT/GGA/ GGT/GTG/GTT/TAG/TGC | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |
| TAA/TTC | 6 | 6 | 6 | 6 | 6 | 6 | 6 | 6 | 6 | 6 | |
| TAT/TTA | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | |
| AAAT | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | |
| Tetra | AATA/ATAA/TTGA/ TTTA/TTTC | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Penta | AAAAT | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| TTTGA | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |
| Hexa | ATCTAT | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 |
| Type | R01 | R04 | R03 | R05 | R02 | R15 | R16 | R17 | M01 | M02 |
|---|---|---|---|---|---|---|---|---|---|---|
| F | 21 | 14 | 14 | 14 | 21 | 15 | 15 | 15 | 15 | 15 |
| P | 25 | 26 | 26 | 26 | 24 | 25 | 25 | 25 | 25 | 25 |
| R | 6 | 5 | 6 | 6 | 0 | 6 | 0 | 6 | 6 | 6 |
| C | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Total | 52 | 45 | 46 | 46 | 45 | 46 | 40 | 46 | 46 | 46 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Coulibaly, D.; Huang, X.; Ting, S.; Iqbal, S.; Ni, Z.; Ouma, K.O.; Hayat, F.; Tan, W.; Hu, G.; Ma, C.; et al. Comparative Analysis of Complete Chloroplast Genome and Phenotypic Characteristics of Japanese Apricot Accessions. Horticulturae 2022, 8, 794. https://doi.org/10.3390/horticulturae8090794
Coulibaly D, Huang X, Ting S, Iqbal S, Ni Z, Ouma KO, Hayat F, Tan W, Hu G, Ma C, et al. Comparative Analysis of Complete Chloroplast Genome and Phenotypic Characteristics of Japanese Apricot Accessions. Horticulturae. 2022; 8(9):794. https://doi.org/10.3390/horticulturae8090794
Chicago/Turabian StyleCoulibaly, Daouda, Xiao Huang, Shi Ting, Shahid Iqbal, Zhaojun Ni, Kenneth Omondi Ouma, Faisal Hayat, Wei Tan, Guofeng Hu, Chengdong Ma, and et al. 2022. "Comparative Analysis of Complete Chloroplast Genome and Phenotypic Characteristics of Japanese Apricot Accessions" Horticulturae 8, no. 9: 794. https://doi.org/10.3390/horticulturae8090794









